Abstract
Shiga toxin-producing Escherichia coli (STEC) in northern Alberta was detected using two enzyme immunoassays and an in-house real-time PCR. Of 2,328 stool samples, 8 were positive for O157:H7 STEC and 13 were positive for non-O157 STEC. No significant gender (P = 0.17) or age (P = 0.81) differences between groups were seen. Most positive diarrheal stool samples were nonbloody.
TEXT
Shiga toxin-producing Escherichia coli (STEC) is an emerging pathogen responsible for sporadic infections and outbreaks. The most common serotype of STEC is O157:H7. However, some non-O157 STEC strains, including the recently identified O104:H4 serotype, have also been associated with outbreaks, the development of hemolytic-uremic syndrome, and deaths (3, 4, 8, 9). To date, a great majority of clinical laboratories have focused on the detection of O157:H7 STEC. In 2009, the Centers for Disease Control and Prevention (CDC) released an updated guideline recommending the simultaneous detection of STEC O157 using traditional culture and of non-O157 STEC using specific testing for Shiga toxins or their genetic determinants (stx genes) for all cases of acute, community-acquired diarrhea (http://www.cdc.gov/mmwr/preview/mmwrhtml/rr5812a1.htm).
In this study, we sought (i) to determine the prevalence of non-O157 STEC strains in northern Alberta and (ii) to assess the performance of the Premier enterohemorrhagic E. coli (EHEC) toxin enzyme immunoassay (EIA) (Meridian Bioscience Inc., Cincinnati, OH) and an in-house real-time PCR using the ABI Prism 7500 FAST sequence detection system (Applied Biosystems, Life Technologies Corporation, Carlsbad, CA) with clinical stool samples following overnight incubation in MacConkey broth. ImmunoCard STAT! (Meridian Bioscience Inc.) was also used for all positive enriched cultures, as tested by the Premier EHEC toxin EIA, to further differentiate among toxin types.
(This study was presented in part at the Canadian Association for Clinical Microbiology and Infectious Diseases-Association of Medical Microbiology and Infectious Diseases conferences on 6 to 8 May 2010 and 6 to 9 April 2011.)
The limit of detection (LOD) of the Premier EHEC EIA and of our in-house real-time PCR was determined by running both assays in replicates of four with cell suspensions containing 108 to 100 CFU/ml. The LOD for STEC toxin using the Premier EHEC assay was 25 × 104 CFU/reaction, and that of our in-house real-time PCR targeting the stx1 and stx2 genes was 5 and 50 bacterial cells/reaction, respectively.
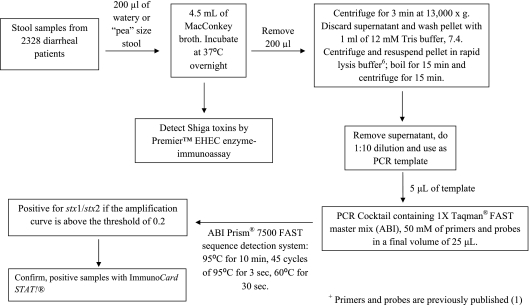
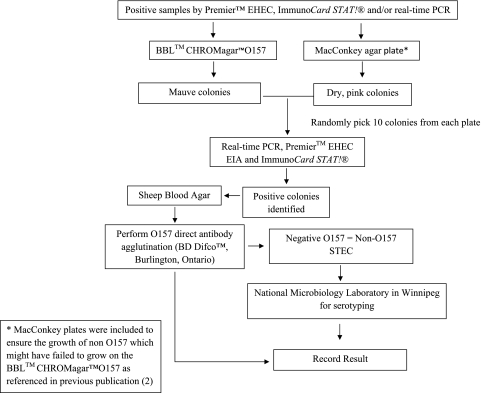
A total of 2,328 stool specimens were collected from 1 May to 15 August 2009 and 2010, and the testing algorithm is presented in Fig. 1 (1, 6). For the isolation of STEC organisms, all samples positive by the Premier EHEC EIA and/or TaqMan real-time PCR analysis for stx1 and/or stx2 were plated onto selective plates as illustrated in Fig. 2. All picked colonies were then tested by real-time PCR, Premier EHEC EIA, and the ImmunoCard STAT! assay, which can further differentiate Shiga toxins 1 and 2. An unpaired t test was used to evaluate the statistical significance of the age variation between gender groups among patients whose stool samples tested positive for STEC (O157:H7 and non-O157). Statistical significance was defined as a P value of <0.05.
Fig. 1.
Algorithm for the detection of stx genes by an in-house real-time PCR using the ABI Prism 7500 FAST sequence detection system and the presence of Shiga toxins using the Premier EHEC EIA.
Fig. 2.
STEC-positive sample isolation.
Among the 2,328 clinical samples tested, 21 (0.9%) STEC-positive samples were identified by our in-house real-time PCR, including 8 (38.1%) O157 and 13 (61.9%) non-O157 strains. In 2009, two samples positive by real-time PCR were missed by the Premier EHEC EIA (Table 1). One was confirmed as E. coli O157:H7, and the other was O103:H25. In 2010, no discordant results were observed among the three assays.
Table 1.
Information on patients with Shiga toxin-positive stool samples
| E. coli serotype | Age (yr) | Gender | Stool sample description | stx1/stx2b |
|---|---|---|---|---|
| O157:H7 | 5 | Female | Liquid | Pos/pos |
| O157:H7a | 23 | Female | Liquid, bloody, mucoid | Pos/pos |
| O157:H7 | 26 | Female | Mucoid, bloody | Neg/pos |
| O157:H7 | 37 | Female | Liquid | Pos/pos |
| O157:H7 | 66 | Female | Bloody, liquid | Pos/pos |
| O157:H7 | 15 | Male | Blood speckled, liquid | Pos/pos |
| O157:H7 | 22 | Male | Bloody, liquid | Pos/pos |
| O157:H7 | 51 | Male | Liquid | Pos/pos |
| O26:H11 | 3 | Female | Liquid | Pos/neg |
| O6:H2 | 20 | Female | Bloody, liquid | Pos/neg |
| O111:HNT | 21 | Female | Bloody, mucoid | Pos/pos |
| O5:HNM | 28 | Female | Liquid | Pos/neg |
| O111:HNM | 34 | Female | Bloody | Pos/pos |
| O103:H25a | 45 | Female | Mucoid | Pos/neg |
| O121:H19 | 54 | Female | Liquid | Neg/pos |
| O111:HNM | 57 | Female | Liquid | Pos/neg |
| O121:H19 | 17 | Male | Semiformed | Pos/neg |
| O121:H19 | 17 | Male | Liquid | Neg/pos |
| O5:HNM | 21 | Male | Liquid, bloody | Pos/neg |
| O145:HNM | 21 | Male | Bloody, liquid | Neg/pos |
| ONT:H25 | 34 | Male | Mucoid | Pos/neg |
Samples were not detected by EIA from enriched broth culture.
Pos, positive; neg, negative.
The different serotypes of non-O157 STEC detected in 2009 and 2010 are shown in Table 1. Of these isolates, 38%, 19%, and 43% were positive by real-time PCR for stx1, stx2, and a combination of stx1 and stx2, respectively. The results of the ImmunoCard STAT! assay were in concordance with the real-time PCR data.
The distribution of STEC in our patients is shown in Table 1. There were 13 female (3 to 66 years of age) and 8 male patients (15 to 55 years of age). The median age for the O157 STEC patient group was 24.5 years (average age, 30.6 ± 19.9 years). Although the female-to-male ratio was 1.7, the age variation between the gender groups did not differ significantly (P = 0.90). In comparison, the median age of the non-O157 STEC patient group was 21 years (average age, 28.6 ± 15.7 years), with a female-to-male ratio of 1.6 and a nonsignificant age variation between the gender groups (P = 0.17). There was also no significant age variation between the patients in both the O157:H7 and non-O157 groups (P = 0.81). Sixty-three percent of the patients (5/8) with O157 STEC, compared to 38% of the patients (5/13) with non-O157 STEC, had documented bloody diarrhea at the time of stool sample testing. Although the question of abdominal pain may not have been posed directly, this symptom appeared to be more common in patients with non-O157 infection than among those with O157 infection, as documented by the attending physicians, i.e., 38% (5/13) versus 25% (2/8), respectively. Given the small sample size in our study, the difference did not reach statistical significance (P = 0.54).
Clearly, non-O157 STEC infections are being underdiagnosed by the current conventional culture method. In our study, only 38.1% (8/21) of the STEC isolates (i.e., all O157) were detected by the current diagnostic approach. The Premier EHEC EIA can easily be implemented in routine diagnostic microbiology laboratories with minimal training. However, its sensitivity is lower than that of our real-time PCR; this might have accounted for the 2 missed STEC-positive samples. Moreover, although EIA could be used for direct testing of stool samples, the lower sensitivity of the assay may lead to more false-negative results. Furthermore, the ImmunoCard STAT! assay is required for the differentiation of Shiga toxins once they are identified by the EIA. In our study, all positive real-time PCR results correlated with both immunoassays when isolates were tested, as well as with the clinical presentation. All patients were symptomatic with diarrhea.
The 2009 CDC guideline on STEC testing also included the identification of non-O157 serogroups in all positive stool samples. The random picking of dry, pink colonies from MacConkey plates and retesting by EIA or PCR is labor-intensive. We have also noted that the BBL CHROMagar O157 from BD did not support the growth of all non-O157 STEC isolates. Therefore, these plates could be used only to screen for O157 STEC.
STEC O26, O103, O111, O121, and O145 seen in our study matched the top 7 serogroups in the United States according to the FoodNet report in 2009 (http://www.cdc.gov/foodnet/), as well as the top 4 serogroups in Switzerland (7). Other serogroups, such as O121:H19, O6:H2, O145:HNM, ONT:H25, and O5:HNM, were also identified.
We also noted that the majority of the STEC-positive samples were not bloody but either watery or mucoid, as described in an earlier publication (2). This was also observed in 3 of our patients who had E. coli O157 infection. If “bloody stool” was used as the sole criterion for STEC testing, 37.5% (3/8) O157 and 61.5% (8/13) non-O157 isolates or a combined total of 52.4% (11/21) STEC isolates would have been missed in our study. It is therefore crucial to include different types of stool samples for STEC testing.
In this study, all isolates producing Shiga toxins were culture positive, contrary to our previous published results (2). The viability of STEC seemed to drop over time when the samples were stored frozen at −20°C. A similar observation was reported when archival stool samples were used for a retrospective study (5).
In light of the CDC guideline and our present observations, alternate methods such as EIA or immunochromatographic assays for the detection of STEC (O157 and non-O157) might be easier and less costly to implement in routine microbiology laboratories than nucleic acid testing. Individual laboratories would need to evaluate the options available and perform a validation study that includes both cost and work flow analyses to find one that suits their practice. It is essential to note that false-negative results could occur when the bacterial load is low in certain stool samples. The most demanding part of STEC detection still remains the final identification of E. coli colonies, in which no differentiation can be made between toxin producers and toxin nonproducers. This will remain a challenge until an easier, faster, less tedious, and more specific method for the identification of STEC becomes available.
Acknowledgments
This project was funded by the University of Alberta Hospital Foundation. Lillian Lim was the recipient of an Alberta Innovates-Health Solutions summer student award and a Women's and Children's Health Research Institute summer student award. Daniel Fok was the recipient of a Northern Alberta Clinical Trials and Research award.
Special thanks to the microbiology department at DynaLIFEDx for the provision of stool specimens from the community and community hospitals for this study and Helen Tabor at the National Microbiology Laboratory for performing the serotyping.
Footnotes
Published ahead of print on 21 September 2011.
REFERENCES
- 1. Chui L., et al. 2010. Comparison of Shiga toxin-producing Escherichia coli detection methods using clinical stool samples. J. Mol. Diagn. 12:469–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Couturier M. R., Lee B., Zelyas N., Chui L. 2011. Shiga-toxigenic Escherichia coli detection in stool samples screened for viral gastroenteritis in Alberta, Canada. J. Clin. Microbiol. 49:574–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Elliott E. J., et al. 2001. Contributors to the Australian Paediatric Surveillance Unit. Nationwide study of haemolytic uraemic syndrome: clinical, microbiological, and epidemiological features. Arch. Dis. Child. 85:125–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. GoldWater P. N., Bettelheim K. A. 1994. The role of enterohaemorrhagic E. coli serotypes other than 0157:H7 as causes of disease, p. 57–60 In Karmali M. A., Goglio A. G. (ed.), Recent advances in verocytotoxin-producing Escherichia coli infections. Elsevier Science, Amsterdam, The Netherlands [Google Scholar]
- 5. Grys T. E., Sloan L. M., Rosenblatt J. E., Patel R. 2009. Rapid and sensitive detection of Shiga toxin-producing Escherichia coli from nonenriched stool specimens by real-time PCR in comparison to enzyme immunoassay and culture. J. Clin. Microbiol. 47:2008–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Holland J. L., Louie L., Simor A. E., Louie M. 2000. PCR detection of Escherichia coli O157:H7 directly from stools: evaluation of commercial extraction methods for purifying fecal DNA. J. Clin. Microbiol. 38:4108–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Käppeli U., Hachler H., Giezendanner N., Beutin L., Stephan R. 2011. Human infections with non-O157 Shiga toxin-producing Escherichia coli, Switzerland, 2000-2009. Emerg. Infect. Dis. 17:180–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Karmali M. A. 1989. Infection by verocytotoxin-producing Escherichia coli. Clin. Microbiol. Rev. 2:15–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Karmali M. A., et al. 1985. The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli. J. Infect. Dis. 151:775–782 [DOI] [PubMed] [Google Scholar]