Abstract
DNA methylation and histone acetylation/deacetylation are distinct biochemical processes that control gene expression. While DNA methylation is a common epigenetic signal that inhibits gene transcription, histone deacetylation similarly represses transcription but can be both an epigenetic and nonepigenetic phenomenon. Here we report that the histone deacetylase SIRT1 regulates the activities of DNMT1, a key enzyme responsible for DNA methylation. In mass spectrometry analysis, 12 new acetylated lysine sites were identified in DNMT1. SIRT1 physically associates with DNMT1 and can deacetylate acetylated DNMT1 in vitro and in vivo. Interestingly, deacetylation of different lysines on DNMT1 has different effects on the functions of DNMT1. For example, deacetylation of Lys1349 and Lys1415 in the catalytic domain of DNMT1 enhances DNMT1's methyltransferase activity, while deacetylation of lysine residues in the GK linker decreases DNMT1's methyltransferase-independent transcriptional repression function. Furthermore, deacetylation of all identified acetylated lysine sites in DNMT1 abrogates its binding to SIRT1 and impairs its capability to regulate cell cycle G2/M transition. Finally, inhibition of SIRT1 strengthens the silencing effects of DNMT1 on the expression of tumor suppressor genes ER-α and CDH1 in MDA-MB-231 breast cancer cells. Together, these results suggest that SIRT1-mediated deacetylation of DNMT1 is crucial for DNMT1's multiple effects in gene silencing.
INTRODUCTION
Posttranslational modification of proteins often alters protein function. Common modifications include phosphorylation, ubiquitination, sumoylation, methylation, and acetylation. Acetylation is reversible and occurs on lysine side chains. Histone acetyltransferases such as p300 (KAT3B) and PCAF (KAT2B) add acetyl groups to histones, their canonical substrates, and nonhistone proteins. Histone deacetylases (HDACs) remove acetyl groups: deacetylation of histones in active chromatin represses transcription, and deacetylation of nonhistone proteins has a variety of functional consequences (e.g., changes in protein stability, DNA binding, and catalytic activities) (26, 39, 77).
The human HDAC family has 18 members, which are grouped into four classes: I, II, III, and IV (78). The sirtuins (class III) differ from class I, II, and IV HDACs in three respects: they are homologous to yeast Sir2 rather than Rpd3 or Hda1, they require NAD (NAD+) for activity, and they are refractory to inhibition by the class I and II inhibitors such as trichostatin A (TSA) (26, 61). As the founding member of Sir2 proteins (sirtuins), SIRT1 has been implicated in caloric restriction-related longevity, insulin sensitivity, DNA damage, and tumorigenesis. In addition to histones, SIRT1 also deacetylates and functionally modifies numerous proteins, including p53, FOXO3, and NBS1 (10, 28, 35, 69, 80).
DNA methyltransferases (DNMTs) add methyl groups to the 5′ position of cytosine residues of CpG dinucleotides. Methylated CpG islands interfere with transcription factor binding and recruit repressor complexes (13, 43). Actively transcribed DNA is typically hypomethylated; conversely, hypermethylated DNA is silenced. Hypermethylation promotes chromosome instability and tumorigenesis and influences genomic imprinting and chromatin remodeling (4, 5, 9, 33, 34, 36, 45, 46, 72).
The first identified and most abundant and ubiquitous DNMT is DNMT1 (6, 8, 29). DNMT1 is a large protein (185 kDa) that preferentially methylates hemimethylated DNA. It consists of an N-terminal regulatory domain, which mediates nuclear localization and targeting to replication foci and discriminates between unmethylated and hemimethylated DNA; a C-terminal catalytic domain; and a central region, which contains a cysteine-rich Zn-binding motif, a polybromo motif, and a series of repeating glycine-lysine dipeptides (the GK linker) (7, 42, 44, 67). DNMT1 is essential for maintenance of methylation patterns, silencing of tumor suppressor genes (TSGs), and cell survival (19). In addition to methylating DNA, DNMT1 also represses transcription by methylation-independent mechanisms, for example, by recruiting transcription corepressor DNMT1-associated protein 1 (DMAP1), HDAC1, HDAC2, and methyl-CpG-binding protein to DNA (25, 38, 59).
The mechanisms by which DNMT1 controls gene expression have been much explored. However, less is known about the mechanisms that regulate DNMT1 abundance and activity, although a role for posttranslational modification is emerging. Recent studies demonstrate that sumoylation, phosphorylation, methylation, and ubiquitination of DNMT1 may associate with changes in catalytic activity, DNA binding activity, and/or stability (21, 41, 63, 81). In addition, acetylation of DNMT1 has been suggested in two global proteomics analyses (14, 37). The functional significance of DNMT1 acetylation, however, is not known. Here we report acetylation of DNMT1 at 12 lysines in its N- and C-terminal regions and deacetylation of DNMT1 by SIRT1 in vivo and in vitro. Acetylation/deacetylation of DNMT1 changes its enzymatic activity, its capacity to silence TSG expression, and its ability to regulate the cell cycle. These findings reveal a novel cross talk between two important epigenetic effectors, DNMT1 and SIRT1.
MATERIALS AND METHODS
Plasmids, antibodies, and reagents.
Full-length DNMT1 (NP_001124295.1) was subcloned from pMyc-DNMT1 into EcoRI/NotI-digested pcDNA3-HA (hemagglutinin) to generate the HA-DNMT1 expression plasmid. HA-DNMT1 deletion mutants were produced by subcloning PCR fragments in frame into pcDNA3-HA using appropriate primer sets. HA-DNMT1 point mutants were generated with the QuikChange multisite-directed mutagenesis kit (Stratagene). Expression plasmids for green fluorescent protein (GFP)-SIRT1 and deletion mutants were generated using pEGFP-C1 vector with PCR and standard recombinant DNA techniques. Gal4-DNMT1 and mutant expression plasmids were constructed by joining the DNMT1 or mutated coding regions, in frame with the Gal4 DNA-binding domain downstream of the cytomegalovirus (CMV) promoter in pcDNA3-Gal4. Flag-PCAF (76); HA-p300 (2); Flag-HDAC1, -2, and -3 (73); Myc-SIRT1 (40); SIRT1, -6, and -7 (51, 54); His-DNMT1 (79); glutathione S-transferase (GST)–SIRT1 (40); and SIRT1 short hairpin RNA (shRNA) (70) expression plasmids have been described elsewhere. pGal4-TK-Luc and pTK-Luc reporter plasmids have been described previously (68). The following reagents were purchased: pRL-SV40 plasmid from Promega; mouse anti-DNMT1 antibody from Abcam; anti-SIRT1 antibody from Millipore; anti-Flag, anti-HA, anti-β-actin antibodies, acetyl coenzyme A (acetyl-CoA), 5-aza-2′-deoxycytidine (5-aza-dC), TSA, nicotinamide, NAD+, and splitomicin from Sigma; EX-527 from Tocris Bioscience; and antiacetyllysine antibody from Cell Signaling.
Cell culture and transfection.
293T, HeLa, and Sirt1+/+ and Sirt1−/− mouse embryo fibroblasts (MEFs) were cultured in Dulbecco's modified Eagle's medium (DMEM). HCT116, MDA-MB-231, and MCF-7 cells were grown in McCoy's 5A medium, Leibovitz's L-15 medium, and RPMI 1640 medium, respectively. All media were supplemented with 10% fetal calf serum, 100 μg/ml streptomycin, and 100 IU/ml penicillin, and all cells were grown at 37°C in an atmosphere of 5% CO2. Transfections were performed using Lipofectamine 2000 (Invitrogen).
Immunoprecipitation and immunoblotting.
Cells were incubated on ice for 30 min in 300 μl NETN buffer (50 mM Tris-HCl, pH 8.0, 0.5 mM EDTA, 150 mM NaCl, 0.5% NP-40, and protease inhibitor cocktail) supplemented with 20 mM nicotinamide and 20 mM sodium butyrate. Cell lysates were precleared with protein A/G agarose for 1 h and incubated overnight with primary antibody at 4°C. Immunoprecipitated material was washed three times with NETN buffer with 500 mM NaCl, and proteins were resolved by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were incubated sequentially in phosphate-buffered saline (PBS) containing 0.05% Tween 20 and the following supplements as follows: 5% fat-free milk for 1 h at room temperature (blocking), primary antibody overnight at 4°C, and secondary antibody for 1 h at room temperature. Membranes were washed three times with PBS containing 0.05% Tween 20 between each treatment. Bound secondary antibody was detected by chemiluminescence according to the manufacturer's instructions (Pierce).
Mass spectrometry analysis.
Immunoprecipitated proteins were visualized by staining of SDS gels with colloidal blue (Invitrogen). Proteins were excised from the gels and analyzed by tandem mass spectrometry for acetylation as described previously (80).
Immunofluorescence.
Cells on chamber slides (Chamber Slide System Lab-TekII) were treated sequentially as follows: 4% paraformaldehyde for 15 min at room temperature (fixing), 0.5% Triton X-100 in PBS for 5 min at room temperature (permeabilization), 3% bovine serum albumin (BSA) for 30 min at room temperature (blocking), primary antibodies at 4°C overnight, and secondary antibodies for 1 h at room temperature. Cells were washed with PBS between each treatment. Secondary antibodies used were Alexa 488-labeled anti-mouse and Alexa 555-labeled anti-rabbit antibodies. Slides were mounted with mounting medium containing DAPI (4′,6-diamidino-2-phenylindole) according to the manufacturer's instructions (Vecta-Shield). Images were observed and captured on a Leica confocal microscope.
In vitro acetylation assay.
Reactions were performed in HAT buffer (50 mM Tris, pH 8.0, 1 mM EDTA, 1 mM dithiothreitol [DTT], and 10% glycerol) containing 20 mM acetyl-CoA for 2 h at 30°C. Additional ingredients are listed in the figure legends. Immunoprecipitates were washed in HAT buffer before being added to reaction mixtures. Reaction products were resolved by SDS-PAGE and visualized by immunoblotting with antiacetyllysine antibody.
In vitro deacetylation assay.
Reactions were performed in HDAC buffer (10 mM Tris, pH 8.0, 150 mM NaCl, and 10% glycerol) containing 1 or 5 mM NAD+ for 2 h at 30°C. Reaction products were resolved on 8% SDS gels and visualized by immunoblotting with antiacetyllysine antibody.
DNA methyltransferase activity assay.
Assays were performed using the EpiQuik methyltransferase 1 activity/inhibitor screening kit as specified by the manufacturer (Epigentek). In brief, cell lysates were incubated with cytosine-rich DNA substrates applied as a coating on a strip. The strip was washed, and methylated DNA on the strip was detected in a colorimetric enzyme-linked immunosorbent assay (ELISA)-like assay using anti-5-methylcytosine antibody. Experiments were repeated at least three times.
A radioactive DNA methyltransferase assay was performed as described previously (1, 30) with some modification. Briefly, cells were lysed with NETN buffer. Five micrograms of lysate was incubated in reaction buffer containing 0.5 μg of poly(dI-dC)·poly(dI-dC) and 1.5 μCi of S-adenosyl-l-methyl-[3H]methionine ([3H]AdoMet) in a final volume of 20 μl for 2 h at 37°C. After removal of unincorporated isotope, the amount of methyl-[3H]methionine incorporated into cytosine was determined on a liquid scintillation counter. Experiments were repeated at least three times.
DNMT1 enzyme kinetics assay.
HA-tagged wild-type and 2KR DNMT1 were transiently overexpressed in 293T cells and immunopurified with anti-HA agarose. Purified proteins were resolved on SDS-PAGE and quantified by comparison to titrated bovine serum albumin (BSA) with Coomassie blue staining and by Western blot assays. Steady-state kinetic parameters were determined using previously published protocols with some modifications (3, 23, 24). Briefly, wild-type and 2KR DNMT1 were preincubated with [3H]AdoMet for 10 min at room temperature. Enzymatic reactions were initiated by adding DNA of poly (dI-dC)·(dI-dC). After incubation for 60 min at 37°C, reaction mixtures were spotted onto DE81 filters and dried. After being washed three times with 5% trichloroacetic acid (TCA), once with 95% ethanol, and once with 100% ethanol, methyl groups transferred onto DNA were determined by detecting the counts per minute (cpm) of 3H with liquid scintillation counting. The velocity of DNA methylation is described as micromoles of methyl group transferred per hour by 1 nM enzyme. All steady-state data were fitted to Michaelis-Menten kinetics. Nonlinear regression analysis was performed, and Km and kcat were calculated using GraphPad Prism 5 software (GraphPad Inc.). Double reciprocal (Lineweaver-Burk) plots were also performed using the Prism 5 software.
Luciferase reporter assay.
293T cells were seeded at a density of 2 × 105 cells in 12-well plates. One day later, cells were transfected with Gal4-TK-Luc (0.5 μg), pRL-SV40 (Renilla) (internal control, 0.018 μg), and other plasmids. Thirty-six hours after transfection, cells were harvested, lysed, and analyzed for luminescence with the dual-luciferase assay system (Promega). Luciferase activity was normalized to pRL-SV40 activity. Experiments were repeated at least three times.
Fluorescence-activated cell sorting (FACS) analysis.
Cells were seeded at a density of 7 × 105 cells in 6-cm plates and, 1 day later, washed once with ice-cold phosphate-buffered saline (PBS) and fixed overnight in cold ethanol. After washing with PBS, cells were harvested by centrifugation and cell pellets were resuspended in 1 ml of cold PBS containing 10 mg/ml RNase A (Novagen) and incubated for 30 min at 37°C. Twenty microliters of 1-mg/ml propidium iodide was added at room temperature, and resuspended pellets were incubated for 10 min. Cells were sorted and analyzed for cell cycle position on a Becton D-FACSstarPLUS cell sorter.
Real-time PCR.
RNA was extracted from cells, reverse transcribed to cDNA, and analyzed for expression of specific gene products using the TaqMan gene expression assay kit according to the manufacturer's instructions (Applied Biosystems). Experiments were repeated three times.
RESULTS
DNMT1 is acetylated in vivo and in vitro.
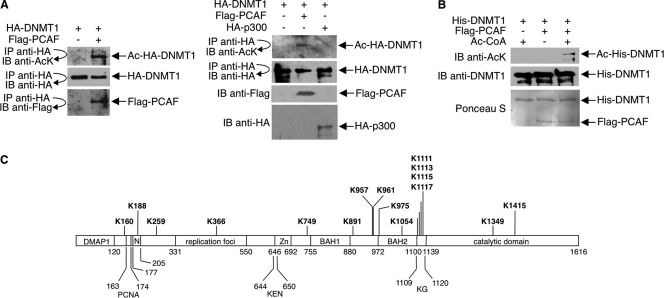
For in vivo analysis, we coexpressed HA-tagged DNMT1 and either PCAF or p300 in 293T cells; anti-HA immunoprecipitates were immunoblotted with an antiacetyllysine antibody. Acetylated HA-DNMT1 was significantly increased in cells cotransfected with PCAF over that in cells cotransfected with p300 or vector alone (Fig. 1 A). Amounts of HA-DNMT1 were similar under all three conditions. For in vitro analysis, we incubated baculovirus-expressed, purified His-tagged DNMT1 with immunopurified Flag-tagged PCAF; reaction mixtures were immunoblotted with antiacetyllysine antibody. PCAF acetylated His-DNMT1 in the presence but not the absence of acetyl-CoA (Fig. 1B). Consistent with the finding that PCAF acetylates DNMT1, the two proteins coimmunoprecipitated in cells (Fig. 1A; see also Fig. S1 in the supplemental material).
Fig. 1.
Acetylation of DNMT1 in vivo and in vitro. (A) 293T cells were transfected with HA-DNMT1 and either Flag-PCAF, HA-p300, or empty Flag vector. Cell lysates were immunoprecipitated (IP) with antibody to HA, and immunoprecipitates were immunoblotted (IB) with antibody to HA, Flag, or acetyllysine (AcK). Ac, acetylated. Direct immunoblot assays were performed with anti-Flag and anti-HA to assess expressions of Flag-PCAF and HA-p300, respectively. (B) His-DNMT1 was expressed in insect cells using the baculovirus system and purified on nickel affinity columns. Flag-PCAF was expressed in 293T cells and immunopurified from cell lysates with an anti-Flag antibody. In vitro acetylation reaction mixtures contained His-DNMT1, Flag-PCAF, and acetyl-CoA as indicated. Reaction products were detected with an antiacetyllysine antibody. Anti-DNMT1 Western blots and Ponceau S staining show equal amounts of His-DNMT1 and Flag-PCAF in reaction mixtures. (C) 293T cells expressing HA-DNMT1 were treated with 15 mM nicotinamide plus 100 ng/ml TSA overnight. HA-DNMT1 was immunoprecipitated from cell lysates with antibody to HA, and immunoprecipitated material was analyzed by mass spectrometry. Acetylated lysines identified in the analysis, plus previously reported acetylation sites (Lys111, Lys1113, Lys1115, and Lys1117), are indicated in bold type. The domains of DNMT1 are shown and are drawn in approximate scale. DMAP1, DNA methyltransferase 1-associated protein 1-binding domain; PCNA, proliferating cell nuclear antigen-binding domain; N, nuclear localization signal; replication foci, replication focus-targeting domain; KEN, KEN box (KENXXXR sequence); Zn, zinc finger region; BAH1 and BAH2, bromo-adjacent homology domains; KG, lysine-glycine repeats, also called GK linker.
To identify the acetylated lysines in DNMT1, we expressed HA-DNMT1 in 293T cells and treated cells with HDAC inhibitors. Immunopurified HA-DNMT1 was analyzed by mass spectrometry. In addition to the previously identified acetylated lysines in the GK linker of DNMT1 (14, 37), 12 novel acetylated lysine sites were detected (Fig. 1C).
SIRT1 deacetylates DNMT1.
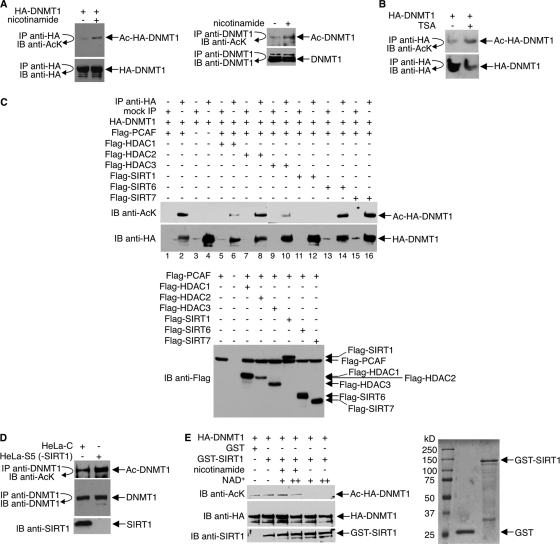
Both endogenous DNMT1 and ectopically expressed DNMT1 undergo hyperacetylation in cells treated with a class III inhibitor, nicotinamide, indicating that the acetylation of DNMT1 is regulated by sirtuin(s) (Fig. 2 A). Like nicotinamide, the class I/II HDAC inhibitor TSA also increased DNMT1 acetylation when added to 293T cells expressing HA-DNMT1 (Fig. 2B). To determine which HDACs deacetylate DNMT1, we transfected 293T cells with HA-DNMT1, Flag-PCAF, and either Flag-HDAC1, -2, or -3 or Flag-SIRT1, -6, or -7. There are two reasons for the choice of HDACs examined. First, we did not test HDAC4, -5, -6, -7, -8, -9, -10, and -11 because, in our hands, these HDACs possess minimal deacetylase activities. Second, although there are reports that under some circumstances DNMT1 may be localized outside the nucleus (16, 32, 48, 49), the majority of DNMT1 is nuclear. Also, although there are reports that many HDACs can be found distributed in the nucleus, in the cytoplasm, or within specific cellular organelles, only HDAC1, -2, and -3 and SIRT1, -6, and -7 have been unequivocally demonstrated to be mostly nuclear proteins and, therefore, more likely to colocalize with and regulate DNMT1. HDAC1, HDAC3, and SIRT1 deacetylated HA-DNMT1 whereas HDAC2, SIRT6, and SIRT7 did not (Fig. 2C; see also Fig. S2 in the supplemental material). Quantitative analysis of the Western blot results with densitometry indicated that of the HDACs that deacetylated DNMT1, SIRT1 was the most robust (see Table S1 in the supplemental material), and for this reason, we chose it for further analysis.
Fig. 2.
Deacetylation of DNMT1 by SIRT1 in vivo and in vitro. (A) Anti-HA (left) or anti-DNMT1 (right) immunoprecipitates from 293T cells, which were transfected with HA-DNMT1 expression plasmid (left panel) or untransfected (right panel) and treated with 15 mM nicotinamide for 12 h, were immunoblotted with antibody to AcK. The membranes were stripped and reprobed with anti-HA or anti-DNMT1. (B) 293T cells expressing HA-DNMT1 received 400 ng/ml of TSA for 2 h. Anti-HA immunoprecipitates were immunoblotted with antibody to acetyllysine or HA. (C) 293T cells were transfected with HA-DNMT1, Flag-PCAF, and Flag-HDAC1, -2, or -3 or Flag-SIRT1, -6, or -7 as indicated. Cell lysates were immunoprecipitated with antibody to HA or were mock precipitated (IgG), and immunoprecipitates were immunoblotted (IB) with antibody to acetyllysine. The membrane was stripped and reprobed with anti-HA. A separate Western blot assay was performed with anti-Flag antibody to assess Flag-HDAC and Flag-SIRT expressions (bottom). (D) HeLa cells were transfected with either shRNA pSuper-SIRT1 (HeLa-S) or scrambled control shRNA (HeLa-C) and grown in 1 μg/ml puromycin for 2 weeks. SIRT1 depletion in HeLa-S cells was assessed by Western blot assays, and one colony (HeLa-S5) was selected for further analysis. Anti-DNMT1 immunoprecipitates were immunoblotted with antibodies to acetyllysine or DNMT1. Immunoblot assays were also performed to assess SIRT1 expression. (E) (Left) HA-DNMT1 was first hyperacetylated in vivo by coexpression with PCAF in 293T cells, and then cell lysates were prepared and immunoprecipitated with antibody to HA. In vitro deacetylation reactions were performed by incubating anti-HA immunoprecipitates (Ac-HA-DNMT1), 1 μg GST or GST-SIRT1, NAD+, and 10 mM nicotinamide as indicated. Reaction products were immunoblotted with antibody to acetyllysine. The membrane was stripped and reprobed with anti-HA or anti-SIRT1. (Right) The quality of bacterially expressed, purified GST and GST-SIRT1 proteins was assessed by SDS-PAGE and Coomassie blue staining.
To confirm that SIRT1 deacetylates DNMT1, HeLa cells were depleted of SIRT1 by RNA interference, and anti-DNMT1 immunoprecipitates were immunoblotted with an antiacetyllysine antibody. Knockdown of SIRT1, assessed by Western blotting with an anti-SIRT1 antibody, increased amounts of acetylated DNMT1 by approximately 3-fold (Fig. 2D).
Next, to demonstrate that SIRT1 deacetylates DNMT1 in vitro, reaction mixtures that contained PCAF-acetylated HA-DNMT1 and GST-SIRT1 were set up. For controls, some reactions omitted NAD+ or included nicotinamide. As shown in Fig. 2E, GST-SIRT1, but not GST alone, efficiently deacetylated DNMT1 in the presence but not the absence of NAD+. Deacetylation of DNMT1 was inhibited in the presence of nicotinamide. Collectively, these results convincingly argue that DNMT1 is a SIRT1 substrate.
SIRT1 associates with DNMT1.
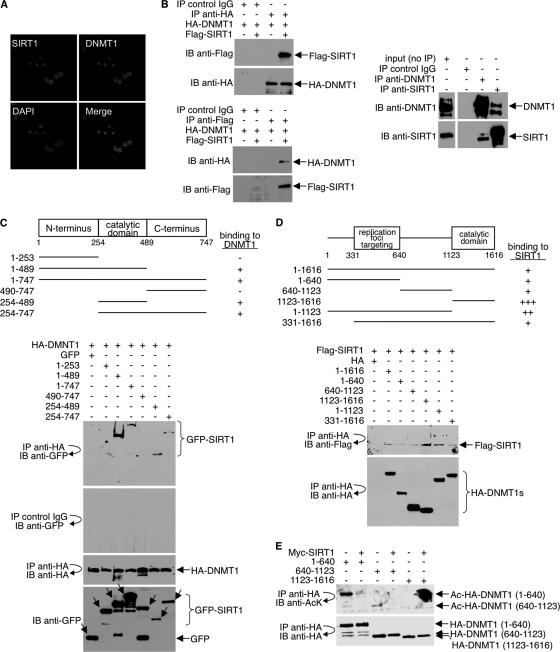
Results from previous studies suggest that DNMT1 may interact with class I HDACs (25, 58, 59). Deacetylation of DNMT1 by SIRT1 suggests that DNMT1 and SIRT1 might also interact. Consistent with this premise, we show colocalization of endogenous DNMT1 and SIRT1 in HeLa cells by immunofluorescence (Fig. 3 A). As observed by confocal microscopy, the colocalized proteins formed a punctate pattern in the nucleus.
Fig. 3.
SIRT1 interacts with DNMT1. (A) To examine the colocalization of endogenous SIRT1 and DNMT1 in the nucleus of HeLa cells, immunofluorescence studies were performed as described in Materials and Methods. Mouse anti-DNMT1 and rabbit anti-SIRT1 antibodies were used. (B) (Left) 293T cells were cotransfected with HA-DNMT1 and Flag-SIRT1. Mock precipitates (IgG control) and anti-HA and anti-Flag immunoprecipitates were immunoblotted with anti-HA or anti-Flag antibody. (Right) IgG, anti-DNMT1, and anti-SIRT1 immunoprecipitates from 293T whole-cell lysates were immunoblotted with antibody to DNMT1 or SIRT1. (C) (Top) Schematic diagram of GFP-SIRT1 and GFP-SIRT1 deletion mutants (not drawn to scale). For simplicity, the GFP portion is not included in the illustration. The ability of wild-type (1–747) and each mutant SIRT1 to bind HA-DNMT1 is indicated (+ or −). (Bottom) 293T cells were cotransfected with full-length HA-DNMT1 and either full-length GFP-SIRT1 (1–747) or the GPF-SIRT1 deletion mutants; IgG and anti-HA immunoprecipitates were immunoblotted with antibody to GFP or HA (top three blots). Cell lysates were immunoblotted with antibody to GFP to show the expression levels of SIRT1 (bottom blot). (D) (Top) Schematic diagram of HA-DNMT1 and HA-DNMT1 deletion mutants (not drawn to scale). For simplicity, the HA portion is not included in the illustration. The ability of wild-type (1–1616) and each mutant DNMT1 to bind Flag-SIRT1 is indicated (+ or −). (Bottom) 293T cells were cotransfected with full-length Flag-SIRT1 and either full-length HA-DNMT1 or HA-DNMT1 deletion mutants. Anti-HA immunoprecipitates were washed exhaustively with a buffer containing 500 mM NaCl and 0.5% NP-40 and immunoblotted with antibody to Flag or HA. (E) 293T cells were cotransfected with the HA-DNMT1 deletion mutants and either Myc-SIRT1 or empty vector. Anti-HA immunoprecipitates were immunoblotted with antibody to antiacetyllysine (AcK) or HA.
To more rigorously demonstrate interaction, 293T cells were cotransfected with HA-DNMT1 and Flag-SIRT1, and coprecipitation assays were performed. Anti-HA antibody coprecipitated Flag-SIRT1 (Fig. 3B, top left), and antibody to Flag coprecipitated HA-DNMT1 (Fig. 3B, bottom left). Antibody to endogenous DNMT1 also coprecipitated endogenous SIRT1 and vice versa (Fig. 3B, right). The association between DNMT1 and SIRT1 was specific because IgG alone (control) did not precipitate either DNMT1 or SIRT1.
To map the domain(s) in SIRT1 that mediates the interaction with DNMT1, we constructed a series of GFP-tagged SIRT1 proteins. 293T cells were cotransfected with HA-DNMT1 (full-length) and either full-length SIRT1 or SIRT1 deletion mutants. As determined by immunoprecipitation/immunoblot analysis, full-length GFP-SIRT1 (1–747) and GFP-SIRT1 mutants lacking the N- and/or C-terminal domain (1–489, 254–489, and 254–747) bound HA-DNMT1, whereas the mutants lacking the central, catalytic domain (1–253 and 490–747) did not (Fig. 3C). These results suggest that the SIRT1 catalytic domain binds to and deacetylates DNMT1. In reciprocal experiments, multiple regions of DNMT1 bound to SIRT1 (Fig. 3D). The immunoprecipitation assays were carried out under high-stringency conditions (10 washes of the immunoprecipitates with a buffer containing 500 mM NaCl and 0.5% NP-40), indicating that the SIRT1-DNMT1 complex is quite stable.
Because multiple regions of DNMT1 associate with SIRT1, we wished to determine if more than one region of DNMT1 is deacetylated by SIRT1. We transfected 293T cells with the HA-DNMT1 deletion mutants (1–640, 640–1123, and 1123–1616) with and without Myc-SIRT1, and anti-HA immunoprecipitates were immunoblotted with antiacetyllysine antibody. In the absence of Myc-SIRT1, the 1–640 mutant and, to a much lesser extent, the 640–1123 mutant were acetylated (Fig. 3E). Both regions were deacetylated in the presence of SIRT1. Basal acetylation of the 1123–1616 mutant was not detected, perhaps because of the presence of only two acetylated lysines in this region (Fig. 1C). Together, the data in Fig. 3 show that SIRT1 interacts with and deacetylates more than one region of DNMT1.
Deacetylation of DNMT1 increases its methyltransferase activity.
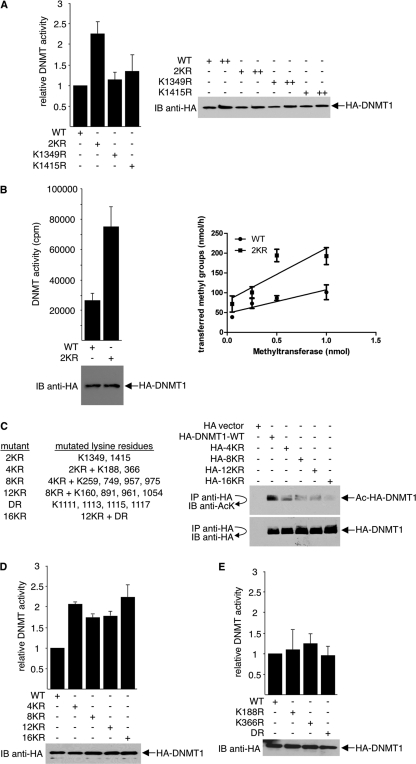
To examine whether acetylation/deacetylation of DNMT1 affects its enzymatic activity, we replaced various lysines of DNMT1 with arginines by site-directed mutagenesis. An arginine mutation retains positive charge and abrogates acetylation and, therefore, mimics lysine deacetylation at the mutated residue. Because mass spectrometry analysis shows that two lysines in the catalytic domain, Lys1349 and Lys1415, are acetylated (Fig. 1C), they were examined first. 293T cells were transfected with wild-type HA-DNMT1 or HA-DNMT1 mutated at Lys1349 (K1349R), Lys1415 (K1415R), or both (2KR). DNA methyltransferase activity was determined by an anti-methyl-cytosine antibody-based ELISA. Double point mutations increased DNMT1 activity approximately 2-fold, whereas the single mutations had no effect (Fig. 4 A). Western blots showed comparable expression levels of wild-type and mutant DNMT1 in cells. Similar results were obtained using an isotope-labeled DNA methyltransferase activity assay (Fig. 4B).
Fig. 4.
Deacetylation of DNMT1 increases its methyltransferase activity. (A) K1349 and K1415 of HA-DNMT1 were mutated to arginine either individually (K1349R and K1415R) or together (2KR). Wild-type DNMT1 (WT) and the DNMT1 mutants were ectopically expressed in 293T cells. Equal amounts of cell lysates were assayed for DNA methyltransferase activity by an anti-methyl-cytosine antibody-based ELISA (left). Twenty micrograms (+) and 60 μg (++) of cell lysates were Western blotted with anti-HA antibody to ensure equal expression of the HA constructs in cells (right). (B) WT and 2KR mutant HA-DNMT1 were overexpressed in 293T cells. Cell lysates were analyzed for DNA methyltransferase activity using the isotope labeling method as described in Materials and Methods. After incubation at 37°C for 2 h, unincorporated nucleotides were removed and the incorporation of radioactivity was determined by liquid scintillation counting. Results of representative Western blot assays to compare WT and 2KR expressions are shown (bottom left). For both purified wild-type and 2KR DNMT1, four different concentrations were compared (right). (C) (Left) List of the lysine residues mutated in DNMT1. (Right) To examine protein acetylation levels, wild-type HA-DNMT1 and the indicated HA-DNMT1 mutants were expressed in 293T cells. Anti-HA immunoprecipitates were immunoblotted with antiacetyllysine antibody, stripped, and reprobed with anti-HA antibody. (D) The indicated wild type and HA-DNMT1 mutants were expressed in 293T cells, and cell lysates were assayed for methyltransferase activity (top). HA-DNMT1 expressions were determined using Western blot assays with anti-HA antibody, and representative results are shown (bottom). (E) The indicated wild type and HA-DNMT1 mutants were expressed in 293T cells, and cell lysates were assayed for methyltransferase activity (top). Protein expressions were determined using Western blot assays with anti-HA antibody, and representative results are shown (bottom).
The 2KR mutant was then serially mutated at 2, 6, 10, and 14 additional lysines (4KR, 8KR, 12KR, and 16KR, respectively) (Fig. 4C, left). Lys1111, Lys1113, Lys1115, and Lys1117, which were previously reported to be acetylated in the GK linker (14, 37), were also mutated (the DR mutant). As expected, increased lysine-to-arginine mutations decreased the overall acetylation levels of the DNMT1 protein (Fig. 4C, right). However, the methyltransferase activity of DNMT1 did not increase further: all 2KR mutants with additional lysine mutations (4KR, 8KR, 12KR, and 16KR) were approximately 2-fold more active than wild-type DNMT1 (Fig. 4D). Also, arginine substitution for four lysines in the GK linker (DR), leaving Lys1349 and Lys1415 intact, as well as mutations in the nuclear localization or the replication targeting domain (K188R and K366R), had no effect on DNMT1 activity (Fig. 4E). Thus, of the acetylated lysines, only deacetylation of lysine 1349 and 1415 together, located in the DNMT1 catalytic domain, appears to regulate DNMT1 activity.
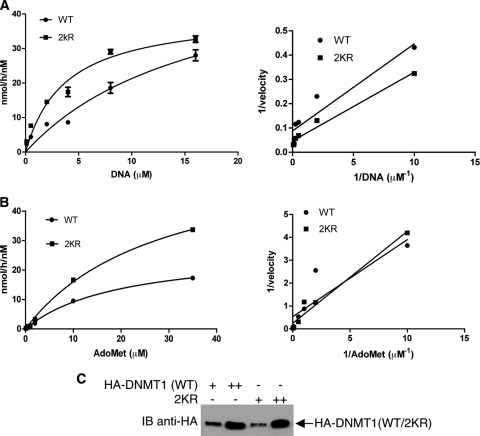
Kinetics assays confirm that the 2KR mutant possesses higher methyltransferase activity.
Because our results suggest that, when supplied with enough substrates in vitro, the 2KR mutant is more efficient in catalyzing DNA methylation than is wild-type DNMT1, we further characterized and compared the kinetic properties of wild-type and 2KR on either poly(dI-dC)·(dI-dC) or AdoMet. To determine the Km for DNA, steady-state reactions were performed by incubating 10 nM enzyme with 10 μM AdoMet, while titrating DNA from 0.1 to 16 μM for 1 h at 37°C. The Km for AdoMet was determined in the presence of 10 nM enzyme, 6 μM DNA, and 0.1 to 35 μM AdoMet. Velocity measurements at various substrate concentrations are shown in Fig. 5A and B (left), and the data were fitted to the Michaelis-Menten equation to derive parameters shown in Table 1. Corresponding double reciprocal (Lineweaver-Burk) plots are shown in Fig. 5A and B (right), and nonlinear regression software was used for fitting the Michaelis-Menten equation. For DNA substrate, 2KR has a 4-fold-lower Km (3.759 ± 0.85 versus 16.81 ± 6.65) and 3-fold higher kcat/Km (1.073 versus 0.34) than those of the wild type. For AdoMet, though 2KR has a higher Km than the wild type (28.31 ± 14.89 versus 19.79 ± 2.488), it still shows a higher kcat/Km than that of the wild type (0.216 versus 0.137). Because kcat/Km is usually used for comparison of catalytic efficiencies, our kinetics assays confirm that the 2KR mutant has a higher catalytic activity than does the wild type on DNA methylation.
Fig. 5.
Steady-state kinetics determination of wild type (WT) and 2KR mutant. (A) Kinetics of poly(dI-dC)·(dI-dC). Duplicate reaction mixtures contained either 10 nM WT or 2KR with 10 μM [3H]AdoMet (10 Ci/1 mmol) and various concentrations of DNA at 0.1, 0.5, 2, 4, 8, or 16 μM in 25 μl reaction buffer (50 mM Tris-HCl [pH 8.0], 1 mM EDTA, 5 mM DTT, 10% glycerol). (B) Kinetics for the substrate of AdoMet. Duplicate reactions were performed by incubating 10 nM enzyme, 6 μM poly(dI-dC)·(dI-dC), and titrated [3H]AdoMet at 0.1, 0.5, 1, 2, 10, and 35 μM in a 25-μl reaction volume. All reactions were performed for 1 h at 37°C, and the transferring of methyl groups was measured as described in Materials and Methods. The resultant velocities (nmol/h/nM) were plotted using GraphPad Prism5 software. (Left) Nonlinear regression of Michaelis-Menten kinetics of velocity versus DNA concentration; (right) corresponding double reciprocal (Lineweaver-Burk) plots of velocity versus DNA concentration. (C) Western blot assay to assess the quality and quantity of WT and 2KR DNMT1 used in the kinetics assays.
Table 1.
Kinetic parameters of wild-type DNMT1 and 2KR DNMT1 mutant
| Enzyme and substrate | Km (μM) | kcat (h−1) | kcat/Km (h−1 μM−1) |
|---|---|---|---|
| Poly(dI-dC)·(dI-dC) | |||
| WTa | 16.81 ± 6.65 | 5.713 ± 1.94 | 0.34 |
| 2KR | 3.759 ± 0.85 | 4.032 ± 0.49 | 1.073 |
| AdoMet | |||
| WT | 19.79 ± 2.49 | 2.717 ± 0.18 | 0.137 |
| 2KR | 28.31 ± 14.89 | 6.115 ± 0.55 | 0.216 |
WT, wild type.
Methyltransferase activity increase from DNMT1 deacetylation requires SIRT1.
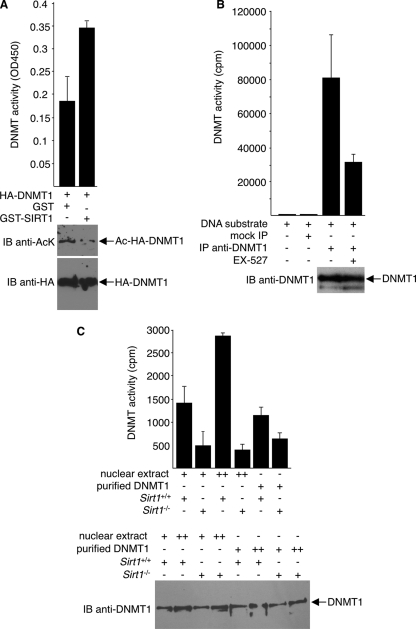
As further evidence of a stimulatory effect of deacetylation on DNMT1 activity and as evidence that the effect is mediated by SIRT1, we examined methyltransferase activity of DNMT1 that is preincubated with GST-SIRT1. Compared to DNMT1 preincubated with GST alone, DNMT1 is more active in vitro when first incubated with GST-SIRT1 (Fig. 6 A). Consistent with the substitution mutation analysis results, the SIRT1-induced increase in activity was approximately 2-fold.
Fig. 6.
Increased methyltransferase activity of SIRT1-deacetylated DNMT1. (A) In vitro methyltransferase assays were performed with an ELISA-like assay. Reaction mixtures contained immunopurified HA-DNMT1s that were preincubated with either GST or GST-SIRT1 in the presence of NAD+. (Bottom) Immunoblot assays with anti-HA and anti-AcK were done to ensure equal HA-DNMT1 quantity in each assay and to assess deacetylation of DNMT1 by GST-SIRT1, respectively. (B) Endogenous DNMT1s immunoprecipitated from 293T cells, treated or untreated with EX-527, were assayed for methyltransferase activity. Negative controls include mock precipitates with IgG or without DNA substrate. Western blot assays with anti-DNMT1 were performed to ensure equal DNMT1 in each assay (bottom). (C) Nuclear extracts or immunopurified endogenous DNMT1s, prepared from Sirt1+/+ or Sirt1−/− MEFs, were assayed for methyltransferase activity. Western blot assays with anti-DNMT1 were performed to ensure equal DNMT1 in each assay (bottom). Representative blot results are shown.
To demonstrate that SIRT1-directed deacetylation of DNMT1 controls endogenous DNMT1 activity, we treated 293T cells with EX-527 under the condition that SIRT1 was selectively inhibited. Endogenous DNMT1 was then immunopurified from EX-527-treated or untreated cells, and DNA methyltransferase assays were performed. Compared to the untreated control, DNMT1 treated with EX-527 exhibited much lower enzymatic activities (Fig. 6B). As a complementary approach, we compared DNA methyltransferase activity using nuclear extracts or purified DNMT1 prepared from Sirt1+/+ versus Sirt1−/− MEFs. Consistent with our model that SIRT1 deacetylates DNMT1 and increases DNMT1 activities, Sirt1-containing extracts are more active than extracts without Sirt1 (Fig. 6C). Likewise, immunopurified endogenous DNMT1 from Sirt1+/+ cells contains higher DNA methyltransferase activity than does DNMT1 derived from Sirt1−/− cells.
Deacetylation of DNMT1 impairs its methyltransferase-independent transcription repression activity.
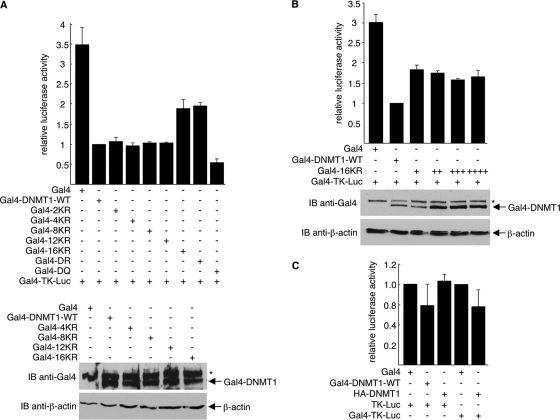
Methylation of CpG islands is the chief means by which DNMT1 silences gene expression. Clearly, the biological functions of DNMT1 are exerted through the methylation of DNA (15). However, DNMT1 can also repress transcription independently of its methyltransferase activity. For example, fragments of DNMT1 (1–1125, 653–730, and 686–812) that did not contain the methyltransferase domain, when fused to the Gal4 DNA-binding domain (Gal4-BD), effectively repressed transcription (25, 59). To determine whether acetylation/deacetylation affects the transcription repression activity of DNMT1, we fused wild-type DNMT1 and the 2KR, 4KR, 8KR, 12KR, and 16KR DNMT1 mutants to the Gal4-BD. These constructs were expressed in 293T cells along with the Gal4-TK-Luc reporter plasmid, and luciferase reporter assays were performed. Results indicated that 2KR, 4KR, 8KR, and 12KR, but not the 16KR mutant, reduced luciferase activity similarly to wild-type DNMT1 (Fig. 7 A). The reduced repression efficiency of 16KR was not due to a lower expression of this mutant. In fact, 16KR did not repress as well as wild-type DNMT1 even in the situation where it was expressed much more highly than wild-type DNMT1 (Fig. 7B). The Gal4-DNMT1-mediated repression was specific because repression by wild-type and mutant Gal4-DNMT1 occurred with the Gal4-TK-Luc reporter but not with the TK-Luc reporter, which lacks Gal4-binding sites (Fig. 7C). Repression was also dependent on the Gal4-BD, as HA-DNMT1 without the Gal4-BD failed to repress (Fig. 7C).
Fig. 7.
Deacetylation of DNMT1 GK linker reduces DNMT1 methyltransferase-independent transcription repression activity. (A and C) 293T cells in 12-well plates were transfected with either pTK-Luc or pGal4-TK-Luc (0.1 μg), pRL-SV40 (internal control, 0.018 μg), and the indicated HA-DNMT1, Gal4, or Gal4-DNMT1 constructs (0.5 μg). (B) 293T cells in 12-well plates were transfected with pGal4-TK-Luc (0.1 μg), pRL-SV40 (internal control, 0.018 μg), and the indicated Gal4-DNMT1-WT (0.5 μg) or Gal4-16KR (0.5, 1, 1.5, and 2 μg). Firefly luciferase activity is normalized to Renilla luciferase activity and depicted as fold over wild-type value with error bars showing standard deviations from three experiments. Anti-Gal4 and anti-β-actin immunoblot assays were performed to assess Gal4-DNMT1 expression and to serve as loading controls. *, nonspecific bands on Western blots.
Unlike the 2KR, 4KR, 8KR, and 12KR mutants, the 16KR mutant contains mutations of the four lysines (Lys1111, Lys1113, Lys1115, and Lys1117) in the GK linker (Fig. 4C). Thus, deacetylation of the GK linker may be responsible for the decrease in transcription repression activity of DNMT1. To further address this possibility, we monitored luciferase activity in cells expressing Gal4-DR constructs in which only the four GK linker lysines were replaced with arginine. Alternatively, to mimic acetylation at these residues, glutamines were substituted for lysines (Gal4-DQ). Like 16KR, the DR mutant reduced the repression activity of DNMT1. In contrast, the DQ mutant increased the DNMT1 repression activity (Fig. 7A).
Deacetylation of the GK linker lysines does not affect methyltransferase activity (Fig. 4D and E); thus, its effects on transcription repression activity are methyltransferase independent. Together, our data suggest that deacetylation of DNMT1 at Lys1349 and Lys1415 (catalytic domain) increases its methyltransferase activity, whereas deacetylation of DNMT1 at Lys1111, Lys1113, Lys1115, and Lys1117 (GK linker) reduces its transcription repression activity.
Deacetylation of DNMT1 affects its cell cycle regulatory function.
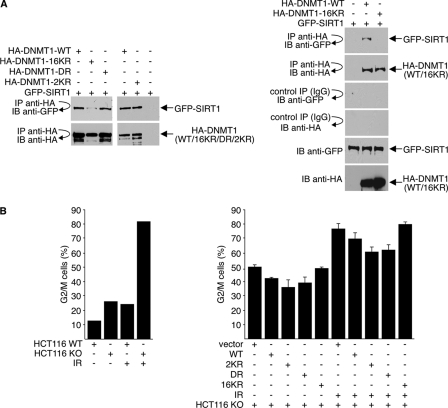
In coimmunoprecipitation studies, we note that the DR and 2KR mutants efficiently interact with SIRT1 (Fig. 8 A, left). Interaction of the 16KR mutant with SIRT1, on the other hand, is severely compromised (Fig. 8A, left and right). This finding suggests that in addition to regulating methyltransferase and transcription repression activities, acetylation of lysines outside the catalytic and the GK region may be important for controlling additional DNMT1 functions.
Fig. 8.
Deacetylation of DNMT1 affects its cell cycle regulatory function. (A) 293T cells were transfected with GFP-SIRT1 and either wild-type or mutant HA-DNMT1 expression plasmids. Cell lysates were immunoprecipitated with antibody to HA, and precipitated material was immunoblotted with antibodies to GFP or HA. GFP-SIRT1 and HA-DNMT1 expressions were monitored by direct Western blotting with anti-GFP and anti-HA antibodies, respectively. (B) Wild-type HCT116 (WT) cells or DNMT1-knockout (KO) HCT116 cells were cotransfected with GFP and either wild-type DNMT1 (WT), mutant HA-DNMT1, or vector plasmid. The ratio of GFP to HA-DNMT1 was 1:10. Cells received 10 Gy of gamma irradiation (IR) 36 h after transfection and were harvested at 48 h posttransfection. Cell cycle position was determined by FACS analysis of propidium iodide-stained cells for GFP-positive cells. The data shown are the average values ± standard deviations from three separate experiments (right).
Complete removal of DNMT1 from cells results in G2/M arrest, mitotic catastrophe, and cell death (12). To assess the function of DNMT1 acetylation in promoting cell cycle progression, rescue experiments were performed. For these experiments, we used DNMT1-knockout (DNMT1-KO) HCT116 colorectal cancer cells containing a deletion of the three exons encoding the DNMT1 regulatory domain. Although originally designed for a complete knockout, generation of a hypomorphic allele by alternative splicing allows expression of minute amounts of DNMT1 in these “DNMT1-null” HCT116 cells (19, 62). Consistent with the earlier report (12), DNMT1-KO cells accumulated in G2 phase in the absence of DNMT1, and treatment of these cells with DNA-damaging agents (gamma irradiation) further increases the percentage of cells in G2/M (Fig. 8B, left). Regardless of irradiation, DNMT1-KO HCT116 cells consistently showed a higher level of G2/M arrest than did wild-type cells. For rescue experiments, we expressed wild-type HA-DNMT1 or either the 2KR, DR, or 16KR mutant in DNMT1-KO HCT116 cells; GFP was also expressed to allow sorting of transfected cells. Cells were either untreated or treated with irradiation, and the cell cycle position of GFP-expressing cells was determined by FACS analysis of propidium iodide-stained cells. Regardless of treatment, expression of wild-type HA-DNMT1 or 2KR or DR mutant reduced the percentage of G2/M cells. Expression of the 16KR mutant, on the other hand, did not and gave results similar to those of vector-transfected cells. These data suggest that cell cycle regulation, at G2/M in particular, by DNMT1 is regulated by acetylated and deacetylation.
The effects of DNMT1 deacetylation on DNMT1 methyltransferase activity, DNMT1 methyltransferase-independent repression activity, SIRT1 interaction, and relief of G2/M arrest are summarized in Table 2. Collectively, our data suggest that reversible acetylations of lysines in different regions of DNMT1 have distinct roles in regulating the activity and protein function of DNMT1. Deacetylations of different lysines also act together to help DNMT1 exert biological functions, depending on cellular context.
Table 2.
Effects of DNMT1 deacetylation mutations
| DNMT1 mutant | Increase in enzymatic activity | Decrease in transcription repression | Decrease in SIRT1 binding | Inability to relieve G2/M arrest |
|---|---|---|---|---|
| 2KR | + | − | − | − |
| DR | − | + | − | − |
| 16KR | + | + | + | + |
SIRT1 regulates DNMT1-mediated silencing of TSGs.
DNMT1 silences TSGs by methylating DNA and recruiting corepressors. As an example, DNMT1 recruits HDAC1 to the estrogen receptor α (ESR1) promoter to turn off expression of ESR1 in MDA-MB-231 breast cancer cells (50, 75). Cotreatment of cells with TSA and the methyltransferase inhibitor 5-aza-dC reactivates transcription of ESR1 (22, 71). In addition, it has been reported that SIRT1 associates with the E-cadherin (CDH1) promoter, which can also be silenced in MDA-MB-231 cells (27, 56).
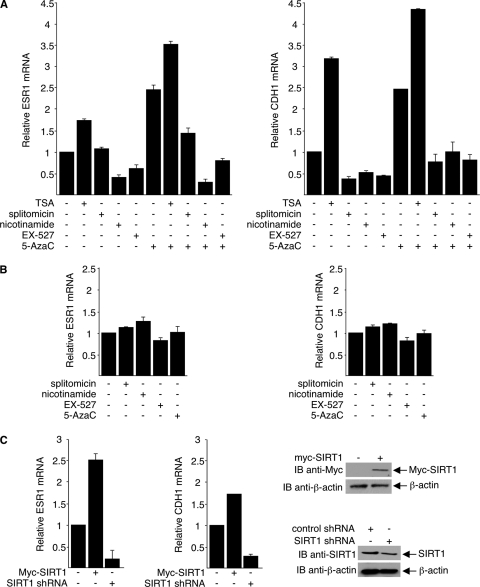
To determine whether inhibition of SIRT1 activity affects repression of the ESR1 and CDH1 promoters, we treated MDA-MB-231 cells with TSA (which inhibits class I and II HDACs), nicotinamide (which inactivates all sirtuins), EX-527 (which selectively inhibits SIRT1 in low concentrations), or splitomicin (which inhibits sirtuin activity), with or without 5-aza-dC. Amounts of ESR1 mRNA and CDH1 mRNA were determined by real-time PCR. As expected, TSA increased the abundance of both ESR1 and CDH1 mRNA, as did 5-aza-dC; cotreatment of cells with the two agents produced an additive effect (Fig. 9 A). Interestingly, all SIRT1 inhibitors examined markedly reduced the abundance of CDH1 mRNA in the absence and the presence of 5-aza-dC. Nicotinamide and EX-527 also repressed expression of ESR1 mRNA both in the presence and in the absence of 5-aza-dC. Splitomicin repressed ESR1 mRNA expression in combination with 5-aza-dC but not alone. As negative controls, we show that the inhibitors had little if any effect on the expression of ESR1 mRNA or CDH1 mRNA in MCF-7 cells, in which ESR1 and CDH1 were not silenced by DNMT1 (55, 74, 75) and were thereby unresponsive to SIRT1 inhibitors (Fig. 9B). It excludes the possibilities that SIRT1 regulates the transcription of ESR1 and CDH1 independently of DNMT1 and that SIRT1 inhibitors are toxic to cells under the conditions used. Together, these data suggest that deacetylation of DNMT1 by SIRT1 impairs silencing of TSGs by DNMT1.
Fig. 9.
SIRT1 regulates DNMT1-mediated silencing of TSGs. (A and B) MDA-MB-231 (A) and MCF-7 (B) cells were treated with 0.3 μM TSA for 12 h, 15 mM nicotinamide for 12 h, 300 μM splitomicin for 24 h, or 1 μM EX-527 for 6 h. Some cultures also received 50 μM 5-aza-dC for 48 h. (C) MDA-MB-231 cells with plasmids that express either Myc-SIRT1 or SIRT1 shRNA. Expressions of ESR1 and CDH1 mRNA were determined by quantitative real-time PCR. The RNA of untreated MCF-7 cells was used as positive control and for generation of a standard curve. 18S RNA was used as the internal control. Amounts of PCR-amplified ESR1 mRNA and CDH1 mRNA were determined from the standard curve and normalized to the amount of 18S RNA. Results from averages of three experiments with standard deviations are depicted as fold of untreated control. SIRT1 protein expressions were assessed using Western blot assays (right).
To further confirm that SIRT1 alters silencing of TSGs by DNMT1, we transfected MDA-MB-231 cells with plasmids that express either Myc-SIRT1 or SIRT1 shRNA. Quantitative real-time PCR then followed to measure ESR and CDH1 mRNA. As shown in Fig. 9C (left), overexpression of SIRT1 led to an increase in both ESR and CDH1 mRNA, and consistently with our results obtained from the use of SIRT1 inhibitors, depletion of SIRT1 repressed transcription of ESR and CDH1. Altogether, our results suggest that SIRT1 can relieve transcriptional repression in a certain cellular context, which might be through deacetylation and inhibition of DNMT1, independently of histone deacetylation.
DISCUSSION
Given its fundamental importance, the key DNA methyltransferase enzyme, DNMT1, is tightly regulated in mammalian cells. Data presented here show that DNMT1 is an acetylated protein. Proteomics analysis identified 12 acetylated lysines in DNMT1: four in the N-terminal region containing the nuclear localization signal and replication focus targeting domain (Lys160, Lys188, Lys259, and Lys266), six further downstream around the BAH1 and BAH2 domains (Lys749, Lys861, Lys957, Lys961, Lys975, and Lys1054), and two in the C-terminal domain (Lys1349 and Lys1415). These findings extend previous reports showing acetylation of four lysines in the GK linker of DNMT1 (Lys1111, Lys1113, Lys1115, and Lys1117) (14, 37).
SIRT1, -6, and -7 are nuclear, and SIRT2 is mostly in the cytoplasm, whereas SIRT3, -4, and -5 are predominantly mitochondrial (51). Of the nuclear SIRTs, only SIRT1 deacetylated DNMT1. SIRT1 colocalized with DNMT1 in the nucleus, and antibody to SIRT1 coprecipitated DNMT1 and vice versa. SIRT1 deacetylated DNMT1 both in vivo and in vitro, and cells depleted of SIRT1 contained hyperacetylated DNMT1 compared to SIRT1-expressing cells. Thus, SIRT1 physically interacts with and directly deacetylates DNMT1 and is a biologically relevant DNMT1 deacetylase. Our results fit well with previous observation of an association of SIRT1 with DNMT1 and its consequent recruitment to rRNA genes in colon cancer cells (20). SIRT1, however, is not the only DNMT1 deacetylase. Our results show that the class I/II HDAC inhibitor TSA also increased DNMT1 acetylation, and ectopic expression of HDAC1 and -3 decreased DNMT1 acetylation. Also, a recent study suggests that HDAC1 deacetylates DNMT1 and protects it from proteasomal degradation (17).
Our results report for the first time that DNMT1 deacetylation by SIRT1 increases its DNA methyltransferase activity and alters its transcription repression function. Previous studies show that different regions of DNMT1 repress transcription by different means. The C terminus of DNMT1 catalyzes the methylation of hemimethylated CpG dinucleotides often present in 5′ regulatory gene regions, which ultimately leads to transcription silencing. In contrast, the N-terminal and central regions recruit transcriptional repressors (DMAP1 and class I/II HDACs, respectively) and direct repressors to DNA and transcription complexes (25, 59), although it is controversial whether the methyltransferase-independent mode of transcription repression by DNMT1 is biologically relevant. We show that SIRT1 interacts with and deacetylates lysines in multiple regions of DNMT1 and suggest that SIRT1-mediated deacetylation has multiple, domain-specific consequences. As determined by arginine substitution of specific lysines, deacetylation of the two lysines in the catalytic domain increased the methyltransferase activity of DNMT1, whereas deacetylation of the four lysines in the GK linker reduced the methyltransferase-independent transcription repression activity of DNMT1. These alterations appear to be antagonistic: deacetylation of the catalytic region and increasing methyltransferase activity would presumably turn off gene expression, whereas deacetylation of the GK linker renders DNMT1 less capable of repression and presumably turns on gene expression independent of methyltransferase activity.
There are several possible explanations for these seemingly contradictory effects. First, multiple different domains independently or coordinately regulate the functions of DNMT1. As shown in this study, the overall effect of DNMT1 is also controlled by deacetylation of lysines outside the catalytic domain and GK linker. Second, lysine deacetylation of distinct domains of DNMT1 may occur temporally and spatially and is dependent on cellular text. Not all domains are simultaneously acetylated and deacetylated. Finally, although SIRT1 interacts with the catalytic domain of DNMT1, it does not necessarily deacetylate K1349 and K1415 in the catalytic domain of DNMT1 alone. Because of the low affinity of panantiacetyllysine antibody and the low density of acetylated lysine residues in the catalytic domain, at this time, we could not detect acetylation of the catalytic domain and thereby its possible deacetylation by SIRT1. The generation of an anti-site-specific acetyl antibody in the future will help resolve this issue.
In addition to catalytic and repression activity, we also examined the effects of DNMT1 deacetylation on other functions. Interestingly, not all acetylated lysine on DNMT1 affects obvious biological functions. For example, deacetylation of K188, which is located in the nuclear localization signal sequence, did not alter DNMT1 nuclear localization (data not shown). Further, deacetylation of DNMT1 most likely does not affect its interaction with PCNA (see Fig. S3 in the supplemental material). We did, however, find that HCT116 cells that are nearly devoid of DNMT1 accumulate in G2/M more readily than do wild-type cells. Ectopic expression of wild-type DNMT1 or of the 2KR or DR mutant in DNMT1-KO cells partially prevented G2/M accumulation, whereas expression of the 16KR mutant did not. Thus, deacetylation of DNMT1 impairs its ability to rescue cells from DNA damage-induced G2/M arrest. These results suggest that the functional consequences of DNMT1 deacetylation extend beyond DNA methylation and gene transcription.
The mechanisms by which DNMT1 inhibits gene expression are complicated and controversial. Methylation of gene promoters and genomic DNA is a well-recognized function of DNMT1; however, whether DNMT1 requires its methyltransferase activity to silence TSGs or to promote cell survival is less clear (18, 52, 60, 65). For example, knockdown of DNMT1 in estrogen receptor-negative breast cancer cells reactivated estrogen receptor expression without affecting DNA methylation. Discrepancies in the literature may reflect an incomplete elimination of DNMT1 from cells; complete genomic depletion of DNMT1 resulted in cell death (12, 31, 47, 66), whereas knockdown of DNMT1 in another study did not significantly affect genomic 5-methylcytosine content or CpG methylation of TSGs (57, 65). In fact, we did not find apparent changes in 5′-methylcytosine content of the whole genome in HCT116 DNMT1-knockout cells transfected with wild-type or mutant DNMT1 (data not shown). As a result, we did not examine DNA methylation of CpG islands on specific gene promoters in cells expressing wild-type DNMT1 or DNMT1 mutants.
It is commonly believed that DNA methylation is tightly linked to other epigenetic signals, particularly histone deacetylation. For example, HDAC1 and HDAC2 complex with the methyl-CpG-binding protein MeCP2 (53). Like histone deacetylation, high levels of methyl-CpG DNA correlate with transcriptional inactivity. Also, hyperacetylated histones are found on hypomethylated CpG islands (64). Indeed, in one study, synergy of demethylation and histone deacetylase inhibition was found in the reexpression of genes silenced in cancer (11). The results presented here suggest that SIRT1 may regulate gene transcription independently of its classic role in histone deacetylation. Conceivably, as a protein deacetylase, SIRT1 deacetylates histones to maintain transcriptionally repressive chromatin and simultaneously functions in transcription repression through deacetylation of nonhistones.
In summary, we show that DNMT1 is acetylated at multiple lysines and that SIRT1 deacetylates DNMT1 in vitro and in vivo. Deacetylation of DNMT1 at specific lysines enhanced its methyltransferase activity, changed its transcription repression activity and cell cycle regulatory function, and impaired its capacity to silence TSGs. In contrast to class I HDACs, which boost the silencing effect of DNMT1 by chromatin modification or stabilization of DNMT1 (59, 81), SIRT1 directly modifies DNMT1 activity. Our study provides new insight into the posttranslational regulation of DNMT1 function and the functional diversity of SIRT1 in gene silencing.
Supplementary Material
ACKNOWLEDGMENTS
We thank Steve Baylin and Bert Vogelstein (Johns Hopkins University) for the DNMT1-null cells, Art Riggs and Gerd Pfeifer (Beckman Research Institute) for the Myc-DNMT1 plasmid, Alejandro Vaquero (Institut Català d'Oncologia) for the Gal4-TK-Luc plasmid, Jia Fang (Moffitt Cancer Center) for the pcDNA3-Gal4 plasmid, and the Moffitt Cancer Center Core Facility for their technical assistance.
This work was supported by grants to E.S. from the NIH (GM081650), the AHA (0755298), and the Kaul Foundation. L.P. was supported by a postdoctoral fellowship from the AHA.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 26 September 2011.
REFERENCES
- 1. Adams R. L., Rinaldi A., Seivwright C. 1991. Microassay for DNA methyltransferase. J. Biochem. Biophys. Methods 22:19–22 [DOI] [PubMed] [Google Scholar]
- 2. Aizawa H., et al. 2004. Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science 303:197–202 [DOI] [PubMed] [Google Scholar]
- 3. Aoki A., et al. 2001. Enzymatic properties of de novo-type mouse DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 29:3506–3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beard C., Li E., Jaenisch R. 1995. Loss of methylation activates Xist in somatic but not in embryonic cells. Genes Dev. 9:2325–2334 [DOI] [PubMed] [Google Scholar]
- 5. Bender C. M., Pao M. M., Jones P. A. 1998. Inhibition of DNA methylation by 5-aza-2′-deoxycytidine suppresses the growth of human tumor cell lines. Cancer Res. 58:95–101 [PubMed] [Google Scholar]
- 6. Bestor T., Laudano A., Mattaliano R., Ingram V. 1988. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J. Mol. Biol. 203:971–983 [DOI] [PubMed] [Google Scholar]
- 7. Bestor T. H. 1992. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO J. 11:2611–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bestor T. H. 2000. The DNA methyltransferases of mammals. Hum. Mol. Genet. 9:2395–2402 [DOI] [PubMed] [Google Scholar]
- 9. Bird A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6–21 [DOI] [PubMed] [Google Scholar]
- 10. Brunet A., et al. 2004. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303:2011–2015 [DOI] [PubMed] [Google Scholar]
- 11. Cameron E. E., Bachman K. E., Myohanen S., Herman J. G., Baylin S. B. 1999. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 21:103–107 [DOI] [PubMed] [Google Scholar]
- 12. Chen T., et al. 2007. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat. Genet. 39:391–396 [DOI] [PubMed] [Google Scholar]
- 13. Chen T., Li E. 2006. Establishment and maintenance of DNA methylation patterns in mammals. Curr. Top. Microbiol. Immunol. 301:179–201 [DOI] [PubMed] [Google Scholar]
- 14. Choudhary C., et al. 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325:834–840 [DOI] [PubMed] [Google Scholar]
- 15. Damelin M., Bestor T. H. 2007. Biological functions of DNA methyltransferase 1 requires its methyltransferase activity. Mol. Cell. Biol. 27:3891–3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Desplats P., et al. 2011. Alpha-synuclein sequesters Dnmt1 from the nucleus: a novel mechanism for epigenetic alterations in Lewy body diseases. J. Biol. Chem. 286:9031–9037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Du Z., et al. 2010. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci. Signal. 3:ra80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eads C. A., et al. 1999. CpG island hypermethylation in human colorectal tumors is not associated with DNA methyltransferase overexpression. Cancer Res. 59:2302–2306 [PubMed] [Google Scholar]
- 19. Egger G., et al. 2006. Identification of DNMT1 (DNA methyltransferase 1) hypomorphs in somatic knockouts suggests an essential role for DNMT1 in cell survival. Proc. Natl. Acad. Sci. U. S. A. 103:14080–14085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Espada J., et al. 2007. Epigenetic disruption of ribosomal RNA genes and nucleolar architecture in DNA methyltransferase 1 (Dnmt1) deficient cells. Nucleic Acids Res. 35:2191–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Esteve P. O., et al. 2009. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 106:5076–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fan J., et al. 2008. ER alpha negative breast cancer cells restore response to endocrine therapy by combination treatment with both HDAC inhibitor and DNMT inhibitor. J. Cancer Res. Clin. Oncol. 134:883–890 [DOI] [PubMed] [Google Scholar]
- 23. Flynn J., Glickman J. F., Reich N. O. 1996. Murine DNA cytosine-C5 methyltransferase: pre-steady and steady-state kinetic analysis with regulatory DNA sequences. Biochemistry 35:7308–7315 [DOI] [PubMed] [Google Scholar]
- 24. Flynn J., Reich N. 1998. Murine DNA (cytosine-5-)-methyltransferase: steady-state and substrate trapping analyses of the kinetic mechanism. Biochemistry 37:15162–15169 [DOI] [PubMed] [Google Scholar]
- 25. Fuks F., Burgers W. A., Brehm A., Hughes-Davies L., Kouzarides T. 2000. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat. Genet. 24:88–91 [DOI] [PubMed] [Google Scholar]
- 26. Glozak M. A., Sengupta N., Zhang X., Seto E. 2005. Acetylation and deacetylation of non-histone proteins. Gene 363:15–23 [DOI] [PubMed] [Google Scholar]
- 27. Graff J. R., et al. 1995. E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res. 55:5195–5199 [PubMed] [Google Scholar]
- 28. Haigis M. C., Guarente L. P. 2006. Mammalian sirtuins—emerging roles in physiology, aging, and calorie restriction. Genes Dev. 20:2913–2921 [DOI] [PubMed] [Google Scholar]
- 29. Hermann A., Gowher H., Jeltsch A. 2004. Biochemistry and biology of mammalian DNA methyltransferases. Cell. Mol. Life Sci. 61:2571–2587 [DOI] [PubMed] [Google Scholar]
- 30. Issa J. P., et al. 1993. Increased cytosine DNA-methyltransferase activity during colon cancer progression. J. Natl. Cancer Inst. 85:1235–1240 [DOI] [PubMed] [Google Scholar]
- 31. Jackson-Grusby L., et al. 2001. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 27:31–39 [DOI] [PubMed] [Google Scholar]
- 32. Jeong Y. S., Oh K. B., Park J. S., Kim J.-S., Kang Y.-K. 2009. Cytoplasmic localization of oocyte-specific variant of porcine DNA methyltransferase-1 during early development. Dev. Dyn. 238:1666–1673 [DOI] [PubMed] [Google Scholar]
- 33. Jones P. A., Baylin S. B. 2002. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3:415–428 [DOI] [PubMed] [Google Scholar]
- 34. Jones P. A., Takai D. 2001. The role of DNA methylation in mammalian epigenetics. Science 293:1068–1070 [DOI] [PubMed] [Google Scholar]
- 35. Kaeberlein M., McVey M., Guarente L. 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13:2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kautiainen T. L., Jones P. A. 1986. DNA methyltransferase levels in tumorigenic and nontumorigenic cells in culture. J. Biol. Chem. 261:1594–1598 [PubMed] [Google Scholar]
- 37. Kim S. C., et al. 2006. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23:607–618 [DOI] [PubMed] [Google Scholar]
- 38. Kimura H., Shiota K. 2003. Methyl-CpG-binding protein, MeCP2, is a target molecule for maintenance DNA methyltransferase, Dnmt1. J. Biol. Chem. 278:4806–4812 [DOI] [PubMed] [Google Scholar]
- 39. Kouzarides T. 2000. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 19:1176–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Langley E., et al. 2002. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 21:2383–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee B., Muller M. T. 2009. SUMOylation enhances DNA methyltransferase 1 activity. Biochem. J. 421:449–461 [DOI] [PubMed] [Google Scholar]
- 42. Leonhardt H., Bestor T. H. 1993. Structure, function and regulation of mammalian DNA methyltransferase. EXS 64:109–119 [DOI] [PubMed] [Google Scholar]
- 43. Leonhardt H., Cardoso M. C. 2000. DNA methylation, nuclear structure, gene expression and cancer. J. Cell. Biochem. Suppl. 35:78–83 [DOI] [PubMed] [Google Scholar]
- 44. Leonhardt H., Page A. W., Weier H. U., Bestor T. H. 1992. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71:865–873 [DOI] [PubMed] [Google Scholar]
- 45. Lewis J., Bird A. 1991. DNA methylation and chromatin structure. FEBS Lett. 285:155–159 [DOI] [PubMed] [Google Scholar]
- 46. Li E., Beard C., Jaenisch R. 1993. Role for DNA methylation in genomic imprinting. Nature 366:362–365 [DOI] [PubMed] [Google Scholar]
- 47. Li E., Bestor T. H., Jaenisch R. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69:915–926 [DOI] [PubMed] [Google Scholar]
- 48. Lodde V., et al. 2009. Localization of DNA methyltransferase-1 during oocyte differentiation, in vitro maturation and early embryonic development in cow. Eur. J. Histochem. 53:199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lundberg J., et al. 2009. Traumatic brain injury induces relocalization of DNA-methyltransferase 1. Neurosci. Lett. 457:8–11 [DOI] [PubMed] [Google Scholar]
- 50. Macaluso M., Cinti C., Russo G., Russo A., Giordano A. 2003. pRb2/p130-E2F4/5-HDAC1-SUV39H1-p300 and pRb2/p130-E2F4/5-HDAC1-SUV39H1-DNMT1 multimolecular complexes mediate the transcription of estrogen receptor-alpha in breast cancer. Oncogene 22:3511–3517 [DOI] [PubMed] [Google Scholar]
- 51. Michishita E., Park J. Y., Burneskis J. M., Barrett J. C., Horikawa I. 2005. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell 16:4623–4635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Milutinovic S., Knox J. D., Szyf M. 2000. DNA methyltransferase inhibition induces the transcription of the tumor suppressor p21(WAF1/CIP1/sdi1). J. Biol. Chem. 275:6353–6359 [DOI] [PubMed] [Google Scholar]
- 53. Nan X., et al. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393:386–389 [DOI] [PubMed] [Google Scholar]
- 54. North B. J., Marshall B. L., Borra M. T., Denu J. M., Verdin E. 2003. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 11:437–444 [DOI] [PubMed] [Google Scholar]
- 55. Ottaviano Y. L., et al. 1994. Methylation of the estrogen receptor gene CpG island marks loss of estrogen receptor expression in human breast cancer cells. Cancer Res. 54:2552–2555 [PubMed] [Google Scholar]
- 56. Pruitt K., et al. 2006. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2:e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rhee I., et al. 2000. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature 404:1003–1007 [DOI] [PubMed] [Google Scholar]
- 58. Robertson K. D., et al. 2000. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 25:338–342 [DOI] [PubMed] [Google Scholar]
- 59. Rountree M. R., Bachman K. E., Baylin S. B. 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 25:269–277 [DOI] [PubMed] [Google Scholar]
- 60. Sato M., et al. 2002. The expression of DNA methyltransferases and methyl-CpG-binding proteins is not associated with the methylation status of p14(ARF), p16(INK4a) and RASSF1A in human lung cancer cell lines. Oncogene 21:4822–4829 [DOI] [PubMed] [Google Scholar]
- 61. Saunders L. R., Verdin E. 2007. Sirtuins: critical regulators at the crossroads between cancer and aging. Oncogene 26:5489–5504 [DOI] [PubMed] [Google Scholar]
- 62. Spada F., et al. 2007. DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J. Cell Biol. 176:565–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sugiyama Y., et al. 2010. The DNA-binding activity of mouse DNA methyltransferase 1 is regulated by phosphorylation with casein kinase 1delta/epsilon. Biochem. J. 427:489–497 [DOI] [PubMed] [Google Scholar]
- 64. Tazi J., Bird A. 1990. Alternative chromatin structure at CpG islands. Cell 60:909–920 [DOI] [PubMed] [Google Scholar]
- 65. Ting A. H., et al. 2004. CpG island hypermethylation is maintained in human colorectal cancer cells after RNAi-mediated depletion of DNMT1. Nat. Genet. 36:582–584 [DOI] [PubMed] [Google Scholar]
- 66. Tucker K. L., et al. 1996. Germ-line passage is required for establishment of methylation and expression patterns of imprinted but not of nonimprinted genes. Genes Dev. 10:1008–1020 [DOI] [PubMed] [Google Scholar]
- 67. Turek-Plewa J., Jagodzinski P. P. 2005. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell. Mol. Biol. Lett. 10:631–647 [PubMed] [Google Scholar]
- 68. Vaquero A., et al. 2004. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell 16:93–105 [DOI] [PubMed] [Google Scholar]
- 69. Vaziri H., et al. 2001. hSIR2 (SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107:149–159 [DOI] [PubMed] [Google Scholar]
- 70. Wang C., et al. 2006. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat. Cell Biol. 8:1025–1031 [DOI] [PubMed] [Google Scholar]
- 71. Wei M., et al. 2008. Estrogen receptor alpha, BRCA1, and FANCF promoter methylation occur in distinct subsets of sporadic breast cancers. Breast Cancer Res. Treat. 111:113–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wu J., et al. 1993. Expression of an exogenous eukaryotic DNA methyltransferase gene induces transformation of NIH 3T3 cells. Proc. Natl. Acad. Sci. U. S. A. 90:8891–8895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yang W. M., Yao Y. L., Sun J. M., Davie J. R., Seto E. 1997. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J. Biol. Chem. 272:28001–28007 [DOI] [PubMed] [Google Scholar]
- 74. Yang X., et al. 2000. Transcriptional activation of estrogen receptor alpha in human breast cancer cells by histone deacetylase inhibition. Cancer Res. 60:6890–6894 [PubMed] [Google Scholar]
- 75. Yang X., et al. 2001. Synergistic activation of functional estrogen receptor (ER)-alpha by DNA methyltransferase and histone deacetylase inhibition in human ER-alpha-negative breast cancer cells. Cancer Res. 61:7025–7029 [PubMed] [Google Scholar]
- 76. Yang X. J., Ogryzko V. V., Nishikawa J., Howard B. H., Nakatani Y. 1996. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382:319–324 [DOI] [PubMed] [Google Scholar]
- 77. Yang X. J., Seto E. 2008. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol. Cell 31:449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yang X. J., Seto E. 2008. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 9:206–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yokochi T., Robertson K. D. 2002. Preferential methylation of unmethylated DNA by mammalian de novo DNA methyltransferase Dnmt3a. J. Biol. Chem. 277:11735–11745 [DOI] [PubMed] [Google Scholar]
- 80. Yuan Z., Zhang X., Sengupta N., Lane W. S., Seto E. 2007. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol. Cell 27:149–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhou Q., Agoston A. T., Atadja P., Nelson W. G., Davidson N. E. 2008. Inhibition of histone deacetylases promotes ubiquitin-dependent proteasomal degradation of DNA methyltransferase 1 in human breast cancer cells. Mol. Cancer Res. 6:873–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.