Abstract
We have characterized group A Streptococcus (GAS) genome-wide responses to hydrogen peroxide and assessed the role of the peroxide response regulator (PerR) in GAS under oxidative stress. Comparison of transcriptome changes elicited by peroxide in wild-type bacteria with those in a perR deletion mutant showed that 76 out of 237 peroxide-regulated genes are PerR dependent. Unlike the PerR-mediated upregulation of peroxidases and other peroxide stress defense mechanisms previously reported in Gram-positive species, PerR-dependent genes in GAS were almost exclusively downregulated and encoded proteins involved in purine and deoxyribonucleotide biosynthesis, heme uptake, and amino acid/peptide transport, but they also included a strongly activated putative transcriptional regulator (SPy1198). Of the 161 PerR-independent loci, repressed genes (86 of 161) encoded proteins with functions similar to those coordinated by PerR, in contrast to upregulated loci that encoded proteins that function in DNA damage repair, cofactor metabolism, reactive oxygen species detoxification, pilus biosynthesis, and hypothetical proteins. Complementation of the perR deletion mutant with wild-type PerR restored PerR-dependent regulation, whereas complementation with either one of two PerR variants carrying single mutations in two predicted metal-binding sites did not rescue the mutant phenotype. Metal content analyses of the recombinant wild type and respective PerR mutants, in addition to regulation studies in metal-supplemented and iron-depleted media, showed binding of zinc and iron by PerR and an iron requirement for optimal responses to peroxide. Our findings reveal a novel physiological contribution of PerR in coordinating DNA and protein metabolic functions in peroxide and identify GAS adaptive responses that may serve to enhance oxidative stress resistance and virulence in the host.
INTRODUCTION
Group A Streptococcus (GAS) (Streptococcus pyogenes) infections vary from superficial infection of the pharynx (strep throat) to serious skin and soft tissue infections that can lead to lethal invasive disease despite antibiotic treatment (10, 51). Resistance mechanisms of GAS to oxidative damage and killing by reactive oxygen species (ROS), which are generated from atmospheric oxygen or are produced by the phagocyte oxidative burst, have been partially characterized. Peroxidases, such as alkyl hydroperoxidase/alkyl hydroperoxide reductase (ahpCF) and glutathione peroxidase (gpoA), neutralize organic and inorganic peroxides and contribute to GAS aerotolerance and ROS detoxification in vitro (5, 33). The significance of these enzymes to bacterial fitness in vivo remains unclear, as ahpC and gpoA mutants show moderate reduction of virulence following subcutaneous and intraperitoneal infection in mice, respectively (5, 6). Resistance to superoxide (O2−) toxicity is mediated by superoxide dismutase (sodA) that converts O2− to H2O2, which is detoxified in turn by peroxidases (17). A critical role for SodA in GAS aerobic growth has been proposed (18). The iron/DNA-binding protein Dpr/MrgA (Dps-like peroxide resistance protein) is also thought to contribute to resistance against oxidative DNA damage by chelating free intracellular iron, thus preventing its reaction with peroxide (Fenton reaction) and leading to generation of highly reactive hydroxyl radicals (5, 65).
Defenses of GAS against ROS are coordinated, in part, by the peroxide stress response transcriptional regulator PerR. Earlier studies recorded significant virulence attenuation of perR mutants following subcutaneous or intraperitoneal inoculation in mice (5, 56), while recent investigations showed a role for PerR-controlled gene expression in GAS resistance to ROS-mediated killing by phagocytes and a critical contribution to pharyngeal colonization (21). To date, the extent of the PerR regulon and its contribution to GAS physiology under oxidative stress remain incompletely defined. In Bacillus and Staphylococcus, PerR activates expression of peroxidases and Dpr/MrgA under peroxide stress and coordinates iron homeostasis in order to minimize ROS formation via the Fenton reaction (12, 25, 27). Transcriptome comparisons between wild-type M-type 5 GAS and an isogenic perR mutant grown in the absence of stress indicated PerR-dependent repression (3-fold or higher) of six genes. These genes included pmtA encoding a putative metal efflux protein and additional loci encoding proteins with predicted function in zinc and/or manganese homeostasis and transport. Whereas pmtA is under direct PerR control, repression of the remaining five genes is thought to be an indirect effect mediated by a second transcriptional regulator, AdcR (7). Similar transcriptome comparisons between wild-type and perR mutant bacteria in M-type 3 GAS confirmed PerR-dependent repression of pmtA and AdcR-regulated loci but also suggested moderate repression of ahpCF and other loci and upregulation of several sugar transport genes (21). Promoter sequences of pmtA and ahpCF carry fully conserved Per boxes and have been shown to bind recombinant PerR in vitro; however, PerR control of ahpCF in vivo remains unclear, as this locus is marginally derepressed in two perR mutant strains (5, 21). These analyses identified GAS loci coordinated by PerR at baseline but did not examine the contribution of this regulator to GAS responses in the presence of a physiological oxidative stimulus.
We now define global responses of wild-type and perR mutant GAS to H2O2 and assess the role of PerR as a ROS sensor in this pathogen. Further genetic and biochemical analyses provide insight into the molecular mechanism of PerR function in streptococci. These studies characterize for the first time the peroxide stimulon of GAS and identify PerR-dependent and -independent defense mechanisms elicited by oxidative stress.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
GAS strain 003Sm is a streptomycin-resistant variant of M-type 3 strain DLS003 isolated from a patient with necrotizing fasciitis (21), whereas strain 854 is an M-type 1 strain isolated from a retroperitoneal abscess (23). Mutant strain 003SmperRΔ (or perRΔ) lacking over 80% of perR has been described previously (21). GAS was grown at 37°C in Todd-Hewitt broth (Difco Laboratories) supplemented with 0.5% yeast extract (THY) or on THY agar or Trypticase soy agar, both supplemented with 5% defibrinated sheep blood. For cloning, Escherichia coli DH5α or XL1-Blue was grown in Luria-Bertani (LB) broth or on LB agar. When appropriate, the following antibiotics were added at the concentrations indicated: ampicillin, 100 μg/ml; chloramphenicol, 20 μg/ml for E. coli and 10 μg/ml for GAS; and kanamycin, 30 μg/ml. In certain studies, THY was supplemented with FeCl3, MnCl2 · 4H2O, or deferoxamine mesylate (Sigma), as indicated. For H2O2 experiments, fresh working solutions (0.1 to 1 M) were prepared in water from a 10 M stock (Sigma).
DNA and RNA techniques.
Plasmid pGEM-T (Promega) is a linear E. coli vector used for cloning of PCR products; pJL1055 is a temperature-sensitive E. coli-Gram-positive shuttle vector used for mutagenesis in Streptococcus (21). Chromosomal DNA isolation and GAS electroporation were performed as described previously (22). Total GAS RNA was isolated using the RNeasy minikit (Qiagen) from bacterial lysates obtained by shaking bacteria with glass beads on a dental amalgamator (22).
PCR and qRT-PCR.
Primers used are shown in Table S1 in the supplemental material and were designed on the basis of the M-type 3 GAS strain MGAS315 or M-type 1 GAS strain SF370 genome sequences (4, 16). Quantitative reverse transcriptase PCR (qRT-PCR) was performed on an ABI PRISM 7300 sequence detection system (Applied Biosystems) using the QuantiTect SYBR green RT-PCR kit (Qiagen). Relative expression levels were calculated by the ΔΔCT (CT stands for threshold cycle) method described previously (22). Expression levels were normalized to recA (spyM3_1800) for studies in the absence of H2O2 and to gyrA (spyM3_0810) encoding DNA gyrase subunit A for studies with H2O2. Expression of both genes remained unaffected by perR mutation, while gyrA expression did not change in peroxide.
Microarray procedures and data analyses.
The GAS DNA microarray represents 1,788 open reading frames (ORFs), including 1,695 ORFs of M-type 1 GAS strain SF370, in addition to 71 M-type 18 and 21 M-type 3 specific ORFs (21). cDNA synthesis and labeling, microarray hybridizations, and data analyses were performed as described previously (22). The wild-type strain 003Sm and perR mutant strain 003SmperRΔ were grown to mid-exponential (ME) phase (A600 of 0.25) or late exponential (LE) phase (A600 of 0.6) at which point they were challenged with H2O2 (final concentration of 0.5 mM) for 15 min at 37°C or with water as a control. For each strain, total bacterial RNA was extracted from three or four independent H2O2- or water-treated control cultures, pooled, and used for cDNA synthesis and labeling. RNA samples to be compared were converted to cDNA, fluorescently labeled with Cy3 or Cy5 in direct (Cy3-Cy5) and dye swap (Cy5-Cy3) labeling reactions to correct for dye-dependent variation of labeling efficiency, and cohybridized on the arrays. Transcriptome comparisons were performed between treated and control wild-type strain 003Sm or perR mutant strain 003SmperRΔ. Statistical significance of the difference in mean fluorescence intensity calculated for each gene in the cDNA samples compared was determined by an unpaired 2-tailed Student's t test. Genes whose relative expression was equal or exceeded 2.0-fold and had P values of ≤0.01 were considered differentially expressed. Validation of the microarray data was performed by qRT-PCR using a minimum of two independent RNA samples obtained from peroxide-treated and untreated cultures of GAS grown to ME or LE phase.
Complementation and site-directed mutagenesis of PerR.
For complementation of strain 003SmperRΔ, wild-type perR was introduced into the native locus on the mutant chromosome by homologous recombination (21). A DNA fragment encompassing the full-length coding sequences of spyM3_0146 and perR (spyM3_0147) and the 3′-end sequence of spyM3_0148 transcribed in the opposite orientation was amplified using primers 0146-F (F for forward) (BamHI) and 0148-R (R for reverse) (EcoRV). This fragment was cloned into BamHI/EcoRV double-digested pJL1055 to obtain plasmid pJLperR. The construct was introduced into strain 003SmperRΔ by electroporation and was inserted into the chromosome upstream of perR by homologous recombination, generating perRΔ::WT (WT stands for wild type) strain. To obtain PerR mutants PerRH44A and PerRC104S, the QuikChange II site-directed mutagenesis kit (Agilent Technologies) was used as instructed with plasmid pJLperR as the template and primer pair perR-H44A-F/perR-H44-R and perR-C104S-F/perR-C104S-R, respectively. The derivative plasmids were introduced into mutant strain 003SmperRΔ by electroporation and then inserted into the chromosome by homologous recombination as described for wild-type perR to generate perRΔ::H44A and perRΔ::C104S strains. The presence of wild-type perR and spyM3_0146 sequences in the perRΔ::WT strain or wild-type spyM3_0146 and the mutations in the perRΔ::H44A and perRΔ::C104S strains were confirmed following sequencing of PCR products encompassing the two genes from each strain.
GAS protein preparation and Western blotting.
Total GAS protein preparations were obtained from cultures grown to ME phase using a combination of mutanolysin and lysozyme (22). Proteins were analyzed by SDS-PAGE under reducing conditions using 10% NuPAGE Bis-Tris gels (Invitrogen). For Western blots, proteins were transferred to nitrocellulose membranes and blocked in phosphate-buffered saline (PBS) containing 5% milk (PBS–5% milk) (Difco Laboratories). Anti-PerR rabbit serum (21) was added to the membranes at 1:5,000 in PBS–5% milk for 1 h, and the membranes were incubated with peroxidase-conjugated secondary antibody (GE Healthcare). Membrane development was carried out using the SuperSignal West Pico chemiluminescent substrate (Thermo Scientific). To confirm equal amounts of loaded proteins, samples were analyzed by SDS-PAGE and stained with Coomassie blue R-250.
Expression and purification of recombinant wild-type and mutant PerR.
The perR sequence without the start codon was amplified using primer pair perR-F (PshAI)/perR-R (HindIII) and was cloned downstream of the glutathione S-transferase (GST) coding sequence in PshAI/HindIII double-digested plasmid pET41a (EMD Chemicals) to generate pET41-perR. Overexpression of the ∼49-kDa recombinant GST-PerR fusion protein, or of GST protein from empty vector as a control, was achieved in E. coli BL21(DE3) using isopropyl-β-d-thiogalactopyranoside (IPTG) induction for 90 min in LB broth supplemented with 10 μM ZnCl2 and 40 μM FeSO4. Cell extracts were obtained by French press lysis, and purification was achieved using GST-binding resin (EMD Chemicals) as instructed by the manufacturer. To obtain GST-PerRH44A and GST-PerRC104S recombinant protein mutants, the QuikChange II site-directed mutagenesis kit (Agilent Technologies) was used with plasmid pET41-perR as the template and with primer pairs perR-H44A-F/perR-H44A-R and perR-C104S-F/perR-C104S-R, respectively. Expression and purification of the two mutant proteins was performed as described for GST-PerR. For certain preparations of GST-PerR, purification was performed in the presence of 1 to 10 mM EDTA. All purified proteins were obtained at higher than 95% purity, as determined by SDS-PAGE analysis. GST-PerR concentrations were determined using a NanoDrop 1000 spectrophotometer (Thermo Scientific) and an extinction coefficient of 55280 M−1 cm−1 at 280 nm.
Metal content analyses of culture media and recombinant PerR.
The metal content of THY broth was determined by inductively coupled plasma mass spectrometry (ICP-MS) using a Thermo Fisher Scientific X-Series instrument at the Department of Earth and Planetary Sciences at Harvard University. In similar analyses, metal bound to purified recombinant wild-type and mutant GST-PerR variant proteins was determined. For these measurements, 4 to 5 mg of each purified product, including recombinant GST expressed from an empty vector as a control, was diluted at ∼0.4 mg/ml and analyzed in buffer with a final composition of 20 mM Tris (pH 8.0), 2% glycerol, 3 mM glutathione, and 0.02% Tween 20. For both THY broth and purified proteins, two or three independent samples were analyzed for eight metal ions (calcium, cobalt, copper, iron, magnesium, manganese, nickel, and zinc), and the averages of these determinations were reported.
Electrophoretic mobility shift assays.
Binding of purified recombinant GST-PerR or GST-PerRH44A to promoter fragments was examined as described previously (21). In brief, GST-PerR or GST-PerRH44A recombinant protein was mixed with DNA at a final concentration of 6 μM or 50 nM, respectively. Reactions were allowed to proceed for 15 min at room temperature, and then the proteins were resolved on 6% native polyacrylamide DNA retardation gels (Invitrogen) and stained with ethidium bromide. The ∼300-bp pmtA promoter fragment carrying a fully conserved Per box was obtained by PCR using primer pair GS-1093-F/GS-1093-R. Negative-control reaction mixtures contained purified recombinant GST or a negative-control (gyrA) promoter fragment obtained with primer pair GS-gyrA-F/GS-gyrA-R (see Table S1 in the supplemental material).
Sequence databases and analyses tools.
Assignment of gene function was obtained from the National Center for Biotechnology Information (NCBI) and the Kyoto Encyclopedia of Genes and Genomes Sequence Similarity Database (KEGG SSDD) (31). Protein sequence alignments were performed using ClustalW2 (35). Searches for Per box-like sequences on promoters were carried out using the PredictRegulon server (67).
Microarray data accession number.
The primary microarray data have been submitted to the ArrayExpress database of the European Bioinformatics Institute under accession number E-MTAB-646.
RESULTS
Genome-wide GAS responses to peroxide.
Wild-type strain 003Sm and isogenic perR mutant strain 003SmperRΔ were grown to mid-exponential (ME) and late exponential (LE) phase in THY broth, and bacteria were challenged with a sublethal concentration of H2O2 (0.5 mM) for 15 min at 37°C. Experimental conditions were chosen on the basis of pilot wild-type and perR mutant cultures challenged with peroxide concentrations ranging from 0.2 to 10 mM (see Fig. S1 in the supplemental material). The 0.5 mM final peroxide concentration used for microarray analyses showed no effect on growth during LE phase and a moderate effect during ME phase, while no significant loss of cell viability was evident in either growth phase as determined by quantitative culture following peroxide challenge. Total RNA samples were collected from untreated control or peroxide-treated wild-type or mutant bacteria, and cDNA was generated and hybridized with GAS genomic DNA microarrays (21). Transcriptome changes in wild-type and perR mutant bacteria were identified by comparing the levels of gene expression in bacterial strains grown in the presence of peroxide with those in untreated samples of each strain. Changes induced by H2O2 in both strains constituted PerR-independent loci, whereas responses identified exclusively in wild-type bacteria constituted PerR-dependent loci.
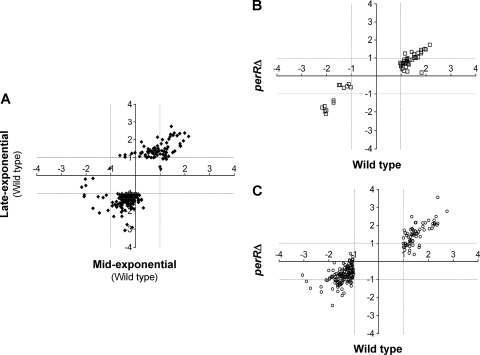
A total of 237 genes, including 40 encoding hypothetical proteins, were differentially regulated (≥2-fold change, P ≤ 0.01) in wild-type bacteria treated with peroxide at ME and/or LE phase: 84 genes were upregulated, and 153 were downregulated (Fig. 1A; see Table S2 in the supplemental material). Of these genes, 51 were regulated at the ME phase, whereas 214 genes were regulated at the LE phase or both growth phases. Interestingly, the majority of genes identified at the ME phase were upregulated (38 of 51, or 75%), whereas approximately two-thirds of the genes identified at the LE phase were downregulated in response to peroxide challenge (Fig. 1B and C).
Fig. 1.
Transcriptome changes elicited by H2O2 in GAS. (A) Gene expression changes (log2) in wild-type strain 003Sm following H2O2 challenge at mid-exponential and late-exponential phase growth. (B and C) Comparison of gene expression changes (log2) between wild-type and perR mutant strain 003SmperRΔ at each of the two growth phases. Data for the wild type and mutant are shown on the x axis and y axis, respectively. Broken vertical and horizontal lines represent 2-fold change in expression (log2 = 1) that was used as a threshold value for regulated genes.
qRT-PCR verification of transcriptome events.
Validation of the microarray data was carried out by quantitative reverse transcriptase PCR (qRT-PCR) analyses of RNA samples obtained from both wild-type and perR mutant bacteria exposed to H2O2 independent from those used for microarray studies (Table 1). A total of 40 genes representing ∼30 putative or known operons that include approximately 80 genes were examined. These genes represented both moderately and highly up- and downregulated PerR-dependent and -independent genes identified by microarray. We observed a high correlation (R = 0.92) between qRT-PCR results and microarray data (see Fig. S2 in the supplemental material). Additional qRT-PCR analyses examined independent samples obtained 5 and 15 min after challenge with 0.2, 0.5, or 1 mM H2O2 in order to evaluate both the strength and kinetics of peroxide-elicited GAS responses. These analyses including 11 or more genes showed dose-dependent responses in peroxide-treated bacteria and suggested optimal regulation at 15 min versus 5 min (data not shown).
Table 1.
qRT-PCR analysis of GAS responses to H2O2
| SPy no.a | Gene | Functionb | Microarray/qRT-PCRc |
Growthd | |
|---|---|---|---|---|---|
| Wild type | perRΔ mutant | ||||
| PerR-dependent genes | |||||
| 0092 (0069) | adcR | Putative transcriptional repressor | +1.8/+2.3 | +1.1/1.0 | ME |
| 0094 (0071) | adcB | Zinc/manganese ABC transporter | +2.4/+1.7 | +1.2/1.0 | ME |
| 0454 (0319) | mtsB | Iron ABC transporter | −2.7/−2.3 | −1.4/−2.2f | ME |
| 1198 (0840) | Putative transcriptional repressor | +3.5/+6.0 | +1.1/−1.3 | ME | |
| 0016 (0013) | Putative amino acid permease | −3.5/−2.2 | −1.4/−1.5 | LE | |
| 0295 (0217) | oppC | Oligopeptide permease | −3.4/−2.6 | −1.7/−1.4 | LE |
| 0416 (0298) | prtS | Interleukin 8 protease | −3.7/−2.4 | −1.7/−1.2 | LE |
| 0740 (0482) | sagC | Streptolysin S-associated ORF | −2.3/−1.6 | −1.1/1.0 | LE |
| 1618 (1363) | cysM | Putative O-acetylserine lyase | +2.7/1.0e,f | +1.3/−1.3 | LE |
| 1791 (1557) | siaD | Putative ABC transporter | −2.9/−1.9 | −1.5/−1.1 | LE |
| 1796 (1560) | shp | Heme/ferrichrome-binding protein | −3.3/−2.0 | −1.6/−1.1 | LE |
| 1922 (1658) | lacB2 | Galactose-6-phosphate isomerase | −8.3/−2.4 | −1.7/−1.2 | LE |
| 1923 (1659) | lacA2 | Galactose-6-phosphate isomerase | −6.7/−3.7 | −2.8/−1.5 | LE |
| 2110 (1794) | nrdD | Ribonucleoside triphosphate reductase | −4.0/−4.4 | −1.4/−1.7 | LE |
| PerR-independent genes | |||||
| n/a (0097) | nra | Pilus transcriptional repressor | ND/+4.4 | ND/nd | ME |
| n/a (0098) | cpa | Pilus ancillary protein | ND/+6.8 | ND/nd | ME |
| n/a (0100) | fctA | Pilus major subunit | ND/+7.1 | ND/nd | ME |
| n/a (0101) | srtC2 | Sortase B family | +2.6/+3.7 | +2.0/nd | ME |
| n/a (0102) | fctB | Pilus minor subunit | ND/+5.8 | ND/nd | ME |
| n/a (0103) | msmR | Pilus transcriptional activator | ND/+6.6 | ND/nd | ME |
| 0146 (0113) | sloR | Putative regulatory protein | −4.1/−6.7 | −4.4/nd | ME |
| 0290 (0212) | sufB | Putative Fe-S cluster assembly protein | +3.0/+4.0 | +2.0/nd | ME |
| 1134 (0792) | proX | l-Proline glycine betaine ABC transport | +2.5/+1.9 | +1.9/nd | ME |
| 1212 (0852) | cls | Cardiolipin synthetase | +3.5/+3.5 | +2.4/nd | ME |
| 1214 (0854) | lplA | Putative lipoate-protein ligase | +4.4/+3.8 | +3.3/nd | ME |
| 1217 (0857) | gcvH | Glycine cleavage system H protein | +3.2/+5.2 | +2.4/nd | ME |
| 1406 (1071) | sodA | Superoxide dismutase | +2.2/+3.5 | +2.4/nd | ME |
| 1835 (1586) | trx | Putative thioredoxin | +2.3/+1.6 | +1.8/nd | ME |
| 1849 (1596) | pfl | Putative pyruvate formate lyase | −3.3/−9.2 | −2.6/nd | ME |
| 1856 (1601) | norA | Putative antibiotic resistance protein | −4.2/−3.7 | −3.2/nd | ME |
| 2080 (1771) | ahpF | Alkyl hydroperoxide reductase | +2.1/+3.2 | +1.8/nd | ME |
| 0100 (0077) | Putative DNA-binding protein | +5.2/+8.5 | +4.4/nd | LE | |
| 0165 (0128) | nga | NAD glycohydrolase | −3.0/−3.6 | −2.1/nd | LE |
| 0386 (0283) | fhuA | Heme/ferrichrome ABC transporter | −3.0/−2.6 | −2.5/nd | LE |
| 0412 (0296) | exoA | Putative 3′-exo-DNase | +2.8/+3.2 | +4.4/nd | LE |
| 0454 (0319) | mtsB | Iron/manganese ABC transporter | −4.3/−2.5 | −2.0/−2.0 | LE |
| 0813 (0546) | gor | Glutathione reductase | +2.4/+3.7 | +1.9/nd | LE |
| 0914 (0629) | Hypothetical protein | +5.2/+7.1 | +11.8/nd | LE | |
| 1131 (0790) | Na+-driven multidrug efflux pump | +4.7/+3.5 | +4.9/nd | LE | |
| 1206 (0847) | Putative ABC transporter | −4.9/−6.8 | −3.2/nd | LE | |
| 1219 (0859) | Putative trimethylamine dehydrogenase | +6.7/+3.3 | +7.0/nd | LE | |
| 1607 (1355) | recX | RecA function regulator | +5.0/+3.5 | +5.6/nd | LE |
| 1680 (1464) | Putative transcriptional regulator | +3.7/+3.6 | +4.4/nd | LE | |
| 1681 (1465) | Putative NADH peroxidase | +2.2/+1.6 | +3.0/nd | LE | |
| 1816 (1570) | scrB | Putative sucrose-6-phosphate hydrolase | +2.8/+1.4e | +3.2/nd | LE |
| 1825 (1577) | uvrA | Excinuclease ABC subunit A | +2.6/+2.6 | +3.3/nd | LE |
| 2162 (1818) | cadD | Putative cadmium resistance protein | −2.6/−1.6e | −1.6/−1.4 | LE |
The SPy numbers of the M-type 1 strain SF370 (16) are shown. The spyM3 numbers of the M-type 3 strain MGAS315 (4) are shown in parentheses. n/a, nonapplicable.
NCBI or KEGG annotation.
Fold change expression in the wild type or perRΔ mutant following challenge with H2O2 (0.5 mM) versus H2O control. Data obtained by microarray and qRT-PCR analyses are shown before and after the slashes, respectively. nd, not done; ND, not detected due to lack of sequence homology between M-type 3 strain 003Sm and M-type 1 sequences on microarrays.
ME, mid-exponential phase; LE, late exponential phase.
Regulation not confirmed by qRT-PCR.
PerR dependence not confirmed by qRT-PCR.
Responses to H2O2 are partially dependent on PerR.
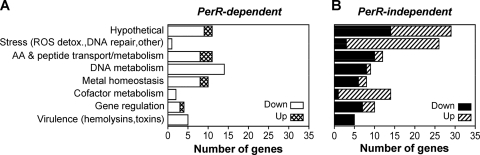
PerR-regulated responses were identified as transcriptome changes in wild-type bacteria that were not observed in the perR mutant. To ensure stringent selection of PerR-dependent loci, regulated genes in wild-type bacteria (≥2-fold change, P ≤ 0.01) that showed a similar regulation trend (≥1.6-fold, P ≤ 0.05) in the perR mutant were not considered PerR dependent. Moreover, peroxide-induced regulation of several genes that showed dependence on PerR in microarrays was verified by qRT-PCR (Table 1). These genes included certain loci for which PerR dependence was not clear by microarray. On the basis of the results of both analyses, 76 of 237 genes (32%) including 11 genes encoding hypothetical proteins were identified as PerR dependent. The great majority of these genes (67 of 76) were downregulated by PerR (directly or indirectly) at LE phase growth and mainly encoded products involved in purine and deoxyribonucleotide synthesis, amino acid/oligopeptide synthesis and transport, and heme acquisition among others (Fig. 2A; see Table S2 in the supplemental material). Upregulated loci encoded proteins involved in Mn2+/Zn2+ import and methionine uptake but also included a putative transcriptional regulator (SPy1198) that was among the most highly regulated PerR-dependent loci (Table 1 and Fig. 3B).
Fig. 2.
PerR-dependent (A) and PerR-independent (B) GAS responses elicited by H2O2. The numbers of downregulated and upregulated genes in each of eight main categories are shown. Note that PerR-dependent genes mainly encode DNA metabolism, metal homeostasis, and amino acid (AA)/peptide transport. The majority of genes with functions in DNA repair, ROS detoxification (detox.), cofactor metabolism and gene regulation as well as genes encoding hypothetical proteins are PerR independent.
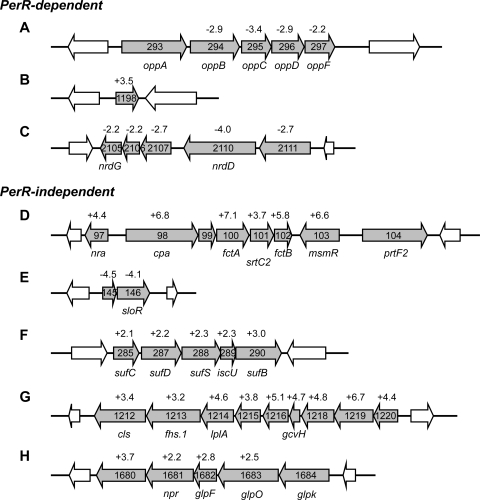
Fig. 3.
Chromosomal organization and regulation levels of GAS loci in peroxide. (A to C) PerR-dependent loci encompassing the oligopeptide permease locus (A), a putative transcriptional regulator (B), and a deoxynucleoside triphosphate (dNTP) synthesis locus (C) are shown. (D to H) PerR-independent loci include the pilus biosynthesis operon and regulator genes in FCT-3 region (D), two independent loci involved in Fe-S (F) and other cofactor (lipoic acid) metabolism (G), and two loci encoding putative regulators SloR (SPy0146) (E) and SPy1680 (H). The gene numbers in each panel represent ORFs of M-type 1 GAS strain SF370 (16), except for panel D, which shows ORFs of M-type 3 GAS strain MGAS315 (4). The numbers above genes indicate relative expression levels (fold change) in wild-type strain 003Sm treated with peroxide as obtained in microarray studies, except for FCT-3 regulation (panel D) that was characterized by qRT-PCR.
PerR-independent responses to peroxide encompassed 161 genes (68% of total) of which 86 were downregulated and 75 were upregulated (Fig. 2B). Repressed genes encoded proteins with functions similar to those in PerR-mediated responses, including iron and heme/ferrichrome uptake, amino acid/peptide transport and metabolism, pyrimidine synthesis, as well as a significant number of hypothetical proteins. Upregulated genes showed very little or no overlap in function with PerR-dependent responses and encoded proteins involved in DNA damage repair, ROS detoxification, lipoic acid protein modification, iron-sulfur cluster (Fe-S) synthesis/repair, pilus biosynthesis, and almost all (15 of 17) upregulated hypothetical proteins.
Overall, the greatest numbers of PerR-dependent loci were identified in the DNA metabolism, metal homeostasis, and amino acid/peptide transport categories, with regulation of over half of the genes in these three groups (35 of 64, or 55%) showing dependence on PerR (Fig. 2). These data demonstrate a significant, but also nonexclusive, role of PerR in GAS responses to peroxide stress.
DNA damage repair, ROS detoxification, and other stress responses.
All but one of 27 genes in the category of DNA damage repair, ROS detoxification, and other stress responses exhibited PerR-independent regulation, with the great majority (23 of 27) upregulated in the presence of peroxide (Fig. 2). Highly upregulated genes encoded proteins with predicted functions in DNA repair, such as excinuclease subunits A (uvrA, SPy1825) and C (uvrC, SPy1068) involved in base excision repair, exo-DNase A (exoA, SPy0412) implicated in oxidative damage repair in Bacillus subtilis (28), and MutY (SPy1833) that is a putative adenine glycosylase that removes misincorporated adenine opposite from 8-oxo-7,8-dihydroguanine, a toxic product resulting from oxidative DNA damage (42). recX (SPy1607) encoding a regulator of recombinase A (recA) function was strongly upregulated (∼5-fold) in PerR-independent fashion, while upregulation of recA itself was 2-fold in peroxide-treated GAS. RecA and RecX, in addition to UvrA and UvrC, are important components of the SOS response in several bacterial species required for repair of DNA damage caused by ROS and other chemical or physical agents (13, 66).
Additional upregulated genes encoded peroxide and other ROS detoxifying enzymes, such as alkyl hydroperoxidase/alkyl hydroperoxide reductase AhpC/AhpF (SPy2079-SPy2080), a putative NADH peroxidase (npr, SPy1681), superoxide dismutase (sodA, SPy1406) and glutathione reductase (gor, SPy0813). The latter converts oxidized glutathione (GSSG) to its reduced form (GSH) that can itself reduce oxidized thiol groups directly or contribute to peroxide detoxification indirectly through the action of glutathione peroxidase, which degrades H2O2 to water with concomitant oxidation of GSH (6). Upregulation of sodA was not surprising, as it is also upregulated by peroxide in B. subtilis (48) and has been reported to contribute to peroxide resistance in streptococci by counteracting oxidative DNA damage (54).
A five-gene locus with a putative role in Fe-S assembly was also activated in the presence of peroxide (Fig. 3F). While certain bacteria encode more than one Fe-S synthesis system (Isc, Nif, Suf), streptococci encode a single system comprised mainly of Suf-like enzymes (SufC/SufD/SufS/SufB) (55). Upregulation of a rhodanese-related sulfurtransferase (SPy0915), which is thought to have a SufS-like function and to contribute to oxidative stress defenses, was also detected (32).
DNA metabolism.
Regulation of a total of 23 genes in the DNA metabolism category was detected exclusively at LE phase growth, and all but one was repressed in peroxide. It was striking that 14 of 23 genes were PerR dependent and included mostly purine (purM, purN, purE, purB, and guaB) and deoxyribonucleotide synthesis loci. The latter were clustered in a single operon or two adjacent potential operons (Fig. 3C) encompassing nrdG (SPy2105) and nrdD (SPy2110), which encode putative class III ribonucleotide reductases involved in deoxyribonucleotide synthesis under anaerobic conditions (19). PerR-independent loci encoded DNA replication machinery components (dnaA, dnaN, dnaX, topA, and priA) as well as pyrimidine biosynthesis (pyrB, pyrF, and pyrE).
Amino acid/peptide transport and metabolism.
The amino acid/peptide transport and metabolism category encompassed mostly repressed genes (18 of 23 total) that included the oligopeptide permease operon oppABCDF (52) and three ABC transport loci with predicted function in polar amino acid transport (SPy0276-SPy0277, SPy1274-SPy1276, and SPy1506-SPy1507). Eight repressed genes including the opp operon were regulated in a PerR-dependent manner. Of the five upregulated genes, SPy0320-SPy0321 are PerR dependent and encode a putative methionine ABC transporter, while SPy1990-SPy1991 seem to encode a protein(s) involved in aromatic amino acid biosynthesis. Among amino acids, methionine is highly reactive with ROS; therefore, increased uptake would counteract peroxide-induced protein damage.
Metal homeostasis and transport.
Twelve of 18 genes represented the three known iron and heme acquisition systems of GAS: the MtsABC iron transport system (SPy0453, SPy0454, and SPy0456), the FhuGBDA or FtsDCBA (SPy0383-SPy0386) heme/ferrichrome transport system, and Shr/Shp/SiaABCD (Spy1791, SPy1793-SPy1796, and SPy1798; siaABC is also known as htsABC) involved in hemoglobin acquisition (3, 24, 30, 39, 40, 62). All three operons were repressed in peroxide with PerR mediating repression of at least the Shr/Shp/SiaABCD gene locus, as confirmed by qRT-PCR (Table 1). Repression of mtsABC at ME phase growth was also suggested to be PerR dependent by microarray analysis. This result could be verified by qRT-PCR in the RNA preparations used for microarrays, but it could not be reproduced using independent RNA preparations; therefore, the role of PerR in mtsABC repression remains unclear. Of the four upregulated genes, three represented AdcRCB (SPy0092-SPy0094), a transport complex linked to Mn2+/Zn2+import in streptococci (14, 41). Upregulation of this locus was moderate but consistently dependent on PerR at ME phase (Table 1).
Cofactor metabolism.
Similar to the first category (DNA damage repair, ROS detoxification, and other stress responses), the majority of genes in the cofactor metabolism category (14 of 16) were upregulated by peroxide in a PerR-independent manner. Nine genes were activated to similar levels at both ME and LE growth phase and seemed to be organized in an operon (SPy1212-SPy1220) that included some of the most highly upregulated genes in the peroxide stimulon (Fig. 3G). While several genes in this locus are genes encoding hypothetical proteins, SPy1214 encodes a predicted lipoate-protein ligase (lplA) that uses exogenous lipoic acid to lipoylate components of keto acid dehydrogenase complexes (20, 47, 61), such as the E2 subunits of pyruvate dehydrogenase and protein H of the glycine cleavage system, with protein H encoded by SPy1217 (gcvH) in the same operon. Lipoic acid (6,8-thiooctanoic acid), a disulfide-containing cofactor, is essential for the decarboxylation activity of these complexes (61), but it has also been reported to contribute to bacterial defenses against oxidative stress (8).
Interestingly, the last gene in this locus (cls, SPy1212) encodes a synthetase involved in production of cardiolipins, anionic glycerophospholipids that have been reported to be upregulated under osmotic stress (57). Although cardiolipins are not essential for GAS growth under standard culture conditions (58), cls activation in peroxide could be important for repair of ROS-inflicted membrane damage or for increasing cardiolipin membrane levels that may enhance bacterial fitness under oxidative stress. Consistent with increased cardiolipin synthesis, glpF (SPy1682) and glpO (SPy1683) involved in glycerol uptake and glycerophospholipid metabolism were also upregulated in the presence of peroxide (Fig. 3H).
Pilus biosynthesis.
The microarray data initially suggested upregulation of srtC2 (spyM3_0101) encoding a type B sortase that is required for pilus assembly in GAS (49). This enzyme is expressed from the pilus biosynthesis operon, also encoding three pilus structural proteins (fctA, fctB, and cpa), which is part of the fibronectin-binding, collagen-binding, T antigen (FCT) region in the GAS chromosome (Fig. 3D). The FCT locus is conserved in GAS strains of M-types 3, 5, 18, and 49 (FCT-3), but it shows variable gene content and poor sequence homology with FCT regions from other strains, such as M-type 1 GAS representing FCT-2 (15, 46). srtC2 regulation was detectable in M-type 3 strain 003Sm, because our microarrays carry probes for the highly homologous srtC2 from M-type 18 GAS (spyM18_0129) (60), whereas regulation of the pilus structural genes was not detectable, since the microarrays contained probes specific to M-type 1 GAS. To further assess pilus operon regulation in strain 003Sm, we designed primers for the three structural genes and srtC2 based on FCT-3 sequences from M-type 3 GAS (4) and measured expression in response to peroxide by qRT-PCR. As shown in Table 1, these studies confirmed upregulation of the sortase and showed 6- to 7-fold increased expression of the three structural genes.
PerR complementation restores PerR-regulated gene expression.
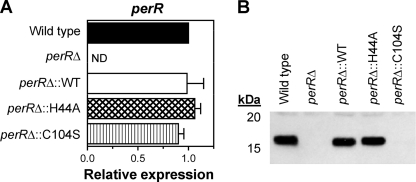
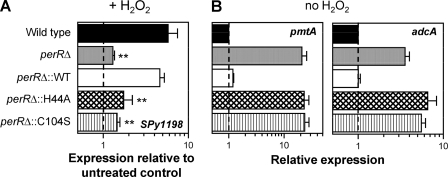
To verify the transcriptome events mediated by PerR in response to peroxide, as well as those identified in previous studies by comparing the perR mutant transcriptome with the wild-type transcriptome in the absence of ROS (7, 21), PerR-dependent regulation was evaluated both in the presence and absence of peroxide in a complemented perR deletion mutant strain 003SmperRΔ. Complementation was achieved by introducing wild-type perR into its native locus on the chromosome using plasmid pJLperR and a homologous recombination strategy identical with that used for constructing the original deletion mutant. The resulting strain designated the perRΔ::WT strain expressed wild-type perR mRNA levels and fully restored PerR protein expression, as indicated, respectively, by qRT-PCR and Western blot analyses using PerR antiserum against cell lysates of the perRΔ::WT strain as well as of the wild type and perR deletion mutant (perRΔ) as controls (Fig. 4). PerR regulation in the perRΔ::WT strain and in control wild-type and perRΔ cultures was then assessed by qRT-PCR analyses of RNA samples isolated from bacteria grown to ME phase with or without subsequent H2O2 challenge. PerR function in peroxide was evaluated by comparing expression levels of SPy1198 in peroxide-treated versus untreated controls, whereas PerR regulation in the absence of peroxide was determined by comparing expression of pmtA (SPy1434 or spyM3_1093) and adcA (SPy0714 or spyM3_0466) that were previously identified as two PerR-repressed genes in the absence of stress (7, 21). SPy1198 was chosen as a marker for peroxide-mediated gene expression, because it represents the most strongly regulated PerR-dependent gene following qRT-PCR verification and encodes a predicted transcriptional regulator that could further influence PerR-mediated adaptive responses to ROS. These studies showed clear restoration of PerR function in the perRΔ::WT strain, leading to ∼5-fold upregulation of SPy1198 in the presence of peroxide versus ∼6-fold in the wild-type parent strain (Fig. 5A) and repression of both pmtA and adcA to wild-type levels in the absence of peroxide (Fig. 5B). As expected, derepression of pmtA and adcA in the absence of stress and no SPy1198 upregulation in peroxide were recorded in the perR deletion mutant.
Fig. 4.
Complementation analyses of perR deletion mutant strain 003SmperRΔ expressing wild-type PerR (perRΔ::WT strain) or one of two mutant variants PerRH44A (perRΔ::H44A strain) and PerRC104S (perRΔ::C104S strain). (A) PerR mRNA levels in each strain relative to the wild-type parent control, as determined by qRT-PCR. Data represent means plus standard deviations (error bars) obtained from at least three independent RNA preparations for each strain. ND, not detected. (B) Western blot analysis of total protein preparations obtained from strains shown in panel A using PerR antiserum.
Fig. 5.
PerR-regulated gene expression in mutant strain 003SmperRΔ expressing wild-type PerR or one of two mutant variants, PerRH44A and PerRC104S. (A) Regulation of SPy1198 after peroxide challenge, as determined by qRT-PCR. Relative expression equal to 1, indicated by the broken line, represents no change in expression in the presence of peroxide compared to expression in untreated control cultures. **, P ≤ 0.009 for comparison of SPy1198 upregulation in perRΔ::WT versus perRΔ, perRΔ::H44A, or perRΔ::C104S (Student's t test). (B) Regulation of pmtA and adcA in the absence of peroxide, as determined by qRT-PCR. The level of expression in the wild-type strain, strain 003Sm, is indicated by the broken line (relative expression equals 1). Data in both panels represent means plus standard deviations (error bars) obtained from at least three independent RNA preparations for each strain in each growth condition.
Mutagenesis of predicted metal-binding sites abrogates PerR function.
In the well-characterized Bacillus system, PerR binds one Fe2+ (or Mn2+) atom at a regulatory site encompassing a histidine (H37) as well as four other amino acid residues, and one Zn2+ atom at a structural site encompassing four cysteines, including C96 (37, 38). Eight of these nine residues are conserved between GAS and Bacillus PerR. Fe2+-bound PerR is responsive to peroxide, as Fe2+ is oxidized to Fe3+ with concomitant oxidation of H37 to oxo-H37 due to Fenton chemistry, leading to the loss of PerR regulatory (repressor) function. In contrast, Mn2+-bound PerR is much less sensitive to oxidation and retains DNA binding in high peroxide concentrations (38, 64).
To gain insight into the mechanism of PerR function in GAS, we studied two additional perR deletion mutant-derived strains complemented with PerR variants carrying single point mutations at H44 or C104 corresponding to H37 and C96 of Bacillus. The wild-type perR gene on the pJLperR plasmid was mutated at H44 to alanine and at C104 to serine, generating plasmids pJLperRH44A and pJLperRC104S, respectively. Each of the two perR mutant variants was introduced into the perRΔ chromosome by homologous recombination as described previously for wild-type perR to produce perRΔ::H44A and perRΔ::C104S strains. The levels of PerR mRNA and protein as well as PerR-regulated gene expression were then compared to those in the wild-type perR-complemented strain (perRΔ::WT strain) by qRT-PCR and Western blot analyses. As shown in Fig. 4A, perR transcript levels of the two mutants were restored to the levels of the wild type, similar to the result obtained for the perRΔ::WT strain. PerR protein expression in the perRΔ::H44A strain was indistinguishable from that of the wild type, but it was not detectable in the perRΔ::C104S strain (Fig. 4B) despite wild-type mRNA levels detected by qRT-PCR. Longer exposure of Western blots revealed very low PerRC104S expression levels, indicating that lack of detection by anti-PerR serum was not due to altered conformation of the mutant protein. This explanation was supported by the finding that the mutant protein could be detected by the same antiserum as a recombinant fusion protein to glutathione S-transferase described below (data not shown). Consistent with rapid degradation of PerRC104S, regulation of SPy1198 in the presence of peroxide and of pmtA and adcA in its absence remained unchanged in the perRΔ::C104S strain compared to the perRΔ strain. Moreover, the complete loss of pmtA and adcA repression in the absence of peroxide as well as loss of SPy1198 upregulation in peroxide-treated bacteria was also observed in the perRΔ::H44A strain, even though PerRH44A expression was at wild-type levels (Fig. 5). These results show the requirement of both H44 and C104 for PerR function in GAS and indicate metal-binding sites analogous to those reported in Bacillus.
PerR-regulated gene expression in vivo is dependent on iron and manganese.
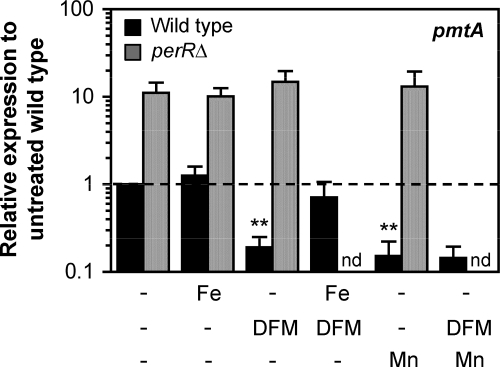
Metal dependence of PerR function was evaluated by comparing pmtA expression levels in bacteria grown in THY broth supplemented with iron, manganese, or the iron chelator deferoxamine mesylate (DFM) with expression levels in unsupplemented broth. Prior measurements of metal concentrations in unsupplemented THY broth indicated approximately 10 μM iron, 11 μM zinc, and 0.25 μM manganese (see Table S3 in the supplemental material). The wild-type strain or mutant strain 003SmperRΔ were grown to ME phase in unsupplemented media or in media supplemented with 40 μM FeCl3, 40 μM MnCl2, or 40 μM DFM. Total RNA was isolated for use in qRT-PCR analyses. As shown in Fig. 6, growth of wild-type bacteria in Mn- or DFM-supplemented medium led to further repression of pmtA compared to expression in unsupplemented broth (∼5- and ∼6-fold, respectively), whereas growth in Fe-supplemented medium did not have any effect. Moreover, the pmtA repression effect of DFM could be reversed using an equimolar concentration of iron, but not manganese. In contrast, expression of pmtA in the perR mutant remained unchanged (>10-fold derepressed) under all conditions, a result indicating a PerR requirement for the enhanced repression detected in wild-type bacteria during growth in excess Mn or added DFM.
Fig. 6.
Effects of iron and manganese on PerR function in the absence of peroxide. Wild-type or perR mutant bacteria were grown in unsupplemented broth or broth supplemented with FeCl3 (Fe), MnCl2 (Mn), DFM, or with equimolar concentrations of DFM and Fe or Mn. Total RNA was obtained from each strain in each growth condition, and regulation of pmtA was quantified by qRT-PCR. Data represent means plus standard deviations obtained from at least three independent RNA preparations for each strain in each growth condition. Wild-type expression in unsupplemented medium is indicated by the broken line (relative expression equals 1). **, P ≤ 0.005 for comparison of pmtA expression in wild-type bacteria grown with added Fe versus DFM or added Mn (Student's t test). nd, not done.
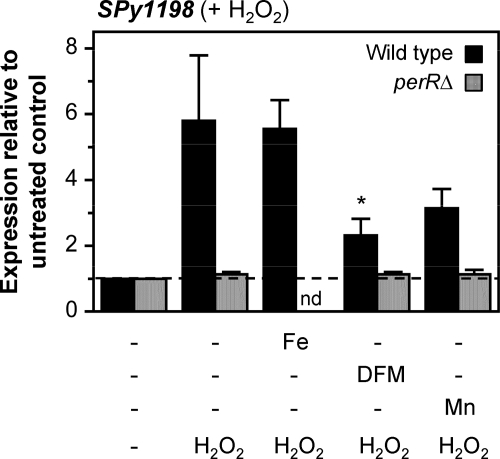
Further studies examined SPy1198 upregulation in response to peroxide following growth in metal- or DFM-supplemented medium. Wild-type and perR mutant cultures were grown as described above and then challenged at ME phase growth with H2O2. For the wild type, SPy1198 was upregulated ∼6-fold in Fe-supplemented medium similar to that of the unsupplemented medium control, but this decreased substantially to 2.3-fold in the presence of DFM and to 3.1-fold in added Mn (Fig. 7). As expected, upregulation of SPy1198 was abolished in the perR mutant grown in unsupplemented broth, but more interestingly, expression remained unchanged in supplemented media, a finding that also implicates PerR in the decreased upregulation levels recorded in wild-type cultures grown with added Mn or DFM. Taken together, these findings suggest iron and/or manganese binding by PerR in vivo and an iron requirement for maximal PerR sensitivity to peroxide.
Fig. 7.
Effects of metal ions on PerR function in the presence of peroxide. Wild-type or perR mutant bacteria were grown in unsupplemented media or media supplemented with FeCl3 (Fe), MnCl2 (Mn), or DFM and then challenged with 0.5 mM H2O2 for 15 min. Total RNA was obtained from each strain in each growth condition, and regulation of SPy1198 was quantified by qRT-PCR. Data represent means plus standard deviations obtained from at least three independent RNA preparations for each strain in each growth condition. No change in expression in the presence of peroxide compared to expression in untreated GAS is indicated by the broken line (relative expression equals 1). *, P < 0.05 for comparison of SPy1198 upregulation elicited by peroxide in unsupplemented broth versus DFM-supplemented broth (Student's t test). nd, not done.
Recombinant PerR binds iron and zinc.
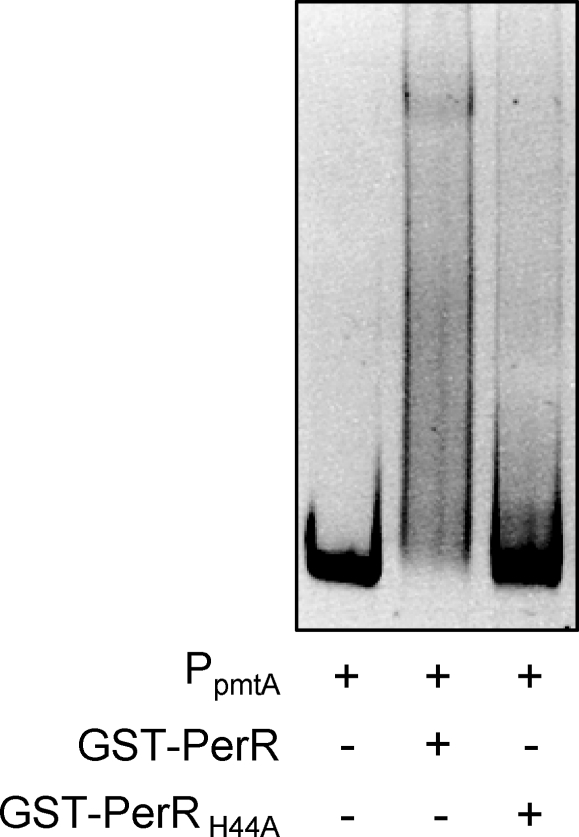
To verify PerR metal binding, fusions of wild-type PerR to glutathione S-transferase (GST-PerR) as well as of the two respective H44A and C104S mutants, GST-PerRH44A and GST-PerRC104S, were expressed and purified from E. coli by glutathione affinity chromatography. This purification approach does not require metal-containing resin and was chosen over nickel affinity chromatography, as it has been suggested that nickel binds to recombinant PerR (26). Moreover, the possible misfolding and degradation of PerRC104S in E. coli, suggested by its degradation in GAS shown above, could potentially be prevented by its fusion to GST. All three recombinant proteins were purified at equivalent yields, and their metal content was obtained by inductively coupled plasma mass spectrometry. As shown in Table 2, these analyses revealed binding of zinc and iron to GST-PerR at 0.81 and 0.58 atom per PerR monomer, respectively, but not to purified GST protein control. Although not fully saturated with either metal, this finding is consistent with zinc and iron binding to the predicted structural and regulatory site, respectively. In contrast, GST-PerRC104S showed dramatic loss of both zinc and iron (80% and 87% reduction, respectively), whereas GST-PerRH44A retained binding of both metals at wild-type levels. Additional purification of GST-PerR was performed in the presence of EDTA, which has been suggested to prevent iron binding and therefore prevent oxidation of histidine residues at the regulatory site that leads to loss of PerR function (38). Metal content analyses of these GST-PerR preparations yielded 0.8 zinc atom, but total loss of iron (Table 2). To assess the functions of recombinant proteins, their DNA binding capacity was evaluated in electrophoretic mobility shift assays using the pmtA promoter, which has been previously shown to encompass a fully conserved Per box and to bind recombinant PerR in vitro (21). GST-PerR purified in the absence of EDTA did not exhibit binding to pmtA, or any other promoter tested, possibly due to partial or full oxidation during purification. GST-PerR purified in the presence of EDTA, however, demonstrated reproducible binding and generated a high-molecular-weight DNA-protein complex, whereas GST-PerRH44A purified under identical conditions lacked binding capacity (Fig. 8). Interestingly, supplementation of GST-PerR with iron or manganese did not improve binding of the protein. It is proposed that although H44 is dispensable for iron binding, it is essential for PerR promoter binding and regulatory function in GAS. Moreover, C104 is required for zinc binding to the putative structural site, thereby promoting correct protein conformation and iron binding at the regulatory site.
Table 2.
Metal content of recombinant PerR proteins
| Protein | Molar ratio (metal/protein) |
|
|---|---|---|
| Zinc | Iron | |
| GST-PerR | 0.812 | 0.580 |
| GST-PerRa | 0.800 | 0.02 |
| GST-PerRH44A | 0.829 | 0.567 |
| GST-PerRC104S | 0.167 | 0.074 |
| GST (control) | 0.042 | 0.016 |
GST-PerR purified in the presence of EDTA.
Fig. 8.
Interaction of recombinant PerR with the pmtA promoter (PpmtA). Purified wild-type GST-PerR or the H44A mutant protein GST-PerRH44A was mixed at a final concentration of 6 μM with 50 ng of promoter DNA and incubated at room temperature for 15 min. Following native polyacrylamide gel electrophoresis, electrophoretic mobility shift of the promoter was visualized by ethidium bromide staining.
Adaptive responses to peroxide include uncharacterized and known regulators of GAS.
In addition to SPy1198, the peroxide stimulon encompassed PerR-independent putative regulators that have not been previously implicated in GAS defenses against ROS (see Table S2 in the supplemental material). Indeed, our finding that a major portion of the response(s) to peroxide does not require PerR suggests the existence of additional regulatory mechanism(s) coordinating GAS defenses against peroxide (and perhaps other ROS) that could encompass PerR-independent transcriptome events identified here. Two highly regulated genes in this category were (i) SPy0146 (sloR) encoding a PfoR-like regulator that was 4- to 7-fold repressed at the ME phase (Table 1 and Fig. 3E) and (ii) SPy1680 encoding a putative transcriptional activator that was ∼4-fold upregulated at the LE phase (Table 1 and Fig. 3H). SPy1680 seems to encode a typical transcriptional regulator and maps to a potential operon with four additional genes, including npr immediately upstream encoding a predicted NADH peroxidase. SloR exhibits high homology with orthologs in Streptococcus and Clostridium. Although previously reported to repress expression of streptolysin O in one of two GAS strains studied (59), SloR has recently been implicated in serine metabolism and/or transport, and it has been proposed that it is under direct control of the serine catabolism regulator, SerR (36).
Upregulation of the pilus biosynthesis operon discussed above prompted us also to investigate potential effects of peroxide on msmR (spyM3_0103) and nra (spyM3_0097), which encode two transcriptional regulators expressed independently from FCT-3 with opposite effects on pilus expression. MsmR activates the pilus operon and nra (50), whereas Nra represses the pilus operon and its own expression (34, 53). Regulation of the two genes was analyzed by qRT-PCR using RNA samples obtained from peroxide-treated and untreated wild-type strain 003Sm and primers designed on the basis of M-type 3 genome sequences. As shown in Table 1, msmR and nra were similarly upregulated in H2O2 by 6.6- and 4.4-fold, respectively. However, CT values obtained in qRT-PCR analyses indicated dramatically higher msmR mRNA levels, suggesting higher protein levels compared to Nra (baseline CT of ∼21 for msmR versus ∼28 for nra) (see Fig. S3 in the supplemental material). These data are consistent with MsmR involvement in ROS-dependent regulation of the pilus.
To further test the putative link between MsmR and pilus upregulation, the effects of peroxide were also examined in M-type 1 GAS strain 854 representing FCT-2, which encompasses the pilus synthesis operon and its transcriptional activator RofA but lacks both msmR and nra (see Fig. S4 in the supplemental material). Primers for rofA (SPy0124) and the pilus structural genes fctA (SPy0128) and fctB (SPy0130) were designed on the basis of M-type 1 strain SF370 (16), and regulation was investigated following peroxide challenge of strain 854 at ME phase growth. Unlike the strong upregulation of msmR and fct genes observed in M-type 3 strain 003Sm, a moderate (2.2-fold) upregulation of rofA and no change in expression of the pilus biosynthesis operon was recorded in M-type 1 GAS. In contrast, strong peroxide-mediated regulation was recorded for FCT-2-independent genes in this strain, such as a 10-fold repression of sloR and 15-fold activation of SPy1198 (Fig. S4).
DISCUSSION
The data presented here support a distinct physiological role for PerR in GAS responses to peroxide. Unlike the upregulation of catalase, ahpCF, mrgA, and heme biosynthesis mediated by PerR in B. subtilis (25, 48), PerR in GAS mediates downregulation of DNA and protein metabolism as well as iron/heme uptake and does not upregulate any known peroxidase(s) or mrgA. Such a decrease in gene expression may be transient, but it would allow for DNA and protein damage repair before normal expression levels resume, similar to decreased DNA replication and cell division reported during the SOS response in other species (48, 66). The finding that a great number of repressed PerR-independent loci also encode proteins involved in DNA and protein metabolism, including rRNA/tRNA synthesis, further supports a model of decreased metabolic function upon oxidative challenge in GAS. The reduced iron/heme uptake coupled to increased Mn2+ import predicted by the adc operon upregulation in our studies is thought to lead to replacement of Fe2+ as a cofactor in enzymes with the Fenton-insensitive Mn2+, thereby allowing optimal bacterial cell function in oxidative environments (2). Higher intracellular Mn2+ would also lead to increased levels of Mn2+-bound PerR and therefore, a gradual decrease in responses initially driven by oxidation of Fe2+-bound PerR species. Lack of downregulation of these cellular functions in a perR mutant would not allow time for damage repair and would result in accumulation of toxic products, which may be the reason, in part, for the attenuated capacity of perR mutant bacteria to resist oxidative killing by phagocytes and to colonize the pharynx (21). Moreover, the strong PerR-dependent regulation of SPy1198 encoding a transcriptional regulator suggests that part of the PerR-mediated responses detected in our studies may be due to SPy1198 upregulation. Initial sequence examination predicts that SPy1198 encodes both a LexA-like peptidase (C terminus) and a helix-turn-helix motif (N terminus), which is characteristic of the domain organization seen in the LexA repressor of the SOS regulon in bacteria (66). This hypothetical function of SPy1198 is in agreement with our current model of PerR involvement in regulating DNA metabolism, and its upregulation could serve to dampen the initial burst in DNA damage repair and/or restore DNA replication. Regulation of this locus seems to be a conserved response in GAS, as peroxide elicited strong upregulation in two independent isolates.
Our findings suggest that iron binding to the putative PerR regulatory site allows responses through iron oxidation but also indicate that binding of manganese and/or other metal(s) occurs in GAS in vivo. Manganese is known to compete with iron for a single regulatory site in Bacillus subtilis PerR (PerRBsu); however, the possibility that it binds to an independent GAS PerR (PerRGAS) site to modulate its function cannot be ruled out. The regulatory site requires H44 and possibly one or more of the three other conserved residues, D93, H99, and D112. Loss of regulation in PerRH44A-expressing GAS and conservation of residues in the predicted regulatory site suggest that the latter is essential to PerRGAS function. Moreover, the conserved structural site requires C104 for stability and coordination of zinc binding, as indicated by the rapid degradation of PerRC104S in vivo and the dramatic reduction in zinc (and iron) content of the recombinant mutant protein. Our result that GST-PerRH44A lacks promoter binding even though it contains wild-type iron levels suggests that H44 is essential for wild-type protein conformation and regulatory function. This mutant seems to be different from PerRH37A in Bacillus, which exhibits an iron binding affinity severalfold lower than that of the wild-type protein (64). Promoter binding of EDTA-treated GST-PerR is consistent with the hypothesis that iron is not required for binding per se, but seems to be required for optimal PerR-regulated responses to peroxide and would serve to mediate PerR oxidation levels and function. Recent evidence indicates that three additional residues (Y92, E114, and H128) coordinate iron binding and stabilize an active PerRBsu conformation (43); however, these residues are not conserved in PerRGAS. Moreover, the apparent limited overlap between the genes controlled by PerRGAS in peroxide with those identified by comparison of wild-type and perR mutant transcriptomes in the absence of stress (see Table S2 in the supplemental material) further indicates an overall unique molecular function mechanism of PerR in Streptococcus. Oxidation of PerRGAS may not lead to its release from bound promoters, a model that would explain the lack of peroxide-mediated derepression of pmtA and other PerR-dependent genes identified previously in the absence of peroxide. Histidine- or GST-tagged recombinant PerR obtained in current or previous studies (21) did not bind to promoters of PerR-dependent loci identified here (adcRCB, SPy1198, and SPy2111-nrdD) although scanning of promoter sequences revealed Per box-like motifs upstream of SPy1198 and SPy2111 (I. Gryllos, unpublished data). It remains possible that regulation of these genes is not direct or that PerRGAS requires other factors and/or conditions for binding to certain promoters.
In addition to newly characterized PerR-regulated gene expression, identification of PerR-independent upregulated responses in peroxide provides further insight into GAS defense strategies against oxidative stress. The stronger activation of DNA damage repair mechanisms over ROS neutralizing enzymes observed in the current study implies a key role of the former in GAS survival under oxidative conditions. DNA repair is considered the primary defense mechanism against ROS in certain bacterial species including Staphylococcus (9, 11). Moreover, strong upregulation of cofactor metabolism and lplA (SPy1214) indicates a contribution of lipoic acid protein modification in streptococcal resistance to oxidative damage. GAS carries a gene that encodes a second LplA homologue (SPy1033) (61) that is not upregulated by peroxide, a finding that implicates SPy1214 in independent or overlapping lipoylation reactions required for optimal survival in oxidative environments. Reduced lipoic acids (dihydrolipoic acids) on pyruvate dehydrogenase complexes have been shown to contribute to mycobacterial defenses against oxidative stress by providing the reducing capacity to AhpC for detoxification of peroxides and peroxynitrites (8, 63). Additionally, lipoic acids can directly (i) detoxify ROS, (ii) chelate iron, which generates ROS via the Fenton reaction, and (iii) reduce thioredoxins as well as GSSG to GSH (45). Induction of the suf locus is likely to be essential for Fe-S maintenance and therefore optimal GAS resistance to oxidative damage. In E. coli, OxyR-mediated activation of the Suf system by peroxide is critical for both de novo synthesis and repair of oxidized Fe-S, even though this species carries a gene that encodes a protein involved in a second Fe-S biosynthesis mechanism (29).
Furthermore, the strong PerR-independent regulation of putative or known regulatory loci recorded in our studies is thought to be of significance, as each of these could control expression of one or more products modulating GAS defenses against ROS. Peroxide-mediated activation of MsmR and the pilus biosynthesis operon is expected to enhance GAS virulence. Pili have been associated with adherence to human tonsil epithelium and to primary human epithelial cells in vitro (1, 44); thus, increased expression elicited by ROS during throat colonization would augment GAS adherence to the pharyngeal mucosa and promote establishment of infection. Upregulation of MsmR could have extra implications on colonization, as it is known to activate expression of fibronectin-binding proteins and other adhesins, in addition to the pilus (50). Increased FCT locus expression in M-type 3, but not M-type 1, GAS predicts strain-dependent effects of peroxide on pilus and other MsmR-regulated loci.
In summary, identification of global transcriptional changes elicited by peroxide reveals uncharacterized streptococcal defense mechanisms and a unique physiological role of PerR in bacterial responses to oxidative stress. The differential regulatory capacity of PerR influenced by environmental metal concentrations and ROS predicts a dynamic pattern of regulation in vivo that is likely to be adaptive to the host environment and the innate immune responses encountered. Future examination of newly identified PerR-dependent and -independent defense mechanisms will expand our knowledge of streptococcal virulence strategies and pathogenesis in the human host.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by NIH grant AI080802 to I.G.
We thank Charles Langmuir and Zhongxing Chen at the Department of Earth and Planetary Sciences, Harvard University, for help with mass spectrometry analyses and Alessandro Muzzi for help with bioinformatics. We also thank John Helmann and Gregory Smaldone for helpful discussions.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 23 September 2011.
REFERENCES
- 1. Abbot E. L., et al. 2007. Pili mediate specific adhesion of Streptococcus pyogenes to human tonsil and skin. Cell. Microbiol. 9:1822–1833 [DOI] [PubMed] [Google Scholar]
- 2. Anjem A., Varghese S., Imlay J. A. 2009. Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol. Microbiol. 72:844–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bates C. S., Montanez G. E., Woods C. R., Vincent R. M., Eichenbaum Z. 2003. Identification and characterization of a Streptococcus pyogenes operon involved in binding of hemoproteins and acquisition of iron. Infect. Immun. 71:1042–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beres S. B., et al. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. U. S. A. 99:10078–10083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brenot A., King K. Y., Caparon M. G. 2005. The PerR regulon in peroxide resistance and virulence of Streptococcus pyogenes. Mol. Microbiol. 55:221–234 [DOI] [PubMed] [Google Scholar]
- 6. Brenot A., King K. Y., Janowiak B., Griffith O., Caparon M. G. 2004. Contribution of glutathione peroxidase to the virulence of Streptococcus pyogenes. Infect. Immun. 72:408–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brenot A., Weston B. F., Caparon M. G. 2007. A PerR-regulated metal transporter (PmtA) is an interface between oxidative stress and metal homeostasis in Streptococcus pyogenes. Mol. Microbiol. 63:1185–1196 [DOI] [PubMed] [Google Scholar]
- 8. Bryk R., Lima C. D., Erdjument-Bromage H., Tempst P., Nathan C. 2002. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science 295:1073–1077 [DOI] [PubMed] [Google Scholar]
- 9. Buchmeier N. A., et al. 1995. DNA repair is more important than catalase for Salmonella virulence in mice. J. Clin. Invest. 95:1047–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carapetis J. R., Steer A. C., Mulholland E. K., Weber M. 2005. The global burden of group A streptococcal diseases. Lancet Infect. Dis. 5:685–694 [DOI] [PubMed] [Google Scholar]
- 11. Chang W., Small D. A., Toghrol F., Bentley W. E. 2006. Global transcriptome analysis of Staphylococcus aureus response to hydrogen peroxide. J. Bacteriol. 188:1648–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen L., Keramati L., Helmann J. D. 1995. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc. Natl. Acad. Sci. U. S. A. 92:8190–8194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cox M. M. 2007. Regulation of bacterial RecA protein function. Crit. Rev. Biochem. Mol. Biol. 42:41–63 [DOI] [PubMed] [Google Scholar]
- 14. Dintilhac A., Alloing G., Granadel C., Claverys J. P. 1997. Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and Mn resulting from inactivation of putative ABC metal permeases. Mol. Microbiol. 25:727–739 [DOI] [PubMed] [Google Scholar]
- 15. Falugi F., et al. 2008. Sequence variation in group A Streptococcus pili and association of pilus backbone types with Lancefield T serotypes. J. Infect. Dis. 198:1834–1841 [DOI] [PubMed] [Google Scholar]
- 16. Ferretti J. J., et al. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. U. S. A. 98:4658–4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gerlach D., Reichardt W., Vettermann S. 1998. Extracellular superoxide dismutase from Streptococcus pyogenes type 12 strain is manganese-dependent. FEMS Microbiol. Lett. 160:217–224 [DOI] [PubMed] [Google Scholar]
- 18. Gibson C. M., Caparon M. G. 1996. Insertional inactivation of Streptococcus pyogenes sod suggests that prtF is regulated in response to a superoxide signal. J. Bacteriol. 178:4688–4695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gon S., Beckwith J. 2006. Ribonucleotide reductases: influence of environment on synthesis and activity. Antioxid. Redox Signal. 8:773–780 [DOI] [PubMed] [Google Scholar]
- 20. Green D. E., Morris T. W., Green J., Cronan J. E., Jr., Guest J. R. 1995. Purification and properties of the lipoate protein ligase of Escherichia coli. Biochem. J. 309:853–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gryllos I., et al. 2008. PerR confers phagocytic killing resistance and allows pharyngeal colonization by group A Streptococcus. PLoS Pathog. 4:e1000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gryllos I., et al. 2007. Mg2+ signalling defines the group A streptococcal CsrRS (CovRS) regulon. Mol. Microbiol. 65:671–683 [DOI] [PubMed] [Google Scholar]
- 23. Gryllos I., et al. 2008. Induction of group A Streptococcus virulence by a human antimicrobial peptide. Proc. Natl. Acad. Sci. U. S. A. 105:16755–16760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hanks T. S., Liu M., McClure M. J., Lei B. 2005. ABC transporter FtsABCD of Streptococcus pyogenes mediates uptake of ferric ferrichrome. BMC Microbiol. 5:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Helmann J. D., et al. 2003. The global transcriptional response of Bacillus subtilis to peroxide stress is coordinated by three transcription factors. J. Bacteriol. 185:243–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Herbig A. F., Helmann J. D. 2001. Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol. Microbiol. 41:849–859 [DOI] [PubMed] [Google Scholar]
- 27. Horsburgh M. J., Clements M. O., Crossley H., Ingham E., Foster S. J. 2001. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 69:3744–3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ibarra J. R., et al. 2008. Role of the Nfo and ExoA apurinic/apyrimidinic endonucleases in repair of DNA damage during outgrowth of Bacillus subtilis spores. J. Bacteriol. 190:2031–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jang S., Imlay J. A. 2010. Hydrogen peroxide inactivates the Escherichia coli Isc iron-sulphur assembly system, and OxyR induces the Suf system to compensate. Mol. Microbiol. 78:1448–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Janulczyk R., Pallon J., Bjorck L. 1999. Identification and characterization of a Streptococcus pyogenes ABC transporter with multiple specificity for metal cations. Mol. Microbiol. 34:596–606 [DOI] [PubMed] [Google Scholar]
- 31. Kanehisa M., Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28:27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kessler D. 2006. Enzymatic activation of sulfur for incorporation into biomolecules in prokaryotes. FEMS Microbiol. Rev. 30:825–840 [DOI] [PubMed] [Google Scholar]
- 33. King K. Y., Horenstein J. A., Caparon M. G. 2000. Aerotolerance and peroxide resistance in peroxidase and PerR mutants of Streptococcus pyogenes. J. Bacteriol. 182:5290–5299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kreikemeyer B., et al. 2007. The Streptococcus pyogenes serotype M49 Nra-Ralp3 transcriptional regulatory network and its control of virulence factor expression from the novel eno ralp3 epf sagA pathogenicity region. Infect. Immun. 75:5698–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Larkin M. A., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 36. LaSarre B., Federle M. J. 2011. Regulation and consequence of serine catabolism in Streptococcus pyogenes. J. Bacteriol. 193:2002–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee J. W., Helmann J. D. 2006. Biochemical characterization of the structural Zn2+ site in the Bacillus subtilis peroxide sensor PerR. J. Biol. Chem. 281:23567–23578 [DOI] [PubMed] [Google Scholar]
- 38. Lee J. W., Helmann J. D. 2006. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440:363–367 [DOI] [PubMed] [Google Scholar]
- 39. Lei B., et al. 2003. Identification and characterization of HtsA, a second heme-binding protein made by Streptococcus pyogenes. Infect. Immun. 71:5962–5969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lei B., et al. 2002. Identification and characterization of a novel heme-associated cell surface protein made by Streptococcus pyogenes. Infect. Immun. 70:4494–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Loo C. Y., Mitrakul K., Voss I. B., Hughes C. V., Ganeshkumar N. 2003. Involvement of the adc operon and manganese homeostasis in Streptococcus gordonii biofilm formation. J. Bacteriol. 185:2887–2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lu A. L., Li X., Gu Y., Wright P. M., Chang D. Y. 2001. Repair of oxidative DNA damage: mechanisms and functions. Cell Biochem. Biophys. 35:141–170 [DOI] [PubMed] [Google Scholar]
- 43. Ma Z., Lee J. W., Helmann J. D. 2011. Identification of altered function alleles that affect Bacillus subtilis PerR metal ion selectivity. Nucleic Acids Res. 39:5036–5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Manetti A. G., et al. 2007. Streptococcus pyogenes pili promote pharyngeal cell adhesion and biofilm formation. Mol. Microbiol. 64:968–983 [DOI] [PubMed] [Google Scholar]
- 45. Moini H., Packer L., Saris N. E. 2002. Antioxidant and prooxidant activities of alpha-lipoic acid and dihydrolipoic acid. Toxicol. Appl. Pharmacol. 182:84–90 [DOI] [PubMed] [Google Scholar]
- 46. Mora M., et al. 2005. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc. Natl. Acad. Sci. U. S. A. 102:15641–15646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Morris T. W., Reed K. E., Cronan J. E., Jr 1994. Identification of the gene encoding lipoate-protein ligase A of Escherichia coli. Molecular cloning and characterization of the lplA gene and gene product. J. Biol. Chem. 269:16091–16100 [PubMed] [Google Scholar]
- 48. Mostertz J., Scharf C., Hecker M., Homuth G. 2004. Transcriptome and proteome analysis of Bacillus subtilis gene expression in response to superoxide and peroxide stress. Microbiology 150:497–512 [DOI] [PubMed] [Google Scholar]
- 49. Nakata M., et al. 2009. Mode of expression and functional characterization of FCT-3 pilus region-encoded proteins in Streptococcus pyogenes serotype M49. Infect. Immun. 77:32–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nakata M., Podbielski A., Kreikemeyer B. 2005. MsmR, a specific positive regulator of the Streptococcus pyogenes FCT pathogenicity region and cytolysin-mediated translocation system genes. Mol. Microbiol. 57:786–803 [DOI] [PubMed] [Google Scholar]
- 51. O'Loughlin R. E., et al. 2007. The epidemiology of invasive group A streptococcal infection and potential vaccine implications: United States, 2000-2004. Clin. Infect. Dis. 45:853–862 [DOI] [PubMed] [Google Scholar]
- 52. Podbielski A., et al. 1996. Molecular characterization of group A streptococcal (GAS) oligopeptide permease (opp) and its effect on cysteine protease production. Mol. Microbiol. 21:1087–1099 [DOI] [PubMed] [Google Scholar]
- 53. Podbielski A., Woischnik M., Leonard B. A., Schmidt K. H. 1999. Characterization of nra, a global negative regulator gene in group A streptococci. Mol. Microbiol. 31:1051–1064 [DOI] [PubMed] [Google Scholar]
- 54. Poyart C., et al. 2001. Contribution of Mn-cofactored superoxide dismutase (SodA) to the virulence of Streptococcus agalactiae. Infect. Immun. 69:5098–5106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Riboldi G. P., Verli H., Frazzon J. 2009. Structural studies of the Enterococcus faecalis SufU [Fe-S] cluster protein. BMC Biochem. 10:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ricci S., Janulczyk R., Bjorck L. 2002. The regulator PerR is involved in oxidative stress response and iron homeostasis and is necessary for full virulence of Streptococcus pyogenes. Infect. Immun. 70:4968–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Romantsov T., Guan Z., Wood J. M. 2009. Cardiolipin and the osmotic stress responses of bacteria. Biochim. Biophys. Acta 1788:2092–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rosch J. W., Hsu F. F., Caparon M. G. 2007. Anionic lipids enriched at the ExPortal of Streptococcus pyogenes. J. Bacteriol. 189:801–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Savic D. J., McShan W. M., Ferretti J. J. 2002. Autonomous expression of the slo gene of the bicistronic nga-slo operon of Streptococcus pyogenes. Infect. Immun. 70:2730–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Smoot J. C., et al. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. U. S. A. 99:4668–4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Spalding M. D., Prigge S. T. 2010. Lipoic acid metabolism in microbial pathogens. Microbiol. Mol. Biol. Rev. 74:200–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sun X., Ge R., Chiu J. F., Sun H., He Q. Y. 2008. Lipoprotein MtsA of MtsABC in Streptococcus pyogenes primarily binds ferrous ion with bicarbonate as a synergistic anion. FEBS Lett. 582:1351–1354 [DOI] [PubMed] [Google Scholar]
- 63. Tian J., et al. 2005. Mycobacterium tuberculosis appears to lack alpha-ketoglutarate dehydrogenase and encodes pyruvate dehydrogenase in widely separated genes. Mol. Microbiol. 57:859–868 [DOI] [PubMed] [Google Scholar]
- 64. Traore D. A., et al. 2009. Structural and functional characterization of 2-oxo-histidine in oxidized PerR protein. Nat. Chem. Biol. 5:53–59 [DOI] [PubMed] [Google Scholar]
- 65. Tsou C. C., et al. 2008. An iron-binding protein, Dpr, decreases hydrogen peroxide stress and protects Streptococcus pyogenes against multiple stresses. Infect. Immun. 76:4038–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Walker G. C. 1985. Inducible DNA repair systems. Annu. Rev. Biochem. 54:425–457 [DOI] [PubMed] [Google Scholar]
- 67. Yellaboina S., Seshadri J., Kumar M. S., Ranjan A. 2004. PredictRegulon: a web server for the prediction of the regulatory protein binding sites and operons in prokaryote genomes. Nucleic Acids Res. 32:W318–W320 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.