Abstract
The production of N-acyl homoserine lactones (AHLs) is widely distributed within the marine Roseobacter clade, and it was proposed that AHL-mediated quorum sensing (QS) is one of the most common cell-to-cell communication mechanisms in roseobacters. The traits regulated by AHL-mediated QS are yet not known for members of the Roseobacter clade, but production of the antibiotic tropodithietic acid (TDA) was supposed to be controlled by AHL-mediated QS in Phaeobacter spp. We describe here for the first time the functional role of luxR and luxI homologous genes of an organism of the Roseobacter clade, i.e., pgaR and pgaI in Phaeobacter gallaeciensis. Our results demonstrate that the AHL synthase gene pgaI is responsible for production of N-3-hydroxydecanoylhomoserine lactone (3OHC10-HSL). Insertion mutants of pgaI and pgaR are both deficient in TDA biosynthesis and the formation of a yellow-brown pigment when grown in liquid marine broth medium. This indicates that in P. gallaeciensis the production of both secondary metabolites is controlled by AHL-mediated QS. Quantitative real-time PCR showed that the transcription level of tdaA, which encodes an essential transcriptional regulator for TDA biosynthesis, decreased 28- and 51-fold in pgaI and pgaR genetic backgrounds, respectively. These results suggest that both the response regulator PgaR and the 3OHC10-HSL produced by PgaI induce expression of tdaA, which in turn positively regulates expression of the tda genes. Moreover, we confirmed that TDA can also act as autoinducer in P. gallaeciensis, as previously described for Silicibacter sp. strain TM1040, but only in the presence of the response regulator PgaR.
INTRODUCTION
Quorum sensing (QS) is a population-density-dependent chemical communication system used by bacteria to control cellular functions through excreted small signaling molecules that interact directly to regulate the expression of sets of genes within certain bacterial species (10). By far the most intensively investigated family of intercellular signal molecules are the N-acylhomoserine lactones (AHLs), which are most common among Gram-negative bacteria and have become a paradigm for bacterial intercellular signaling. AHLs are synthesized by a LuxI type AHL synthase and directly bind to their cognate LuxR type transcriptional regulator proteins, thus activating the expression of target genes mediating a specific response (18, 51). In Roseobacter genomes a considerably high complexity of these canonical luxI/luxR-like genes, as well as “orphan” luxR genes, are present (50), which is consistent with the experimental evidence for AHL production by many members of the Roseobacter clade (6, 23, 32, 34, 46, 52, 53, 56). AHLs have also been detected in roseobacters obtained from samples of marine snow (23) and marine sponge (34, 52), emphasizing AHL-mediated QS as one of the most common intercellular communication mechanisms in roseobacters (20).
The various AHL compounds described for roseobacters differ from one another in chain length, ranging from C8-HSL to C18-HSL, and modifications on their acyl side chain by one or two double bounds or by a 3-hydroxyl or 3-oxo group (8, 56). Often multiple AHLs are produced by a single strain, such as C18en-HSL and N-3-hydroxydecanoyl-homoserine lactone (3OHC10-HSL) excreted by Phaeobacter inhibens strain T5 (56). A new class of HSLs, the p-coumaryl-HSL, is produced by Silicibacter pomeroyi in the presence of p-coumaric acid (46) and seems also to be important for the host-symbiont relationship of Phaeobacter gallaeciensis (49).
Members of the Roseobacter clade and especially Phaeobacter spp. are often associated with diverse marine eukaryotes or organic particles (9), suggesting that surface association and colonization is central to the ecology of clade members (20, 50). Generally, the production of signaling molecules is, in addition to motility, chemotaxis, and the production of antimicrobial metabolites, a key factor in colonization success (50). However, despite the widespread occurrence of AHLs within the Roseobacter clade, to our knowledge, no traits regulated by AHL-mediated QS have been described thus far for these organisms.
Production of the antibiotic tropodithietic acid (TDA) is a trait of a subgroup of marine Rhodobacteraceae composed of the genera Phaeobacter, Silicibacter, Ruegeria, and Pseudovibrio. In all TDA-producing strains investigated thus far, the production of TDA co-occurred with the formation of a yellow-brown pigment (5–8, 19, 21–23, 38). The pigment was produced under the same conditions as TDA (6–8, 24, 38) and depends on the same genes (22). Therefore, the production of the pigment was used as a reliable indicator for TDA production in various studies (5–8, 19, 21–23, 38). For Phaeobacter sp. strain 27-4 it was speculated that the pigment could be a polyphenolic polymer, while in contrast to TDA the pigment has no antimicrobial activity (8). In Phaeobacter sp. strain Y4I a bipyridyl pigment was identified, whose production correlated with antagonistic behavior (50), but it is unknown whether this is the yellow-brown pigment produced by the other organisms mentioned above.
TDA seems to play an important role in colonization success (42) due to the competitive advantage over other species (5, 8, 19, 37) and was proposed to be important for the symbiosis of P. gallaeciensis and Silicibacter sp. strain TM1040 with microalga, where TDA protects the algal host from bacterial pathogens (19, 49). The competitive advantage also has practical applications, since the presence of Phaeobacter and Ruegeria spp. in turbot larval farms was found to suppress growth of marine pathogens such as Vibrio anguillarum (37) and enhanced survival of the larvae (45). Thus, the use of these roseobacters as probiotics in aquaculture facilities (25, 36, 38) or as antifouling agents in marine systems (15, 40) is of great interest.
Due to the correlation of TDA and AHL production in several organisms, it was speculated that the expression of TDA is controlled by AHL-mediated QS (8, 23, 32). This was supported by detection of antimicrobial activity only at high cell densities in the AHL producing Phaeobacter strain 27-4 (6, 7). In contrast, Silicibacter sp. strain TM1040 lacks genes coding for known QS systems and does not produce common AHL molecules (6, 35), but it was suggested that TDA can act as autoinducer of its own synthesis in this organism (19). Biosynthesis of TDA in Silicibacter sp. strain TM1040 requires the tda genes tdaA, tdaB, tdaC, tdaD, tdaE, and tdaF that are specifically involved in TDA biosynthesis (22), with TdaA acting as a positive regulator of tdaCDE gene expression (21).
In the present study we investigated the role of AHL-mediated QS in secondary metabolite production of Phaeobacter gallaeciensis DSM 17395. We identified genes with homology to known luxR-luxI quorum-sensing systems and could show that these are essential for the synthesis of TDA. Furthermore, the results revealed that the LuxR-type transcriptional regulator in strain DSM 17395 (PgaR) and 3OHC10-HSL or, alternatively, TDA are required to induce the expression of tdaA and subsequently for the production of TDA.
MATERIALS AND METHODS
Bacterial strains and media.
The strains and plasmids used in the present study are listed in Table 1. P. gallaeciensis strains were routinely grown on marine broth 2216 medium (MB; BD Biosiences, Franklin Lakes, NJ). Since the pH increased in MB cultures to pH 8.6 and AHLs are not stable at high pH (58), the MB was buffered with 0.3 M HEPES (Carl Roth, Karlsruhe, Germany) and adjusted to pH 7.0 (8). Unless otherwise stated, liquid cultures of Phaeobacter strains were grown in Erlenmeyer flasks at 28°C on a rotary shaker at 90 rpm and were inoculated with 2% preculture grown in the same medium and under the same conditions. When required, antibiotics were added to a final concentration of 60 μg of kanamycin/ml or 25 μg of gentamicin/ml. Escherichia coli strains were used for mutagenesis and grown in Luria-Bertani (LB) medium at 37°C with shaking at 100 rpm or on corresponding solid agar medium (15 g of agar liter−1). When required, gentamicin, kanamycin, ampicillin, or tetracycline were added to a final concentration of 25, 25, 250, or 10 μg ml−1, respectively.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype, phenotype, and/or characteristic(s) | Source or referenceb |
|---|---|---|
| Strains | ||
| Phaeobacter gallaeciensis | ||
| DSM 17395 | Wild-type strain | DSMZ |
| WP38 | DSM 17395 pgaI::Gm; Gmr | This study |
| WP52 | DSM 17395 pgaR::EZTn5; Gmr | This study |
| WP75 | DSM 17395 tdaA::EZTn5; Gmr | This study |
| Escherichia coli | ||
| DH5α | F−endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG φ80dlacZΔM15 Δ(lacZYA-argF)U169 hsdR17(rK− mK+) λ− | 24 |
| TransforMax EC100D pir-116 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ−rpsL (Strr) nupG pir-116(DHFR) | Epicentre |
| Pseudoalteromonas tunicata | DSM 14096 | DSMZ |
| Agrobacterium tumefaciens NTL4 | AHL biosensor strain | 12 |
| Plasmids | ||
| pBBR1MCS-2 | Broad-host-range vector; Kmr | 28 |
| pBBR1MCS-5 | Source of gentamicin resistance cassette; Gmr | 28 |
| pBluescript KS(+) | Cloning vector; Ampr | MBI Fermentas |
| pMB21 | PCR product pgaI-f pgaR-r cloned into EcoICRI site of pBluescript KS(+); Ampr | This study |
| pMB22 | Gm cloned into EcoICRI site of pMB21 | This study |
| pMOD3gm | Gm cloned into SmaI site of EZ-Tn5pMOD<R6Kgori/MCS> | This study |
| pRK415iq | Broad-host-range expression vector; Tcr | 27 |
| pRKpgaR | PCR amplified pgaR ligated into EcoICRI site of pRK415iq; Tcr | This study |
| pBB0808 | PCR amplified lac pgaR from pRK415iqpgaR cloned into pBBR1MCS-2; Kmr | This study |
| EZ-Tn5pMOD<R6Kγgori/MCS> | Transposon construction vector | Epicentre |
Ampr, ampicillin resistance; Kmr, kanamycin resistance; Tcr, tetracycline resistance; Gmr, gentamicin resistance.
DSMZ, German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany.
Bioassay for AHL synthesis.
For the detection of AHLs from P. gallaeciensis DSM 17395 and derived mutants the biosensor strain Agrobacterium tumefaciens NTL4(pZLR4) was used (12). The well diffusion assay was carried out as described by Ravn et al. (43) with some minor modifications. The ABT-agar was supplemented with 40 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; Carl Roth)/ml. Next, 50 μl of the test substance was added to each well. The plates were incubated 24 h at 20°C before the diameters of the AHL-induced zones surrounding the wells were measured. As a negative control, acetonitrile and MB were used. For the R-3OHC10-HSL standard curve, dilutions of R-3OHC10-HSL ranging from 1 to 100 nmol (1, 5, 10, 25, 50, and 100 nmol) were prepared in acetonitrile. The absence of a blue zone in the lawn indicated that the R-3OHC10-HSL concentration was less than 5 nM.
Extraction and identification of AHLs from P. gallaeciensis.
Cells of the P. gallaeciensis wild type and the pgaI- and pgaR-negative mutants were grown, each in 1-liter Erlenmeyer flasks with 100 ml of MB containing 2% Amberlite XAD-16 (Sigma-Aldrich, Germany). Then, 2-ml precultures were added to the medium, followed by incubation for 21 h at 28°C on a rotary shaker (90 rpm) to an optical density at 600 nm (OD600) of ca. 2.0. The AHLs were extracted according to the method of Wagner-Döbler et al. (56), except that a 10:1 dichloromethane-water solvent system was used instead of methanol for the extraction of XAD-16 resin. The two phases were separated, and the organic phase was evaporated to dryness and resolved in low amount of dichloromethane (14). Analysis of the dichloromethane extracts was carried out on a GC System 7890A connected to a 5975C mass selective detector (Agilent Technologies, Palo Alto, CA) with split/splitless injector. The instrument was equipped with a fused-silica-capillary column (HP-5 ms, 30 m by 0.25 mm [inner diameter], 0.25-μm film thickness; Agilent Technologies). The temperature conditions were as follows: 5 min at 50°C, followed by increases at 5°C/min to 320°C and operation in splitless mode. The carrier gas was helium at a flow rate of 1.2 ml min−1. The injection volume was 1 μl, and the front inlet temperature was held at 250°C. The configuration of 3OHC10-HSL was determined as described previously (53).
Characterization of the antibiotic produced by strain DSM 17395.
P. gallaeciensis DSM 17395 was incubated in MB for 20 h at 28°C and 90 rpm. Cells were removed first by centrifugation (6,000 × g, 15 min, 4°C) and then by filtration through a 0.22-μm-pore-size mixed-cellulose-ester membrane (Carl Roth), resulting in cell-free supernatant. The pH of the supernatant was adjusted to 3.0 with 2 M HCl, followed by extraction with 20 ml of ethyl acetate repeated two times. The vacuum-dried ethyl acetate extract was dissolved in 1 ml of acetonitrile and analyzed on a high-pressure liquid chromatography (HPLC) system (Goebel Instrumentelle Analytik, Hallertau, Germany) equipped with a diode array detector and an evaporated light-scattering detector. Samples were separated on a Nucleodur 100–5 C18 EC column (Macherey-Nagel, Düren, Germany) by using an acetonitrile-water gradient system containing 0.1% trifluoroacetic acid, started with 20% acetonitrile, which was increased linearly to 100% in 25 to 30 min. Extraction and HPLC analysis were carried out by BioViotica Naturstoffe GmbH, Göttingen, Germany.
Assay of TDA production.
Antimicrobial activity of P. gallaeciensis DSM 17395 and derived strains against Pseudoalteromonas tunicata DSM 14096 was determined by agar diffusion tests according to the method of Brinkhoff et al. (5) with the following modifications. Exponentially grown cultures of the target organism were adjusted to an OD600 of 0.0025, and 1 ml was spread onto each plate with half-strength MB 2216 agar. Cells were removed from cultures first by centrifugation (6,000 × g, 15 min, 4°C) and then by filtration through a 0.22-μm-pore-size mixed-cellulose-ester membrane (Carl Roth). From this cell-free filtrate, 20 μl was applied onto an antibiotic assay disk (Rotilabo-test leaves; Carl Roth), followed by incubation for 20 h at 20°C. For the generation of standard curves, pure TDA (Bioviotica Naturstoffe GmbH, Göttingen, Germany) dissolved in dimethyl sulfoxide (DMSO) at 1 mM and a standard set containing seven concentrations (1, 2.5, 5, 10, 25, 50, and 100 μM) and DMSO as a negative control were used. For each assay, an internal standard curve was created in triplicates by calculating the ratio of TDA concentrations and the mean diameter of the zones. Only standard curves with a correlation coefficient (r2) of >0.97 were used for quantification. The detection limit of this assay was less than 2.5 μM.
Time course of TDA production.
TDA production by P. gallaeciensis DSM 17395 was measured during growth on MB at 28°C and 90 rpm. Aliquots were collected at various times (Fig. 1) and used for determination of antimicrobial activity (see above). The OD600 was measured by spectroscopy (DU520; Beckman Instruments, Fullerton, CA). Bacterial cell counts were derived from 200 μl of bacterial culture fixed with 2% (final concentration) formaldehyde and stored at −80°C until further processing. Bacteria were enumerated after staining with SybrGreen I by epifluorescence microscopy as described previously (31), after aggregates were dissolved with sonication. Therefore, the bacteria were thawed, diluted 1:10 in phosphate-buffered saline, and lysed by sonication on ice (Bandelin Sonopuls HD2000 with a SH 213 booster horn and an MS 73 tip) three times for 10 s with a 10-s break at 15% power. A 500- to 1,000-μl subsample of the corresponding dilution was filtered through a black 0.22-μm-pore-size polycarbonate filter (GE, 25-mm diameter). SybrGreen I-stained cells were counted with an Axiolab 2 microscope (Zeiss, Jena, Germany). The filtered sample volume yielded 30 to 100 stained cells in the counting grid. For each sample, 15 grids and a minimum of 500 cells per filter were enumerated.
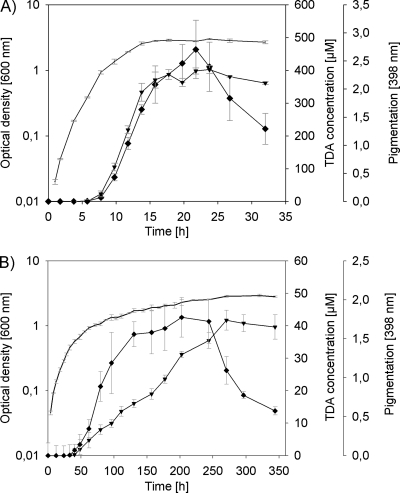
Fig. 1.
(A and B) Time course of TDA and pigment production of P. gallaeciensis grown on MB at 28°C under shaken (A) and stagnant (B) conditions. Bacterial growth (−) is shown as absorption at 600 nm as the average of three parallel cultures. TDA production (♦) of cell-free culture fluids was measured using agar diffusion assays with P. tunicata as target strain (n = 3). The TDA concentration was calculated by the use of an internal standard curve with pure TDA (r2 = 0.98). Pigmentation (▾) of cell-free culture fluids was measured by spectroscopy at 398 nm (n = 3).
Transformation of P. gallaeciensis.
Cells were prepared for electroporation according to the procedure of Dower et al. (14a) with minor changes. P. gallaeciensis cells were grown in MB at 28°C with shaking at 90 rpm to a cell density of 7.5 × 108 cell/ml and washed four times with 10% glycerol. Subsequently, the cell pellet was resuspended in 10% (vol/vol) glycerol to yield an approximate concentration of 1010 to 1011 cells/ml. Aliquots of 40 μl were stored at −80°C until use. For transformation, 40-μl portions of the cells were mixed with 1 μl of DNA, and the mixtures were electroporated at 13.75 kV/cm. Afterward, the cells were suspended in 1 ml of MB and incubated for 4 h at 28°C and 90 rpm. The cell suspension was diluted and plated on half-strength MB agar containing the appropriate antibiotic and incubated for at least 48 h at 28°C.
Construction of strains and plasmids.
To construct the pgaI gentamicin insertion mutant, the gene, including the flanking regions, was amplified from chromosomal DNA of the P. gallaeciensis wild-type strain DSM 17395 in a PCR using the primers 5′-ACAGTGAACACCCCTGAATATGC-3′ and 5′-GCAGCTGCGTAAATGTCAGGC-3′. The resulting PCR product was ligated into EcoICRI-digested pBluescript KS(+) to yield pMB21. The gentamicin resistance gene, amplified from pBBR1MCS-5 using a Gm primer set (28), was ligated into the EcoICRI site of pMB21, yielding plasmid pMB22. Electroporation of P. gallaeciensis wild-type cells with pMB22 was carried out as described above. Subsequent selection on gentamicin yielded the insertion mutant strain WP38 (pgaI::gm, Table 1). The insertion was verified by PCR analysis.
The P. gallaeciensis tdaA mutant strain WP75 and the pgaR mutant strain WP52 were obtained by transposon mutagenesis using the EZ-Tn5 pMOD3<R6Kgori/MCS> transposon construction vector (Epicentre, Madison, WI). For preparation of the EZ-Tn5 transposon a gentamicin-resistant gene was amplified from pBBR1MCS-5 using the Gm primer set (28) and ligated into the SmaI site of the EZ-Tn5 pMOD3<R6Kgori/MCS> transposon construction vector, giving rise to pMOD3gm. The PvuII restriction fragment of pMOD3gm was electroporated, along with the EZ-Tn5 transposase, into electrocompetent cells of wild-type DSM 17395. Transposon mutants were selected on half-strength MB agar with gentamicin. Mutants were screened for altered pigmentation compared to the wild-type by replica plating on MB agar. The locations of the insertions of the transposon were determined by rescue cloning of the candidate mutant as described by Geng et al. (22).
Complementation assays.
For a complementation study, the pgaR gene was PCR amplified from chromosomal DNA of P. gallaeciensis DSM 17395 using the primers 5′-ATGGCAACCAAAGTTAATCTTGATC-3′ and 5′-TTAAATGATGATCAGCCCGAGAC-3′ and cloned into the EcoICRI sites of pRK415iq (27), resulting in pRKpgaR. The open reading frame of pgaR and the lac promoter were PCR amplified from pRKpgaR using the primers 5′-CCAATACGCAAACCGCCTCTC-3′ and 5′-TTAAATGATGATCAGCCCGAGAC-3′ and ligated into the EcoICRI site of pBBR1MCS-2 (28), giving rise to plasmid pBB0808. The resulting plasmid was subsequently electroporated into the pgaR mutant (WP52), and transformants harboring pBB0808 were selected on kanamycin. The antimicrobial activity and AHL production of WP52 pBB0808 were determined in bioassays as described above.
The mutation in pgaI (strain WP38) was tested for complementation by exogenous addition of culture fluid from the wild-type DSM 17395, synthetic AHLs, or pure TDA. Sterile-filtered culture fluid obtained after growth of the wild-type in MB for 14 h under shaking conditions (see Fig. 1A) was added to a culture of WP38 (4% [vol/vol]). The AHLs C4, C8, C12, C14, 3oxoC6-AHL, and R-3OHC10-HSL were prepared as described by Wagner-Döbler et al. (56), and 3OHC12-HSL, C10-HSL, and 3oxoC10-HSL were obtained from Sigma-Aldrich. Stock solutions of the AHLs were prepared in acetonitrile and added to MB with a final concentration of 100 nM unless otherwise stated. Pure TDA was obtained from Bioviotica Naturstoffe GmbH. Final concentrations of 0.1, 1, and 10 μM TDA were used for complementation analysis.
RNA preparation and quantitative real-time PCR.
The wild-type and the derived mutants (WP38, WP52, and WP75) were grown in 20 ml of MB broth supplemented with 0.3 M HEPES, adjusted to pH 7.0, in a 100-ml Erlenmeyer flask at 20°C and 90 rpm. At an OD600 of ca. 1.5, 1 ml of the culture was centrifuged, and the total RNA was extracted from the resulting cell pellet, using a High-Pure RNA isolation kit (Roche Diagnostics GmbH, Mannheim, Germany) according to the manufacturer's instructions. For complete removal of genomic DNA, we performed an additional DNase I degradation step with subsequent inactivation, according to the manufacturer's recommendation (Roche Diagnostics GmbH). cDNA was synthesized from DNA-free RNA with a transcriptor first-strand cDNA synthesis kit (Roche Diagnostics GmbH) using anchored-oligo(dT)18 primer and random hexamer primer. Quantitative reverse transcription-PCR (qRT-PCR) was performed by using probes of the Universal ProbeLibrary (Roche Diagnostics GmbH) and primer pairs (TIP MolBiol, Berlin, Germany) designed for specific genes using the Universal ProbeLibrary Tool (Table 2). qRT-PCR assays were carried out using DNA probe master (Roche Diagnostics GmbH) and the following thermal cycling parameters: preincubation at 95°C for 10 min, followed by amplification in 45 cycles at 95°C for 10 s, 60°C for 30 s, and 72°C for 1 s, and finally 40°C for 30 s (1 cycle) in a LightCycler 480 detection system (Roche Diagnostics GmbH). Target cDNA levels were analyzed with the comparative cycle time (CT) method for real-time RT-PCR, and all values were normalized relative to the expression of rpoB. The reported values show the average and range of three biological replicates assayed each in triplicates. Calibration curves from each gene were created, which cover a range of at least 4 orders of magnitude with a correlation coefficient (r2) of at least 0.98.
Table 2.
Primer and hydrolysis probes used in this study for qRT-PCR
| Gene | Primer | Sequence (5′–3′) | Amplicon (nt) | UPL probe |
|---|---|---|---|---|
| rpoB | rpoB54-f | CTGGACGAAGATGGCAAGTT | 60 | #54 |
| rpoB54-r | GTAGTCGCCGGACTGACG | |||
| pgaI | pgaI26-f | CACGTGATGTGAGTGGTAATGTC | 75 | #26 |
| pgaI26-r | AATGTTGAAATTCCGCATGAT | |||
| tdaA | tdaA47-f | CGGATCTGGAAGTCGCTTT | 63 | #47 |
| tdaA47-r | CGTTGCGAATATCGTCCA | |||
| tdaB | tdaB149-f | GCATGCCACTTGAGGTTATG | 66 | #149 |
| tdaB149-r | AAAGTTTTTCCGGCAGCA | |||
| tdaE | tdaE70-f | ATGGGGCGTCTGTACTATGG | 76 | #70 |
| tdaE70-r | TGGAGAAATCAAATGCCTCA | |||
| tdaF | tdaF152-f | ATCACGATATTCTGGCCATTCT | 60 | #152 |
| tdaF152-r | CATGCGCTATCGCAGACA | |||
| paaG | paaG18-f | CCGAAGGCAACTGGATCA | 76 | #18 |
| paaG18-r | GCCTCATCCTGCACCTTG | |||
| paaZ2 | paaZ32-f | ACCTGCAAGGAAATCACACC | 61 | #32 |
| paaZ32-r | AACAGTCCCAACGCACATCT |
The expression of tda genes in the pgaI-negative background (strain WP38) was also measured by qRT-PCR after exogenous addition of 10 μM TDA, as well as after the addition of 100 nM R-3OHC10-HSL, using the conditions described above. Purified TDA, as well as R-3OHC10-HSL, was individually added to triplicate samples and compared to uninduced samples (also in triplicates). The RNA extraction, cDNA synthesis, and qRT-PCR analysis were all performed as described above.
RESULTS
Growth-phase-dependent TDA production under shaken and stagnant culture conditions.
Strain DSM 17395 produced an antimicrobial compound when grown in MB (Fig. 1). The retention time of 11.8 min and the characteristic UV spectra of the HPLC peak were identical to those of pure TDA published previously (8, 29), demonstrating that TDA is the antibacterial metabolite produced by strain DSM 17395. In this way, TDA was produced when grown in MB under shaken, as well as under stagnant culture conditions, even though TDA was produced in 10-fold-higher concentrations in shaken cultures (Fig. 1). Under both conditions, however, nearly the same maximal cell density of ca. 2 × 109 cells ml−1 was reached. TDA production and concomitant formation of a yellow-brown pigment was undetectable under both culture conditions during the early exponential growth phase and increased considerably during the late exponential phase, reaching maximum levels in the early stationary phase (Fig. 1). In shaken cultures, the production of the brownish pigment was found to be strictly correlated with the TDA synthesis (Fig. 1A), whereas in stagnant cultures the pigment production lagged behind the TDA production but remained below the maximal absorption observed for shaken cultures. Further experiments were carried out with shaken cultures due to the higher TDA production.
Identification of the AHL(s) produced by P. gallaeciensis.
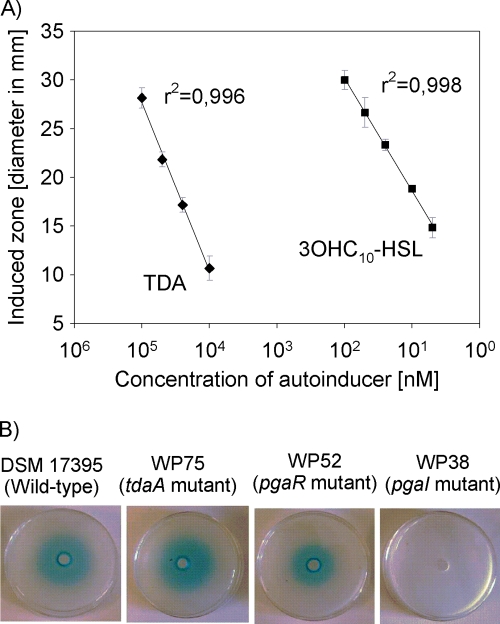
Increasing the amounts of pure 3OHC10-HSL added to the wells in the AHL bioassay with A. tumefaciens NTL4 caused an increase in the diameters of the induced blue zones surrounding the wells, which showed a high correlation coefficient (r2 = 0.998) (Fig. 2A). A concentration-dependent induction of A. tumefaciens was also observed when pure TDA was applied (Fig. 2A). This means that the induction by sterile filtered culture fluid of the wild-type strain DSM 17395 (Fig. 2B) could be due to TDA and/or AHLs. To find out whether AHLs were produced by strain DSM 17395, a sterile-filtered culture fluid of a TDA-negative mutant (strain WP75) was used. A clearly induced zone surrounding the well was visible in the bioassay with this strain (Fig. 2B), verifying that AHLs were produced by P. gallaeciensis.
Fig. 2.
A. tumefaciens NTL4 well diffusion assay. (A) Relationship between the concentration of 3-N-hydroxy-decanoyl-l-homoserine lactone (3OHC10-HSL) and TDA in well diffusion assays and resulting diameters of induced blue zones. Error bars indicate standard deviations (n = 3). (B) Well diffusion assays with sterile-filtered culture fluids from late-exponential-phase cultures of the wild-type strain DSM 17395, the tdaA mutant strain WP75, the pgaR mutant strain WP52, and the pgaI mutant strain WP38. The culture fluids were added to wells in agar containing A. tumefaciens NTL4 and X-Gal. Blue zones surrounding the wells indicate induced β-galactosidase activity.
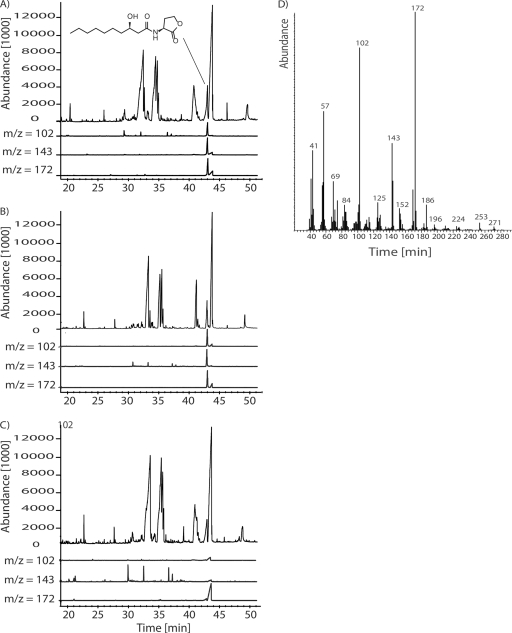
For identification of the produced AHL(s), a gas chromatography-mass spectrometry (GC-MS) analysis of a dichloromethane extract from culture fluid of P. gallaeciensis was performed. The GC-MS analysis was difficult due to presence of large peaks of diketopiperazines from the amino acid-containing MB (56) that masked the AHLs (Fig. 3). The results shown in Fig. 3A revealed, however, presence of a compound showing the typical fragmentation pattern of AHLs: m/z 143 and 102 (11, 56) at 42.5 min. Identification of the AHL as 3-hydroxy-HSL was supported by the prominent ion at m/z 172 at the same retention time (56). The AHL present in the extract was R-3OHC10-HSL, which was unambiguously identified by its mass spectrum and its gas chromatographic retention index, that correspond to those of synthetic 3OHC10-HSL (Fig. 3D). The absolute configuration proved to be pure “(R)-” as described for Phaeobacter inhibens T5.
Fig. 3.
(A to C) Total ion chromatograms and the characteristic ion traces m/z 102, 143, and 172 (slightly offset for improved visibility) of culture extracts of the P. gallaeciensis wild-type strain DSM 17395 (A), pgaR mutant strain WP52 (B), and pgaI mutant strain WP38 (C). (D) Mass spectrum of R-3OHC10-HSL/peak at 42.6 min.
The pgaI gene product catalyzes the synthesis of R-3OHC10-HSL.
The genome of P. gallaeciensis comprises genes coding for a common LuxR-LuxI-type QS system with an AHL synthase PgaI (accession number ZP_02146368) and a LuxR family transcription factor PgaR (accession number ZP_02146367). The derived amino acid sequence of pgaR showed 34% identity to RaiR of Rhizobium etli (accession number O54452), which was predicted to be a typical LuxR-type QS regulator (44) and harbored a characteristic helix-turn-helix domain and a typical autoinducer binding domain. The derived amino acid sequence of pgaI shows 36% identity to the AHL-synthase RaiI of Rhizobium etli (accession number O54451), which is required for the synthesis of autoinducer molecules that are involved in the restriction of nodule number (44).
Culture fluids of mutants lacking pgaI (strain WP38) or pgaR (strain WP52) were analyzed with the AHL assay and GC-MS analysis. It was found that the sterile-filtered supernatant of the pgaR mutant gave a positive signal in the AHL bioassay, whereas the supernatant of the pgaI mutant was negative (Fig. 2B). The GC-MS analysis confirmed these results because the typical fragmentation pattern of AHLs, as found for the wild type (Fig. 3A), was also found in extracts of the pgaR mutant (Fig. 3B), but not for the pgaI mutant (Fig. 3C). Thus, it is obvious that pgaI is responsible for the synthesis of R-3OHC10-HSL, and PgaI can still produce R-3OHC10-HSL in a pgaR-negative background, indicating that PgaR is not essential for the production of R-3OHC10-HSL.
The pgaI-pgaR QS system is essential for TDA production in P. gallaeciensis.
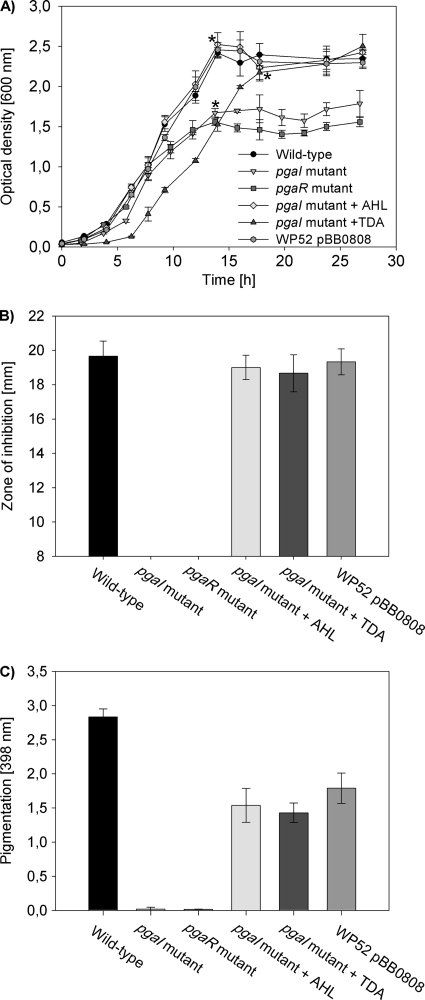
Cell-free culture fluids of the pgaI and the pgaR mutant grown in MB were tested for antimicrobial activity to determine whether TDA production is influenced by the pgaR-pgaI QS system. Culture fluids of both mutants did not inhibit Pseudoalteromonas tunicata in agar diffusion assays, suggesting that they did not produce TDA in contrast to the wild-type (Fig. 4B). Due to the coincident production of TDA with the formation of a yellow-brown pigment, mutations in pgaI and pgaR also resulted in loss of pigmentation (Fig. 4C). TDA and pigment production were restored for the pgaI mutant by the addition of sterile-filtered supernatant from the wild type (Table 3), as well as by the exogenous addition of R-3OHC10-HSL (Table 3 and Fig. 4). The wild-type phenotype was restored dependent on the concentration of R-3OHC10-HSL, i.e., TDA synthesis was restored starting at a concentration of 0.5 nM (Table 3). The exogenous addition of other synthetic AHLs than R-3OHC10-HSL to MB cultures of the pgaI mutant failed to restore TDA and pigment production, indicating that the induction is highly specific to R-3OHC10-HSL (Table 3). However, pigment and TDA production were also restored in cultures of the pgaI mutant by the addition of 1 μM pure TDA (Fig. 4 and Table 3).
Fig. 4.
Growth, antimicrobial activity, and pigmentation of P. gallaeciensis DSM 17395 and derived mutants. The results demonstrate that the PgaI-PgaR QS system of P. gallaeciensis is essential for the TDA and pigment production. (A) Growth curves of P. gallaeciensis wild-type strain DSM 17395, pgaR mutant strain WP52, strain WP52 carrying pBB0808, pgaI mutant strain WP38, and strain WP38 in the presence of either added 100 nM R-3OHC10-HSL or 10 μM TDA. The cultures were grown at 28°C in MB under shaken conditions. Growth was measured by OD600 in triplicate cultures. The asterisks indicate points in time where culture fluids were taken for the analysis shown in panels B and C. (B) Zone of inhibition against P. tunicata determined in agar diffusion assays from cell-free culture fluids of different P. gallaeciensis strains at indicated points in time (n = 3). (C) Pigmentation of different P. gallaeciensis strains measured by spectroscopy at 398 nm (n = 3).
Table 3.
Relative concentration of TDA produced by the pgaI mutant strain WP38 grown in MB medium after the addition of AHLs, TDA, or culture fluids
| Exogenous addition (final concn) | Relative amt (%) of TDAa |
|---|---|
| C4-HSL (100 nM) | 0 |
| 3oxoC6-HSL (100 nM) | 0 |
| C8-HSL (100 nM) | 0 |
| C10-HSL (100 nM) | 0 |
| C12-HSL (100 nM) | 0 |
| 3OHC12-HSL (100 nM) | 0 |
| C14-HSL (100 nM) | 0 |
| 3oxoC10-HSL (100 nM) | 0 |
| R-3OHC10-HSL (0.1 nM) | 0 |
| R-3OHC10-HSL (0.5 nM) | 62 (15) |
| R-3OHC10-HSL (1 nM) | 90 (27) |
| R-3OHC10-HSL (100 nM) | 87 (18) |
| TDA (0.1 μM) | 0 |
| TDA (1 μM) | 72 (21) |
| TDA (10 μM) | 65 (15) |
| DMSO | 0 |
| Acetonitrile | 0 |
| Culture fluids | |
| Wild type | 83 (13) |
| pgaR mutant (WP52) | 72 (18) |
| pgaI mutant (WP38) | 0 |
| MB | 0 |
The values are the average relative percentages of TDA from three independent cultures compared to the wild-type percentage; the range ± is indicated in parentheses.
The fact that PgaR is also required for production of the pigment and TDA was confirmed by recovery of the wild-type phenotype for strain WP52 (pgaR mutant) by a plasmid-borne copy of pgaR (pBB0808). Culture fluid of WP52 carrying pBB0808 showed the same antimicrobial activity as the wild type (Fig. 4B), whereas pigment production was not completely restored (Fig. 4C). In contrast to the pgaI mutant, the wild-type phenotype of the pgaR mutant could not be restored by the exogenous addition of pure R-3OHC10-HSL or TDA.
QS can play a crucial role in bacterial growth, such as was shown for Yersinia pseudotuberculosis or Rhodobacter sphaeroides (1, 39). We measured growth of the wild type, the pgaI mutant (strain WP38), and the pgaR mutant (strain WP52) in MB to test whether QS affects the growth behavior of P. gallaeciensis. Figure 4A shows that the pgaI and pgaR mutants reached lower ODs compared to the wild type, but both strains reached the same cell density as the wild type (ca. 2 × 109 cells ml−1). The difference between the ODs is based on the absorption of the yellow-brown pigment present in the cell-free culture fluid of the wild type. The absorption at 600 nm of a sterile-filtered culture fluid of the wild type after an incubation time of 14 h was 0.6, and the absorption of the cells suspended in fresh MB was 1.8, which is similar to the maximal ODs reached by the pgaI and pgaR mutants. In all complementation experiments in which the TDA and thus pigment production was restored as well (Fig. 4B and C), strains reached the same maximal OD as the wild type (Fig. 4A). The wild-type-like multicellular star-shaped growth behavior, as previously described for Phaeobacter sp. strain 27-4 (8) and other members of the Roseobacter clade (6, 55), was also observed in liquid cultures of the pgaI and pgaR mutants (data not shown). Thus, our results suggest that QS does not affect the growth and aggregation behavior of P. gallaeciensis.
Influence of pgaR on the expression of genes essential for TDA biosynthesis.
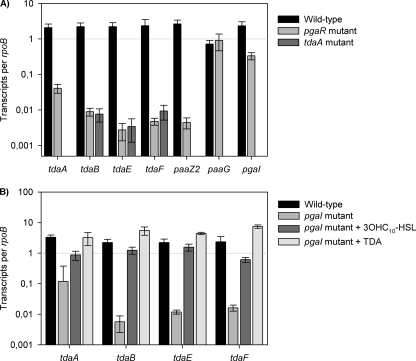
Since the pgaR and pgaI mutants produced no TDA (Fig. 4B), we assumed that the tda genes tdaA to tdaF, whose expression is essential for the synthesis of TDA in Silicibacter sp. strain TM1040 (22), are QS controlled in P. gallaeciensis DSM 17395. The tda genes tdaA, tdaB, tdaE, and tdaF and paaZ2, as well as paaG, are also known to be essential for TDA biosynthesis in strain DSM 17395 (S. Thole et al., unpublished data). Therefore, qRT-PCR was used to compare the transcription levels of the above mentioned genes in the wild-type and the pgaR mutant strain WP52. Measurements of these transcripts showed that the genes tdaB, tdaE, tdaF, and paaZ2 were clearly downregulated in the pgaR mutant compared to the wild type (Fig. 5A). Transcription of paaG was not affected in the pgaR mutant as expected, because paaG is essential for the primary metabolism and not specific to TDA biosynthesis (22). Figure 5A also shows that the transcription level of pgaI was 5-fold higher in the wild type than in the pgaR mutant, suggesting that the transcription of pgaI is slightly positively regulated by PgaR. Interestingly, the expression of tdaA, coding for a transcriptional regulator (19, 21), decreased by a factor of 51 in the pgaR mutant compared to the transcription level of the wild type, indicating that transcription of tdaA is also PgaR dependent in P. gallaeciensis (Fig. 5A).
Fig. 5.
Effects of disrupting pgaR, pgaI, and tdaA genes on the expression of genes essential for TDA biosynthesis at the RNA level. (A) Relative transcription levels of tdaA, tdaB, tdaE, tdaF, paaZ2, paaG, and pgaI from wild-type P. gallaeciensis DSM 17395 (black bars), the pgaR mutant strain WP52, and the tdaA mutant strain WP75 grown in liquid MB and determined by using qRT-PCR analysis of reverse-transcribed RNA samples (see Materials and Methods for details). (B) Relative transcription levels of tda genes of the wild type and the pgaI mutant strain WP38 grown without or in the presence of either exogenously added R-3OHC10-HSL or TDA. The transcription is shown as the relative expression of each target gene compared to rpoB. The error bars are derived from three independent cultures assayed each in triplicate.
Expression of the tda genes was measured in a tdaA-negative background to verify that tdaA is essential for transcription of the tda genes in P. gallaeciensis. Therefore, we used the tdaA mutant strain WP75 (Table 1), which is unable to produce TDA. The results of the qRT-PCR revealed that tdaB, tdaE, and tdaF were strongly downregulated in a tdaA-negative genetic background and confirmed that TdaA is a transcriptional activator for the tda genes in P. gallaeciensis (Fig. 5A). Thus, it is not surprising that a decrease of the tdaA transcription level in the pgaR genetic background results in a decrease of transcripts of the other tda genes (Fig. 5A).
R-3OHC10-HSL or TDA act as autoinducers depending on RaiR.
Expression analyses in the pgaI-negative mutant strain WP38 revealed that the transcription levels of the tda genes, including tdaA (28-fold), were clearly reduced, indicating that R-3OHC10-HSL induces the expression of tdaA (Fig. 5B). Addition of R-3OHC10-HSL to cultures of the pgaI mutant resulted in an increase in tdaA transcription up to the wild-type level (Fig. 5B). We investigated the tdaA expression in the pgaI mutant after the exogenous addition of TDA since the exogenous addition of TDA also restored pigment and TDA production (Fig. 4B and C). The addition of TDA resulted in a tdaA transcription level comparable to that of the wild type (Fig. 5B). Thus, the expression of tdaA requires R-3OHC10-HSL or TDA for maximal transcription.
DISCUSSION
Monitoring TDA production as a function of growth (Fig. 1) showed that TDA accumulated during the transition of the exponential to the stationary phase. This suggests that production of TDA in P. gallaeciensis strain DSM 17395 is cell density dependent and typifies the production of a classical secondary metabolite. Furthermore, this was a first hint that production of TDA could be controlled in a quorum-dependent manner as previously postulated for Phaeobacter sp. strain 27-4, due to the production of AHLs and TDA at high cell densities (8). Growth of strain DSM 17395 under static conditions lead to a decreased growth rate and resulted in 10-fold-lower level of TDA compared to cultivation under shaken conditions (Fig. 1B). It was shown that different Phaeobacter spp. produced TDA when grown under shaken and stagnant conditions, whereas Ruegeria spp. expressed this phenotype only during stagnant conditions (37), which was also observed for Silicibacter sp. strain TM1040 and Phaeobacter sp. strain 27-4 (7, 8). Other than DSM 17395, only Silicibacter pomeroyi DSS-3, of those that have been assessed to date, can produce elevated levels of TDA under shaken conditions (6). The striking discrepancy between these strains in TDA production indicates differences in response to environmental conditions and in the regulation of TDA production.
Production of AHLs was previously shown for several members of the Roseobacter clade (6, 23, 32, 46, 56). An influence of the AHLs on hydrolytic enzymatic activities and antibiotic production in marine snow was already suggested for these bacteria by Gram et al. (23). However, no traits or genetic elements regulated by AHL-mediated QS were identified yet, although the genome sequences provide some obvious clues (50).
In our study we could demonstrate that PgaI is an AHL-synthase that produces R-3OHC10-HSL in P. gallaeciensis (Fig. 3), an AHL that is also produced by other members of the genus Phaeobacter (e.g., Phaeobacter sp. strain 27-4 and Phaeobacter inhibens strain T5), which were shown to produce TDA, too (5, 8, 56). In addition to these strains, AHLs were often detected in other members of the Roseobacter clade that showed antimicrobial activity, like Roseovarius spp., S. pomeroyi DSS-3 (6), P. gallaeciensis 2.10 (41), and Phaeobacter sp. strain Y4I (50). Bacteria of the Silicibacter-Ruegeria subgroup that were isolated from marine sponges were all identified as AHL producers (34). However, there is not always a correlation between AHL production and antimicrobial activity since Silicibacter sp. strain TM1040 produces no common AHL molecules (6, 35) but TDA (22). This suggests a different regulation mechanism for the production of TDA in Silicibacter sp. strain TM1040 compared to the AHL-producing strains, including P. gallaeciensis DSM 17395. Differences between these two organisms are also reflected by production of highest TDA concentrations of strain TM1040 under static conditions, whereas strain DSM 17395 produces 10-fold-higher amounts of TDA at shaken culturing conditions (Fig. 1). As a conclusion, the data mentioned above suggest adaptation of the strains to different ecological niches.
In both QS-negative mutants (WP38 and WP52, Table 1), the expression of tdaA was clearly reduced compared to the wild type (Fig. 5), indicating that the regulatory protein TdaA, which is required for TDA synthesis, was regulated by the pgaR-pgaI QS system. In Silicibacter sp. strain TM1040 it was shown that the expression of tdaAB was not affected by the type of culturing; only the expression of tdaCDEF, and therefore tdaA, was suggested to be expressed constitutively in Silicibacter sp. strain TM1040 (19). Nevertheless, tdaA positively controls the expression of other tda genes, and tdaA is essential to respond to extracellular signaling molecules for the induction of TDA biosynthesis in P. gallaeciensis and Silicibacter sp. strain TM1040.
In the pgaI mutant strain WP38, the wild-type phenotype was restored by addition of R-3OHC10-HSL but not by the exogenous addition of other AHLs (Table 3). This indicates a high level of specificity of the PgaR receptor protein toward R-3OHC10-HSL in P. gallaeciensis. However, TDA production, as well as the expression of tdaA, was also restored in cultures of the pgaI mutant by addition of pure TDA (Fig. 4B and 5B). In Silicibacter sp. strain TM1040 it was shown that the addition of TDA induces expression of tdaC and tdaF, whereas tdaA seems to be expressed constitutively (19). Furthermore, we showed that in P. gallaeciensis TDA production induced by either R-3OHC10-HSL or TDA depends on the response regulator PgaR. Here, PgaR is necessary to induce the expression of tdaA, and TdaA consequently induces the expression of tda genes (Fig. 5A). For Silicibacter sp. strain TM1040 it was hypothesized that TDA is necessary for the maximal activity of TdaA (19), which subsequently induces the expression of tdaCDEF (21). However, the findings that TDA is not required for heterologous tdaC expression in E. coli and that TDA is not necessary for binding of TdaA to tdaC (21) were contradictory to this hypothesis and were already discussed by Geng and Belas (21).
Induction of the monitor system of A. tumefaciens (Fig. 2A) indicates that TDA could also act as an autoinducer in bacteria other than roseobacters. A. tumefaciens is known to respond to many AHLs of different length and substitutions of the acyl-chain moiety and therefore displays a broad detection window, which depends on the specificity of the TraR receptor protein (12, 16, 59). Biosensor strains are often used for AHL detection, but it was also observed that they can be activated by compounds other than AHLs, such as diketopiperazine or mimic compounds (2, 26). Therefore, a direct detection method such as HPLC coupled with MS, as shown in this and previous studies (8, 53, 56), is necessary to unequivocally demonstrate the presence of AHLs. Despite the fact that TDA and R-3OHC10-HSL do not show structural similarity and the only superficial resemblance are the ring structure and the presence of polar and apolar sides in both compounds, it is possible that TDA competes for the same PgaR-binding side as R-3OHC10-HSL. Similar findings were described for diketopiperazine, which antagonize the AHL-mediated induction of bioluminescence (26). TraR of A. tumefaciens as well as PgaR of P. gallaeciensis respond significantly better to 3OHC10-HSL than TDA. In both strains a 2,000-fold-higher concentration of TDA was necessary compared to R-3OHC10-HSL (Table 3; Fig. 2A). This is in accordance with the observation that a LuxR homolog responds best to the AHL produced by its cognate LuxI homolog (26). Furthermore, the threshold concentration of the signal molecule depends on the nature of the signal, the transport and diffusion rate (4), which obviously varies considerably between TDA and R-3OHC10-HSL. It is also possible that both classes of signal molecules, TDA and R-3OHC10-HSL, can act as signals in a hierarchical regulatory cascade that controls the production of TDA. This was often observed such as in the plant pathogen Ralstonia solanacearum, where a non-AHL signal, a 3-hydroxypalmitic acid methyl ester modulates pathogenicity in conjunction with AHLs (13, 17).
In several bacteria production of secondary metabolites is regulated hierarchically and luxR-luxI systems are often part of such networks (30, 33, 47). These QS signaling systems are often global regulators of gene expression that allow ordered expression of hundreds of genes (48). Often global regulatory genes control antibiotic production and cause pleiotropic effects, like afsR, afsB, bldA, and ndgR of Streptomyces coelicolor (3, 57). With our study we demonstrate that the antibiotic and pigment production in P. gallaeciensis is coordinately regulated via the pgaR-pgaI QS system, such as the carbapenem antibiotic and prodigiosin pigment in Serratia sp. strain ATCC 39006 (54). The pleiotropic phenotype could be due to the activation of the tda operon, which encodes proteins involved both in the synthesis of TDA and pigment (22). We hypothesize that the pgaR-pgaI QS system is located relatively high in the hierarchy of regulation in P. gallaeciensis, whereas TDA production directly depends on TdaA. The production of TDA in strain DSM 17395 is obviously regulated by a complex regulatory network, which may act at different levels in a regulatory cascade. Such a complex regulatory web would be consistent with the need of P. gallaeciensis to sense, integrate, and respond to a variety of environmental and physiological signals and to adapt to and colonize different niches and environments.
ACKNOWLEDGMENTS
We thank two anonymous reviewers for their comments on earlier versions of the manuscript.
This study was supported by the Volkswagen-Stiftung, VW-Vorab, Lower Saxony 11-7651-13-4/06 (ZN2235), and by the Transregional Collaborative Research Centre “Roseobacter” (Transregio TRR 51), funded by the Deutsche Forschungsgemeinschaft.
Footnotes
Published ahead of print on 23 September 2011.
REFERENCES
- 1. Atkinson S., Sockett R. E., Camara M., Williams P. 2006. Quorum sensing and the lifestyle of Yersinia. Curr. Issues Mol. Biol. 8:1–10 [PubMed] [Google Scholar]
- 2. Bauer W. D., Mathesius U. 2004. Plant responses to bacterial quorum sensing signals. Curr. Opin. Plant Biol. 7:429–433 [DOI] [PubMed] [Google Scholar]
- 3. Bibb M. 1996. The regulation of antibiotic production in Streptomyces coelicolor A3(2). Microbiology 142:1335–1344 [DOI] [PubMed] [Google Scholar]
- 4. Boyer M., Wisniewski-Dye F. 2009. Cell-cell signaling in bacteria: not simply a matter of quorum. FEMS Microbiol. Ecol. 70:1–19 [DOI] [PubMed] [Google Scholar]
- 5. Brinkhoff T., et al. 2004. Antibiotic production by a Roseobacter clade-affiliated species from the German Wadden Sea and its antagonistic effects on indigenous isolates. Appl. Environ. Microbiol. 70:2560–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bruhn J. B., Gram L., Belas R. 2007. Production of antibacterial compounds and biofilm formation by Roseobacter species are influenced by culture conditions. Appl. Environ. Microbiol. 73:442–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bruhn J. B., Haagensen J. A. J., Bagge-Ravn D., Gram L. 2006. Culture conditions of Roseobacter strain 27-4 affect its attachment and biofilm formation as quantified by real-time PCR. Appl. Environ. Microbiol. 72:3011–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bruhn J. B., et al. 2005. Ecology, inhibitory activity, and morphogenesis of a marine antagonistic bacterium belonging to the Roseobacter clade. Appl. Environ. Microbiol. 71:7263–7270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Buchan A., Gonzalez J. M., Moran M. A. 2005. Overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 71:5665–5677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Camilli A., Bassler B. L. 2006. Bacterial small-molecule signaling pathways. Science 311:1113–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cataldi T. R. I., Bianco G., Frommberger M., Schmitt-Kopplin P. 2004. Direct analysis of selected N-acyl-l-homoserine lactones by gas chromatography/mass spectrometry. Rapid Commun. Mass Spectrom. 18:1341–1344 [DOI] [PubMed] [Google Scholar]
- 12. Cha C., Gao P., Chen Y. C., Shaw P. D., Farrand S. K. 1998. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant-Microbe Interact. 11:1119–1129 [DOI] [PubMed] [Google Scholar]
- 13. Clough S. J., Lee K. E., Schell M. A., Denny T. P. 1997. A two-component system in Ralstonia (Pseudomonas) solanacearum modulates production of PhcA-regulated virulence factors in response to 3-hydroxypalmitic acid methyl ester. J. Bacteriol. 179:3639–3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Decho A. W., et al. 2009. Autoinducers extracted from microbial mats reveal a surprising diversity of N-acylhomoserine lactones (AHLs) and abundance changes that may relate to diel pH. Environ. Microbiol. 11:409–420 [DOI] [PubMed] [Google Scholar]
- 14a. Dower W. J., Miller J. F., Ragsdale C. W. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127–6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Egan S., Thomas T., Kjelleberg S. 2008. Unlocking the diversity and biotechnological potential of marine surface associated microbial communities. Curr. Opin. Microbiol. 11:219–225 [DOI] [PubMed] [Google Scholar]
- 16. Farrand S. K., Qin Y. P., Oger P. 2002. Quorum-sensing system of Agrobacterium plasmids: analysis and utility. Methods Enzymol. 358:452–484 [DOI] [PubMed] [Google Scholar]
- 17. Flavier A. B., Clough S. J., Schell M. A., Denny T. P. 1997. Identification of 3-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum. Mol. Microbiol. 26:251–259 [DOI] [PubMed] [Google Scholar]
- 18. Fuqua C., Winans S. C., Greenberg E. P. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727–751 [DOI] [PubMed] [Google Scholar]
- 19. Geng H., Belas R. 2010. Expression of tropodithietic acid biosynthesis is controlled by a novel autoinducer. J. Bacteriol. 192:4377–4387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Geng H., Belas R. 2010. Molecular mechanisms underlying roseobacter-phytoplankton symbioses. Curr. Opin. Biotechnol. 21:332–338 [DOI] [PubMed] [Google Scholar]
- 21. Geng H., Belas R. 2011. TdaA regulates tropodithietic acid synthesis by binding to the tdaC promoter region. J. Bacteriol. 193:4002–4005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Geng H. F., Bruhn J. B., Nielsen K. F., Gram L., Belas R. 2008. Genetic dissection of tropodithietic acid biosynthesis by marine roseobacters. Appl. Environ. Microbiol. 74:1535–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gram L., Grossart H. P., Schlingloff A., Kiorboe T. 2002. Possible quorum sensing in marine snow bacteria: production of acylated homoserine lactones by Roseobacter strains isolated from marine snow. Appl. Environ. Microbiol. 68:4111–4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 25. Hjelm M., et al. 2004. Selection and identification of autochthonous potential probiotic bacteria from turbot larvae (Scophthalmus maximus) rearing units. Syst. Appl. Microbiol. 27:360–371 [DOI] [PubMed] [Google Scholar]
- 26. Holden M. T., et al. 1999. Quorum-sensing cross talk: isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other gram-negative bacteria. Mol. Microbiol. 33:1254–1266 [DOI] [PubMed] [Google Scholar]
- 27. Kickstein E., Harms K., Wackernagel W. 2007. Deletions of recBCD or recD influence genetic transformation differently and are lethal together with a recJ deletion in Acinetobacter baylyi. Microbiology 153:2259–2270 [DOI] [PubMed] [Google Scholar]
- 28. Kovach M. E., et al. 1995. 4 new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176 [DOI] [PubMed] [Google Scholar]
- 29. Liang L. 2003. Investigation of secondary metabolites of North Sea bacteria: fermentation, isolation, structure elucidation, and bioactivity. Ph.D. thesis University of Göttingen, Göttingen, Germany [Google Scholar]
- 30. Lu J., et al. 2009. The distinct quorum sensing hierarchy of las and rhl in Pseudomonas sp. M18. Curr. Microbiol. 59:621–627 [DOI] [PubMed] [Google Scholar]
- 31. Lunau M., Lemke A., Walther K., Martens-Habbena W., Simon M. 2005. An improved method for counting bacteria from sediments and turbid environments by epifluorescence microscopy. Environ. Microbiol. 7:961–968 [DOI] [PubMed] [Google Scholar]
- 32. Martens T., et al. 2007. Bacteria of the Roseobacter clade show potential for secondary metabolite production. Microb. Ecol. 54:31–42 [DOI] [PubMed] [Google Scholar]
- 33. McGowan S. J., et al. 2005. Carbapenem antibiotic biosynthesis in Erwinia carotovora is regulated by physiological and genetic factors modulating the quorum sensing-dependent control pathway. Mol. Microbiol. 55:526–545 [DOI] [PubMed] [Google Scholar]
- 34. Mohamed N. M., et al. 2008. Diversity and quorum-sensing signal production of Proteobacteria associated with marine sponges. Environ. Microbiol. 10:75–86 [DOI] [PubMed] [Google Scholar]
- 35. Moran M. A., et al. 2007. Ecological genomics of marine roseobacters. Appl. Environ. Microbiol. 73:4559–4569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pintado J., et al. 2010. Monitoring of the bioencapsulation of a probiotic Phaeobacter strain in the rotifer Brachionus plicatilis using denaturing gradient gel electrophoresis Aquaculture 302:182–194 [Google Scholar]
- 37. Porsby C. H., Nielsen K. F., Gram L. 2008. Phaeobacter and Ruegeria species of the Roseobacter clade colonize separate niches in a Danish Turbot (Scophthalmus maximus)-rearing farm and antagonize Vibrio anguillarum under different growth conditions. Appl. Environ. Microbiol. 74:7356–7364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Porsby C. H., Webber M. A., Nielsen K. F., Piddock L. J., Gram L. 2011. Resistance and tolerance to tropodithietic acid, an antimicrobial in aquaculture, is hard to select. Antimicrob. Agents Chemother. 55:1332–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Puskas A., Greenberg E. P., Kaplan S., Schaefer A. L. 1997. A quorum-sensing system in the free-living photosynthetic bacterium Rhodobacter sphaeroides. J. Bacteriol. 179:7530–7537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rao D., et al. 2007. Low densities of epiphytic bacteria from the marine alga Ulva australis inhibit settlement of fouling organisms. Appl. Environ. Microbiol. 73:7844–7852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rao D., Webb J. S., Kjelleberg S. 2005. Competitive interactions in mixed-species biofilms containing the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 71:1729–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rao D., Webb J. S., Kjelleberg S. 2006. Microbial colonization and competition on the marine alga Ulva australis. Appl. Environ. Microbiol. 72:5547–5555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ravn L., Christensen A. B., Molin S., Givskov M., Gram L. 2001. Methods for detecting acylated homoserine lactones produced by Gram-negative bacteria and their application in studies of AHL-production kinetics. J. Microbiol. Methods 44:239–251 [DOI] [PubMed] [Google Scholar]
- 44. Rosemeyer V., Michiels J., Verreth C., Vanderleyden J. 1998. luxI- and luxR-homologous genes of Rhizobium etli CNPAF512 contribute to synthesis of autoinducer molecules and nodulation of Phaseolus vulgaris. J. Bacteriol. 180:815–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ruiz-Ponte C., Samain J. F., Sanchez J. L., Nicolas J. L. 1999. The benefit of a Roseobacter species on the survival of scallop larvae. Mar. Biotechnol. 1:52–59 [DOI] [PubMed] [Google Scholar]
- 46. Schaefer A. L., et al. 2008. A new class of homoserine lactone quorum-sensing signals. Nature 454:595–599 [DOI] [PubMed] [Google Scholar]
- 47. Schuster M., Greenberg E. P. 2006. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 296:73–81 [DOI] [PubMed] [Google Scholar]
- 48. Schuster M., Lostroh C. P., Ogi T., Greenberg E. P. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Seyedsayamdost M. R., Case R. J., Kolter R., Clardy J. 2011. The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat. Chem. 3:331–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Slightom R. N., Buchan A. 2009. Surface colonization by marine roseobacters: integrating genotype and phenotype. Appl. Environ. Microbiol. 75:6027–6037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Swift S., et al. 2001. Quorum sensing as a population-density-dependent determinant of bacterial physiology. Adv. Microb. Physiol. 45:199–270 [DOI] [PubMed] [Google Scholar]
- 52. Taylor M. W., et al. 2004. Evidence for acyl homoserine lactone signal production in bacteria associated with marine sponges. Appl. Environ. Microbiol. 70:4387–4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Thiel V., Kunze B., Verma P., Wagner-Döbler I., Schulz S. 2009. New structural variants of homoserine lactones in bacteria. Chembiochem 10:1861–1868 [DOI] [PubMed] [Google Scholar]
- 54. Thomson N. R., Crow M. A., McGowan S. J., Cox A., Salmond G. P. 2000. Biosynthesis of carbapenem antibiotic and prodigiosin pigment in Serratia is under quorum sensing control. Mol. Microbiol. 36:539–556 [DOI] [PubMed] [Google Scholar]
- 55. Uchino Y., Hirata A., Yokota A., Sugiyama J. 1998. Reclassification of marine Agrobacterium species: proposals of Stappia stellulata gen. nov., comb. nov., Stappia aggregata sp. nov., nom. rev., Ruegeria atlantica gen. nov., comb. nov., Ruegeria gelatinovora comb. nov., Ruegeria algicola comb. nov., and Ahrensia kieliense gen. nov., sp. nov., nom. rev. J. Gen. Appl. Microbiol. 44:201–210 [DOI] [PubMed] [Google Scholar]
- 56. Wagner-Döbler I., et al. 2005. Discovery of complex mixtures of novel long-chain quorum sensing signals in free-living and host-associated marine Alphaproteobacteria. Chembiochem 6:2195–2206 [DOI] [PubMed] [Google Scholar]
- 57. Yang Y. H., et al. 2009. NdgR, an IclR-like regulator involved in amino-acid-dependent growth, quorum sensing, and antibiotic production in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 82:501–511 [DOI] [PubMed] [Google Scholar]
- 58. Yates E. A., et al. 2002. N-acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect. Immun. 70:5635–5646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhu J., Chai Y., Zhong Z., Li S., Winans S. C. 2003. Agrobacterium bioassay strain for ultrasensitive detection of N-acylhomoserine lactone-type quorum-sensing molecules: detection of autoinducers in Mesorhizobium huakuii. Appl. Environ. Microbiol. 69:6949–6953 [DOI] [PMC free article] [PubMed] [Google Scholar]