Abstract
Rhodobacter sphaeroides is able to assemble two different flagella, the subpolar flagellum (Fla1) and the polar flagella (Fla2). In this work, we report the swimming behavior of R. sphaeroides Fla2+ cells lacking each of the proteins encoded by chemotactic operon 1. A model proposing how these proteins control Fla2 rotation is presented.
TEXT
Chemotaxis is a complex response that allows bacteria to swim toward favorable stimuli and away from repellents. A network of proteins which transmit a signal from the chemoreceptors to the flagellar motor controls this process.
In Escherichia coli, membrane chemoreceptors, or MCPs (methyl-accepting chemotactic proteins), are coupled to CheA through CheW. The kinase activity of CheA is controlled by the ligand occupancy state of the MCP (for reviews, see references 1, 7, 17, 28, 46, and 49). When activated, CheA promotes its own phosphorylation; this phosphate is transferred to a specific Asp residue on CheY (18, 36, 53). The phosphorylated form of CheY (CheY-P) suffers a conformational change that increases its affinity for FliM (51, 52). FliM, FliG, and FliN constitute the C-ring or switch apparatus (12, 13, 20–22, 55). Binding of CheY-P to FliM and FliN changes flagellar rotation from counterclockwise (CCW) to clockwise (CW) (5, 37, 53). When the flagellar motor is rotating CCW, cells swim in a linear trajectory, known as a run. However, when the motor switches to the CW direction, swimming is interrupted by a tumbling event (48). Tumbling events are short-lived given the rapid dephosphorylation of CheY-P, caused by the phosphatase activity of CheZ (8, 18, 53).
During adaptation, CheA kinase activity is reset to prestimulus levels. This step involves the reversible methylation of the MCPs that is carried out by the constitutively active methyltransferase CheR (43) and the methylesterase CheB (44). When CheB is phosphorylated by CheA, methylesterase activity increases about 100-fold (3, 23). An increase in methylation counteracts the inhibitory signal generated by attractant binding (6, 14, 26, 47).
Many nonenteric bacterial species show multiple homologues of the chemotactic genes, suggesting that more complex chemosensory pathways are frequently present (16, 54). Rhodobacter sphaeroides has become an interesting model with which to study these pathways. The complete genome sequence of R. sphaeroides revealed the presence of multiple chemotactic genes arranged mainly in three different loci: cheOp1, cheOp2, and cheOp3 (for a review, see reference 32).
R. sphaeroides swims in liquid medium using a single subpolar flagellum (Fla1) that rotates unidirectionally to produce smooth swimming. Reorientation occurs when flagellar rotation stops briefly. Brownian motion and the slow rotation of the filament coil reorient the cell body before smooth swimming is resumed (4, 29). Genetic and biochemical evidence suggests that the proteins encoded by cheOp2 and cheOp3 are responsible for the control of the chemotactic response of Fla1 flagella (15, 24, 25), whereas deletion of cheOp1 does not have any effect in this system (15, 50). In particular, CheY6 plus CheY3 or CheY4 is required to stop the flagellar motor (33).
We have previously shown that pseudorevertants from a Fla1− mutant are able to swim in liquid medium by means of multiple polar flagella whose structural components are encoded by a second set of genes (fla2). This set is not expressed in the wild-type strain under any laboratory conditions tested so far (30). Cells expressing Fla2 flagella form a ring in swimming plates containing different organic acids. This chemotactic response is mediated only by CheY1, CheY2, and CheY5 (11). Given that these proteins are encoded by cheOp1, we hypothesized that this locus must control Fla2 flagella.
In this work, we isolated nonpolar mutants for each of the chemotactic genes present in cheOp1. From the behavioral analysis, we conclude that the majority of the proteins encoded by cheOp1 are involved in controlling Fla2. A model of the signal-processing circuit controlling Fla2 rotation is presented below.
Bacterial strains and plasmids are listed in Table 1. R. sphaeroides (40) was grown in liquid or solid Sistrom's minimal medium (39) as described previously (30).
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| TOP10 | Cloning strain | Invitrogen |
| S17-1 | recA endA thi hsdR RP4-2 Tc::Mu-Kn::Tn7 Tpr Smr | 38 |
| R. sphaeroides | ||
| WS8-N | Wild type, spontaneous Nalr | 40 |
| RS1Y5 | WS8-N derivative, cheY5::aadA | This study |
| RS1B | WS8-N derivative, mcpB::aadA | This study |
| RS1S | WS8-N derivative, tlpS::aadA | This study |
| RS1A | WS8-N derivative, mcpA::aadA | This study |
| RS1D | WS8-N derivative, cheD::aadA | This study |
| RS1Y1 | WS8-N derivative, cheY1::aadA | This study |
| RS1A1 | WS8-N derivative, cheA1::aadA | This study |
| RS1W1 | WS8-N derivative, cheW1::aadA | This study |
| RS1R1 | WS8-N derivative, cheR1::aadA | This study |
| RS1Y2 | WS8-N derivative, cheY2::aadA | This study |
| RS12432 | WS8-N derivative, RSP2432::aadA | This study |
| AM1 | SP13 derivative, fla2+ | 11 |
| RS2Y5 | AM1 derivative, cheY5::aadA | 11 |
| RS2B | AM1 derivative, mcpB::aadA | This study |
| RS2S | AM1 derivative, tlpS::aadA | This study |
| RS2A | AM1 derivative, mcpA::aadA | This study |
| RS2D | AM1 derivative, cheD::aadA | This study |
| RS2Y1 | AM1 derivative, cheY1::aadA | 11 |
| RS2A1 | AM1 derivative, cheA1::aadA | This study |
| RS2W1 | AM1 derivative, cheW1::aadA | This study |
| RS2R1 | AM1 derivative, cheR1::aadA | This study |
| RS2Y2 | AM1 derivative, cheY2::aadA | 11 |
| RS22432 | AM1 derivative, RSP2432::aadA | This study |
| Plasmids | ||
| pTZ19R | Cloning vector; pUC derivative; Apr | Pharmacia |
| pJQ200mp18 | Suicide vector used for gene replacement; Gmr | 34 |
| pRK415 | pRK404 derivative, used for expression in R. sphaeroides; Tcr | 19 |
| pRKtlpS | pRK415 derivative expressing tlpS | This study |
| pRKmcpA | pRK415 derivative expressing mcpA | This study |
| pRKcheD | pRK415 derivative expressing cheD | This study |
| pRKcheA1 | pRK415 derivative expressing cheA1 | This study |
| pRKcheW1 | pRK415 derivative expressing cheW1 | This study |
| pRKcheR1 | pRK415 derivative expressing cheR1 | This study |
| pRK2432 | pRK415 derivative expressing RSP2432 | This study |
Rotation and energy source of Fla2 flagella.
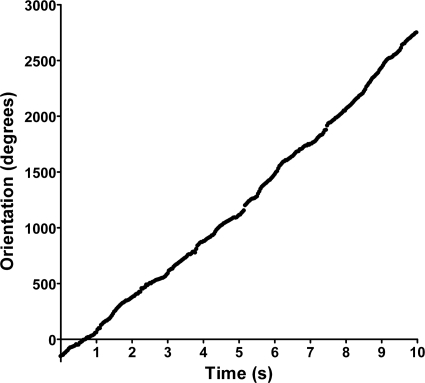
To analyze the rotational bias of Fla2 flagella, AM1 cells (Fla1− Fla2+) were tethered to a microscope slide using antiflagellin antibodies. Cells were recorded using a digital camera attached to a high-intensity dark-field microscope (Olympus BH-2). Video recordings of tethered cells were analyzed using CellTrak 1.5 (Motion Analysis Corp., Santa Rosa, CA). A total of 40 cells were analyzed for at least 10 s, and results from a single representative cell are shown in Fig. 1. The orientation (measured in degrees) accumulates as a function of time, suggesting that Fla2 flagella rotate only in one direction and in a stop-go fashion, as has been shown for the Fla1 system (4). Additionally, we investigated the energy source of the Fla2 motor. AM1 cells were grown on Sistrom's minimal medium devoid of succinate under photoheterotrophic conditions. When the culture reached an optical density at 600 nm of approximately 0.3, a 500-μl aliquot of cells was removed and treated with the protonophore carbonyl cyanide 3-chloro-phenylhydrazone (CCCP) or with the sodium channel blocker amiloride. We added 10 μM CCCP or 0.5 mM amiloride to the cell culture, as described previously (27, 45). Motility was completely inhibited by CCCP (see Movie S1 in the supplemental material). In contrast, cells continued swimming in the presence of amiloride. These results demonstrate that Fla2 flagella rotate unidirectionally and that the proton motive force is responsible for motor rotation.
Fig. 1.
Tethered-cell analysis of Fla2 flagella from Rhodobacter sphaeroides. Cells were analyzed using CellTrak 1.5 to determine the rotational bias of flagella. The direction of orientation of individual cells is measured in degrees and given as a function of time in seconds. As cells rotate in one direction, the orientation value increases linearly. Results for an individual representative cell are shown in this graph.
Isolation and swimming phenotype of cheOp1 mutants.
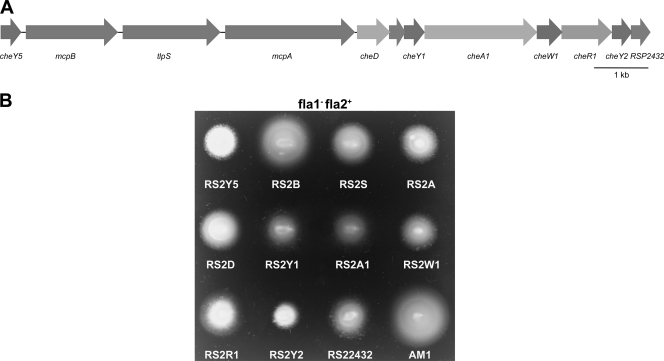
The cheOp1 locus (Fig. 2A) contains 11 genes, including one unassigned open reading frame and genes encoding the following putative proteins: three response regulators, one sensor kinase, one methyltransferase, one adaptor protein, two membrane chemoreceptors, one cytoplasmic chemoreceptor, and one receptor deamidase. We mutated each gene, with the exception of the cheY genes, which were mutated previously (11). Mutants were generated by the amplification of the desired gene followed by the insertion of a nonpolar Spcr cassette (11). These fragments were subcloned into the suicide plasmid pJQ200mp18 (34) and mobilized into WS8-N (Fla1+ Fla2−) and AM1 (Fla1− Fla2+) by conjugation with the S17-1 strain (10, 38). The Spcr Gms transconjugants were selected, and the allelic exchange was confirmed by PCR.
Fig. 2.
(A) Organization of the cheOp1 locus of R. sphaeroides. The arrows represent genes and their direction of transcription. (B) Phenotypic analysis of the cheOp1 mutants in the fla1− fla2+ genetic background. Soft agar motility assays were carried out in 0.25% agar supplemented with 100 μM succinate. Cells were grown under aerobic conditions at 30°C for 48 h.
Analysis of the chemotactic behavior of the cheOp1 mutants in soft agar swimming plates revealed that strains RS2Y5, RS2S, RS2A, RS2D, RS2Y1, RS2A1, RS2W1, RS2R1, RS2Y2, and RS22432 showed a smaller swimming ring than AM1 (Fig. 2B). This result is consistent with the idea that the proteins encoded by cheOp1 control the chemotactic response of Fla2. The same mutations in the genetic background where only the Fla1 flagellum is expressed have no significant effect on chemotaxis, as reported previously (data not shown) (11, 15, 24, 25, 33, 50).
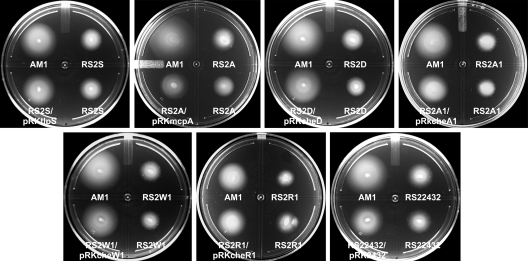
In order to demonstrate that the observed phenotypes in the Fla1− Fla2+ background were due only to the absence of each of the cheOp1 genes, we carried out a complementation test introducing the wild-type gene cloned into pRK415 (19). Figure 3 shows that all the strains were complemented by the corresponding wild-type gene, except for the mutant strain RS2A, which was partially complemented, indicating that the impairment in motility produced by each mutation was recovered.
Fig. 3.
Complementation of each cheOp1 mutant by the corresponding wild-type gene cloned in pRK415. Soft agar motility assays were carried out in 0.25% agar supplemented with 100 μM succinate. Cells were grown under aerobic conditions at 30°C for 48 h.
Characterization of swimming motility.
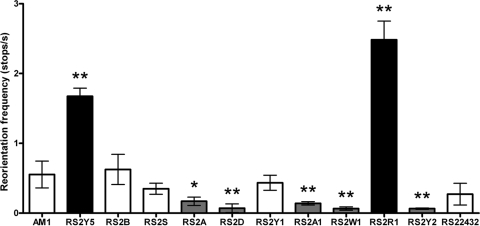
To better characterize the cheOp1 mutants, we analyzed free-swimming behavior. Swimming paths were obtained with CellTrak software by analyzing video recordings of motile cultures of R. sphaeroides strains as described above. The stopping frequency and average run velocity were determined by analyzing for 100 cells from three independent cultures for 2 s. As shown in Fig. 4, AM1 cells reorient under these conditions approximately once every 2 s (0.55 stop/s). Mutant strains RS2A (mcpA), RS2D (cheD), RS2A1 (cheA1), RS2W1 (cheW1), and RS2Y2 (cheY2) showed a lower reorientation frequency, which is equivalent to a smooth swimming bias. In contrast, RS2Y5 (cheY5) and RS2R1 (cheR1) stop at a higher frequency, similar to a tumbling behavior. Furthermore, strains RS2Y1 (cheY1), RS2B (mcpB), RS2S (tlpS), and RS22432 (RSP2432) had essentially the same reorientation frequency as the parent strain. This suggests that under this particular condition, none of these products participate in the Fla2 chemotaxis signaling pathway.
Fig. 4.
Reorientation frequency of free-swimming cells. Values are means ± standard deviations from 100 cells. Significance was assessed by one-way analysis of variance; **, P < 0.01; *, P < 0.05.
From these results we propose that CheY2 is mainly responsible for stopping the Fla2 flagellar motor. Given that RS2A1 and RS2W1 show the same phenotype as RS2Y2, it could be assumed that CheA1, CheW1, and CheY2 comprise the signal transduction pathway that controls Fla2 (Fig. 5). In agreement with this idea, it has been shown in vitro that CheA1 is able to phosphorylate the response regulators encoded by cheOp1 (CheY1, CheY2, and CheY5) (31). Therefore, it is likely that the phosphorylated form of CheY2 (CheY2-P) is the signal that stops the Fla2 flagellar motor. Moreover, we observed that the absence of mcpA also reduces the stopping frequency significantly; hence, it is possible that this MCP together with CheW1 modulates CheA1 activity.
Fig. 5.
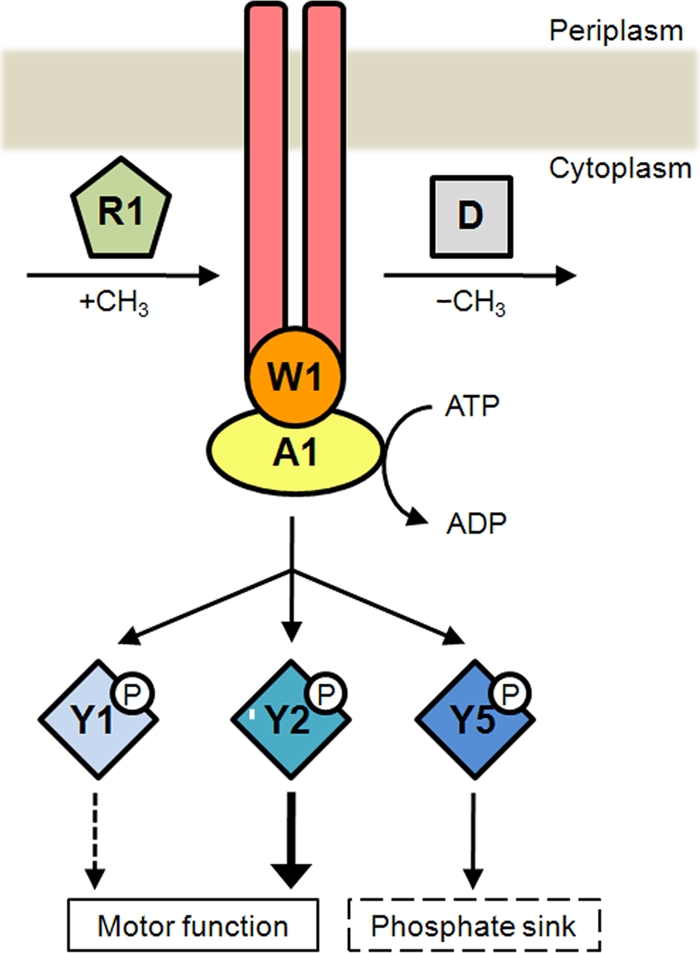
Chemotactic control of Fla2 flagella is mediated by the proteins encoded in cheOp1. In this model, a chemoreceptor, which could be McpA, controls the kinase activity of CheA1. When the MCP is methylated by the product of CheR1, autophosphorylation of CheA1 is inhibited and flagella rotate continuously for a longer time. CheD could demethylate the chemoreceptor and promote CheA1 activation, bringing about the phosphorylation of CheY1, CheY2, and CheY5. The accumulation of CheY2-P, in turn, increases the probability of reorientation, whereas CheY5 could modulate the intracellular levels of CheY2-P by acting as a phosphate sink.
Conversely, in the absence of CheY5, a marked increase of the stopping frequency was detected, suggesting that the intracellular concentration of CheY2-P was increased. Given that the phenotype shown by RS2Y5 cells evokes that observed for cheZ mutants in E. coli, it is tempting to suggest that CheY5 could act as a phosphate sink. A similar situation occurs in Sinorhizobium meliloti, which has two different CheY proteins and no CheZ. In this case, CheA phosphorylates both CheYs, but only CheY2 binds to the flagellar motor to control rotation. The response is terminated when CheY2-P transfers its phosphate group back to CheA, and then it is transferred to CheY1 (41, 42). In this regard, CheY5 in R. sphaeroides could play a role similar to that of CheY1 in S. meliloti (Fig. 5).
Another strain that showed a high reorientation frequency is the cheR1 mutant (strain RS2R1), indicating that CheR1 is a relevant component of the chemotactic pathway that controls the Fla2 motor.
In E. coli, an attractant detected by an MCP inhibits CheA activity, and the reduction in the concentration of CheY-P allows bacteria to swim toward the attractant; in the adaptation phase, CheR methylates the MCP, and in this state, CheA autophosphorylation is reactivated (6, 26). In Bacillus subtilis, a more complex situation has been observed, given that methylation of some residues of certain MCPs inhibits CheA, whereas methylation of other residues activates it (35). In our case, the phenotype of RS2R1 (Fig. 4) leads us to propose that methylation of a yet-unknown receptor (that may be McpA) inhibits CheA1 activity (Fig. 5).
Under our experimental conditions, the absence of CheR and McpA produced a bias in flagellar rotation, whereas the absence of McpB did not. This finding could indicate that McpB senses different signals that were absent under these conditions.
Adaptation also requires a methylesterase that counteracts the methylating activity of CheR. In E. coli, CheB accomplishes this function, although in other bacteria, such as Thermotoga maritima, the protein CheD could act as a methylesterase (9). In R. sphaeroides, the absence of CheD induces a reduction in the stopping frequency, which is in agreement with the possibility that a demethylated receptor would activate CheA1, whereas a methylated receptor would inhibit its activity (Fig. 5).
The average run speed, measured as the average velocity between stops, for each one of these mutants was determined. Table 2 shows that RS2Y5 and RS2R1 were the only strains with a lower swimming speed value than AM1. This reduction in swimming speed correlates with the fact that these strains show a very high stopping frequency, so it is possible that maximal speed cannot be reached between stops. A similar effect was observed in E. coli, where overexpression of CheY-P not only caused an increase in the tumbling frequency but also affected the swimming speed negatively (2).
Table 2.
Free-swimming behavior of R. sphaeroides cheOp1 mutants
| Strain | Run speed (μm/s)a |
|---|---|
| AM1 | 61.35 ± 7.98 |
| RS2Y5 | 51.67 ± 0.91 |
| RS2B | 62.62 ± 5.94 |
| RS2S | 53.92 ± 2.97 |
| RS2A | 55.83 ± 6.13 |
| RS2D | 61.24 ± 8.34 |
| RS2Y1 | 61.40 ± 2.19 |
| RS2A1 | 64.42 ± 2.68 |
| RS2W1 | 62.57 ± 5.53 |
| RS2R1 | 39.51 ± 7.73b |
| RS2Y2 | 57.53 ± 3.39 |
| RS22432 | 55.37 ± 3.19 |
Values are averages ± standard deviations for 100 cells from three independent experiments.
P < 0.01; significance was assessed by one-way analysis of variance.
Intriguingly, the stop frequencies and swimming speeds of RS2Y1 (cheY1), RS2S (tlpS), and RS22432 (RSP2432) are similar to those of AM1 (Fig. 4 and Table 2); nonetheless, in a swimming plate, these mutants showed a different phenotype (Fig. 2B). It is possible that chemotactic signals in soft agar plates could be stronger than in liquid medium (due to different diffusion rates), so it is still possible that CheY1, TlpS, and RSP2432 may control Fla2 flagella when the chemotactic stimulus is strong. Further research will determine if this hypothesis is adequate to explain the observed phenotypes.
Based on the results obtained in this work, we propose the model shown in Fig. 5.
Supplementary Material
Acknowledgments
We thank Aurora Osorio and Javier de la Mora for helpful technical assistance, the IFC Molecular Biology Unit for sequencing facilities, and Bertha González-Pedrajo, Sebastian Poggio, and Diego González-Halphen for critically reading the manuscript.
A.M.-D.C. was supported by a fellowship from CONACyT (215775). This work was supported by grants from CONACyT (106081) and DGAPA/UNAM (IN206811-3).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 23 September 2011.
REFERENCES
- 1. Alexander R. P., Zhulin I. B. 2007. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc. Natl. Acad. Sci. U. S. A. 104:2885–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alon U., et al. 1998. Response regulator output in bacterial chemotaxis. EMBO J. 17:4238–4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anand G. S., Stock A. M. 2002. Kinetic basis for the stimulatory effect of phosphorylation on the methylesterase activity of CheB. Biochemistry 41:6752–6760 [DOI] [PubMed] [Google Scholar]
- 4. Armitage J. P., Macnab R. M. 1987. Unidirectional, intermittent rotation of the flagellum of Rhodobacter sphaeroides. J. Bacteriol. 169:514–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barak R., Eisenbach M. 1992. Correlation between phosphorylation of the chemotaxis protein CheY and its activity at the flagellar motor. Biochemistry 31:1821–1826 [DOI] [PubMed] [Google Scholar]
- 6. Borkovich K. A., Kaplan N., Hess J. F., Simon M. I. 1989. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc. Natl. Acad. Sci. U. S. A. 86:1208–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bourret R. B., Stock A. M. 2002. Molecular information processing: lessons from bacterial chemotaxis. J. Biol. Chem. 277:9625–9628 [DOI] [PubMed] [Google Scholar]
- 8. Bren A., Welch M., Blat Y., Eisenbach M. 1996. Signal termination in bacterial chemotaxis: CheZ mediates dephosphorylation of free rather than switch-bound CheY. Proc. Natl. Acad. Sci. U. S. A. 93:10090–10093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chao X., et al. 2006. A receptor-modifying deamidase in complex with a signaling phosphatase reveals reciprocal regulation. Cell 124:561–571 [DOI] [PubMed] [Google Scholar]
- 10. Davis J., Donohue T. J., Kaplan S. 1988. Construction, characterization, and complementation of a Puf− mutant of Rhodobacter sphaeroides. J. Bacteriol. 170:320–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. del Campo A. M., et al. 2007. Chemotactic control of the two flagellar systems of Rhodobacter sphaeroides is mediated by different sets of CheY and FliM proteins. J. Bacteriol. 189:8397–8401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Francis N. R., Irikura V. M., Yamaguchi S., DeRosier D. J., Macnab R. M. 1992. Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc. Natl. Acad. Sci. U. S. A. 89:6304–6308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Francis N. R., Sosinsky G. E., Thomas D., DeRosier D. J. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235:1261–1270 [DOI] [PubMed] [Google Scholar]
- 14. Goy M. F., Springer M. S., Adler J. 1977. Sensory transduction in Escherichia coli: role of a protein methylation reaction in sensory adaptation. Proc. Natl. Acad. Sci. U. S. A. 74:4964–4968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hamblin P. A., Maguire B. A., Grishanin R. N., Armitage J. P. 1997. Evidence for two chemosensory pathways in Rhodobacter sphaeroides. Mol. Microbiol. 26:1083–1096 [DOI] [PubMed] [Google Scholar]
- 16. Hamer R., Chen P. Y., Armitage J. P., Reinert G., Deane C. M. 2010. Deciphering chemotaxis pathways using cross species comparisons. BMC Syst. Biol. 4:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hazelbauer G. L., Falke J. J., Parkinson J. S. 2008. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem. Sci. 33:9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hess J. F., Oosawa K., Kaplan N., Simon M. I. 1988. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell 53:79–87 [DOI] [PubMed] [Google Scholar]
- 19. Keen N. T., Tamaki S., Kobayashi D., Trollinger D. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70:191–197 [DOI] [PubMed] [Google Scholar]
- 20. Khan I. H., Reese T. S., Khan S. 1992. The cytoplasmic component of the bacterial flagellar motor. Proc. Natl. Acad. Sci. U. S. A. 89:5956–5960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Khan S., Khan I. H., Reese T. S. 1991. New structural features of the flagellar base in Salmonella typhimurium revealed by rapid-freeze electron microscopy. J. Bacteriol. 173:2888–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Khan S., Zhao R., Reese T. S. 1998. Architectural features of the Salmonella typhimurium flagellar motor switch revealed by disrupted C-rings. J. Struct. Biol. 122:311–319 [DOI] [PubMed] [Google Scholar]
- 23. Lupas A., Stock J. 1989. Phosphorylation of an N-terminal regulatory domain activates the CheB methylesterase in bacterial chemotaxis. J. Biol. Chem. 264:17337–17342 [PubMed] [Google Scholar]
- 24. Martin A. C., Wadhams G. H., Armitage J. P. 2001. The roles of the multiple CheW and CheA homologues in chemotaxis and in chemoreceptor localization in Rhodobacter sphaeroides. Mol. Microbiol. 40:1261–1272 [DOI] [PubMed] [Google Scholar]
- 25. Martin A. C., et al. 2001. CheR- and CheB-dependent chemosensory adaptation system of Rhodobacter sphaeroides. J. Bacteriol. 183:7135–7144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ninfa E. G., Stock A., Mowbray S., Stock J. 1991. Reconstitution of the bacterial chemotaxis signal transduction system from purified components. J. Biol. Chem. 266:9764–9770 [PubMed] [Google Scholar]
- 27. Packer H. L., Harrison D. M., Dixon R. M., Armitage J. P. 1994. The effect of pH on the growth and motility of Rhodobacter sphaeroides WS8 and the nature of the driving force of the flagellar motor. Biochim. Biophys. Acta 1188:101–107 [DOI] [PubMed] [Google Scholar]
- 28. Parkinson J. S., Ames P., Studdert C. A. 2005. Collaborative signaling by bacterial chemoreceptors. Curr. Opin. Microbiol. 8:116–121 [DOI] [PubMed] [Google Scholar]
- 29. Pilizota T., et al. 2009. A molecular brake, not a clutch, stops the Rhodobacter sphaeroides flagellar motor. Proc. Natl. Acad. Sci. U. S. A. 106:11582–11587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Poggio S., et al. 2007. A complete set of flagellar genes acquired by horizontal transfer coexists with the endogenous flagellar system in Rhodobacter sphaeroides. J. Bacteriol. 189:3208–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Porter S. L., Armitage J. P. 2002. Phosphotransfer in Rhodobacter sphaeroides chemotaxis. J. Mol. Biol. 324:35–45 [DOI] [PubMed] [Google Scholar]
- 32. Porter S. L., Wadhams G. H., Armitage J. P. 2011. Signal processing in complex chemotaxis pathways. Nat. Rev. Microbiol. 9:153–165 [DOI] [PubMed] [Google Scholar]
- 33. Porter S. L., et al. 2006. The CheYs of Rhodobacter sphaeroides. J. Biol. Chem. 281:32694–32704 [DOI] [PubMed] [Google Scholar]
- 34. Quandt J., Hynes M. F. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15–21 [DOI] [PubMed] [Google Scholar]
- 35. Rao C. V., Glekas G. D., Ordal G. W. 2008. The three adaptation systems of Bacillus subtilis chemotaxis. Trends Microbiol. 16:480–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sanders D. A., Gillece-Castro B. L., Stock A. M., Burlingame A. L., Koshland D. E., Jr 1989. Identification of the site of phosphorylation of the chemotaxis response regulator protein, CheY. J. Biol. Chem. 264:21770–21778 [PubMed] [Google Scholar]
- 37. Sarkar M. K., Paul K., Blair D. 2010. Chemotaxis signaling protein CheY binds to the rotor protein FliN to control the direction of flagellar rotation in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 107:9370–9375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Simon R., Priefer U., Pühler A. 1982. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology 1:784–791 [Google Scholar]
- 39. Sistrom W. R. 1962. The kinetics of the synthesis of photopigments in Rhodopseudomonas spheroides. J. Gen. Microbiol. 28:607–616 [DOI] [PubMed] [Google Scholar]
- 40. Sockett R. E., Foster R. M., Armitage J. P. 1990. Molecular biology of the Rhodobacter sphaeroides flagellum. FEMS Symp. 53:473–479 [Google Scholar]
- 41. Sourjik V., Schmitt R. 1996. Different roles of CheY1 and CheY2 in the chemotaxis of Rhizobium meliloti. Mol. Microbiol. 22:427–436 [DOI] [PubMed] [Google Scholar]
- 42. Sourjik V., Schmitt R. 1998. Phosphotransfer between CheA, CheY1, and CheY2 in the chemotaxis signal transduction chain of Rhizobium meliloti. Biochemistry 37:2327–2335 [DOI] [PubMed] [Google Scholar]
- 43. Springer W. R., Koshland D. E., Jr 1977. Identification of a protein methyltransferase as the cheR gene product in the bacterial sensing system. Proc. Natl. Acad. Sci. U. S. A. 74:533–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stock J. B., Koshland D. E., Jr 1978. A protein methylesterase involved in bacterial sensing. Proc. Natl. Acad. Sci. U. S. A. 75:3659–3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sugiyama S., Cragoe E. J., Jr., Imae Y. 1988. Amiloride, a specific inhibitor for the Na+-driven flagellar motors of alkalophilic Bacillus. J. Biol. Chem. 263:8215–8219 [PubMed] [Google Scholar]
- 46. Szurmant H., Ordal G. W. 2004. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol. Mol. Biol. Rev. 68:301–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Toews M. L., Adler J. 1979. Methanol formation in vivo from methylated chemotaxis proteins in Escherichia coli. J. Biol. Chem. 254:1761–1764 [PubMed] [Google Scholar]
- 48. Turner L., Ryu W. S., Berg H. C. 2000. Real-time imaging of fluorescent flagellar filaments. J. Bacteriol. 182:2793–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wadhams G. H., Armitage J. P. 2004. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell. Biol. 5:1024–1037 [DOI] [PubMed] [Google Scholar]
- 50. Ward M. J., Bell A. W., Hamblin P. A., Packer H. L., Armitage J. P. 1995. Identification of a chemotaxis operon with two cheY genes in Rhodobacter sphaeroides. Mol. Microbiol. 17:357–366 [DOI] [PubMed] [Google Scholar]
- 51. Welch M., Oosawa K., Aizawa S., Eisenbach M. 1993. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc. Natl. Acad. Sci. U. S. A. 90:8787–8791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Welch M., Oosawa K., Aizawa S. I., Eisenbach M. 1994. Effects of phosphorylation, Mg2+, and conformation of the chemotaxis protein CheY on its binding to the flagellar switch protein FliM. Biochemistry 33:10470–10476 [DOI] [PubMed] [Google Scholar]
- 53. Wolfe A. J., Conley M. P., Kramer T. J., Berg H. C. 1987. Reconstitution of signaling in bacterial chemotaxis. J. Bacteriol. 169:1878–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wuichet K., Zhulin I. B. 2010. Origins and diversification of a complex signal transduction system in prokaryotes. Sci. Signal. 3:ra50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhao R., Pathak N., Jaffe H., Reese T. S., Khan S. 1996. FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. J. Mol. Biol. 261:195–208 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.