Abstract
It has been proposed that plumbing systems might serve as a significant environmental reservoir of human-pathogenic isolates of Fusarium. We tested this hypothesis by performing the first extensive multilocus sequence typing (MLST) survey of plumbing drain-associated Fusarium isolates and comparing the diversity observed to the known diversity of clinical Fusarium isolates. We sampled 471 drains, mostly in bathroom sinks, from 131 buildings in the United States using a swabbing method. We found that 66% of sinks and 80% of buildings surveyed yielded at least one Fusarium culture. A total of 297 isolates of Fusarium collected were subjected to MLST to identify the phylogenetic species and sequence types (STs) of these isolates. Our survey revealed that the six most common STs in sinks were identical to the six most frequently associated with human infections. We speculate that the most prevalent STs, by virtue of their ability to form and grow in biofilms, are well adapted to plumbing systems. Six major Fusarium STs were frequently isolated from plumbing drains within a broad geographic area and were identical to STs frequently associated with human infections.
INTRODUCTION
Fungi that cause human mycoses usually exist as saprophytes in the environment. Fusarium is an example; its extreme metabolic diversity reflects its diverse ecology, since these fungi are abundant in soil, as saprophytes, mutualists, and parasites on plants and animals, in the indoors, and in aquatic habitats (12, 16, 28, 46). Like many fungi, Fusarium is responsible for an array of mostly opportunistic mycoses, including localized subcutaneous infections and life-threatening invasive mycoses in immunocompromised and immunosuppressed individuals (52). Unlike many human pathogenic fungi which enter through the respiratory tract, Fusarium typically tends to enter the body through wounds, via catheters and intravenous apparati in the hospital (56), through trauma (20), or through introduction of a biofilm to the eye (8). Fusarium also exhibits very high levels of resistance to the spectrum of antifungal drugs currently available, making these infections particularly difficult to treat (52).
Fusarium consists of over 200 species grouped into at least 10 phylogenetically defined species complexes (45). The 69 known species associated with infections of humans and other animals reflect a great deal of the ecological and phylogenetic diversity in the genus, in that they are distributed among six species complexes. Approximately 80% of all human infections are caused by members of the Fusarium solani and F. oxysporum species complexes (FSSC and FOSC, respectively) (45).
Mycotic keratitis appears to be caused disproportionately by Fusarium species (1). Previous work indicated that F. falciforme, a soil-associated phylogenetic species in the FSSC also known as FSSC 3+4, was the most common species associated with fungal keratitis, which is consistent with soil and plant debris particles being the main source of infection (58). However, in the 2005 to 2006 outbreaks of mycotic keratitis in Southeast Asia and North America associated with contact lens use, the most frequent fusaria associated with eye infections were two unnamed phylogenetic species, FSSC 1 and FSSC 2 (8). The main environmental sources known for these species were plumbing systems, suggesting that this was likely the primary environmental reservoir for eye infections. The results of the outbreak investigation established that fungal biofilms can form on contact lenses and lens cases and that 10 isolates from either corneas or patient environments shared the same multilocus sequence type (ST) (41). Collectively, these results suggest that patients inadvertently inoculated their cornea with contaminated lenses (8). This hypothesis is further supported by the fact that Fusarium has been recovered in many surveys of water systems, including surveys of municipal water system pipe sections (11) and from a hospital survey sampling plumbing fixtures (2). It has also been demonstrated experimentally that fusaria, particularly members of the FSSC and FOSC, can attach to and form biofilms on contact lenses and polyvinyl chloride pipe (14, 25, 32, 51).
Previous studies have shown that a majority of clinically relevant fusaria are represented by six STs representing four different species in three phylogenetically distinct species complexes: one from the FOSC (FOSC ST 33), four from two species in the FSSC (STs a and b in FSSC 1, and STs d and k in FSSC 2), and a single ST of F. dimerum (F. dimerum ST a), in the F. dimerum species complex (FDSC). All six STs have been recovered from hospital environments, including plumbing systems (2, 8, 38, 41, 43, 49, 58). This connection has led to the hypothesis that indoor plumbing-associated biofilms may act as hidden source of infection in hospitals and the community.
Although a number of studies have shown that putatively pathogenic fusaria inhabit a wide variety of manufactured water systems, the frequencies and genetic diversity of common pathogenic STs has not been investigated over a broad geographic range. In the present study we assembled a panel of 297 plumbing-associated fusaria collected on a broad geographic scale and used existing multilocus sequence typing (MLST) schemes to determine their identities at the species and ST levels. The distribution of STs collected from sinks was compared to the distribution of known human pathogenic STs reported in previous studies.
MATERIALS AND METHODS
Sampling from drains.
A total of 471 plumbing drains were sampled, more than 95% of which were bathroom sink drains. To capture as many different sampling points over as broad a geographic range as possible, drains were sampled from 131 buildings including businesses, homes, university dormitories, and public facilities in Pennsylvania, Virginia, Maryland, North Carolina, South Carolina, Georgia, Florida, and California. Buildings that were <10 miles apart were grouped into 57 “sites” for depiction in Fig. 7. To isolate Fusarium, sterile 15-cm cotton-tipped applicators were dipped into bathroom drains, usually through the openings in drain filters, and used to scour the interior surface of drainpipes for approximately 5 s. Cotton-tipped applicators, often with visible detritus, were then immediately streaked onto 5- or 8.5-cm petri plates of Nash-Snyder agar (Nash), a Fusarium semiselective medium that contains the fungicide pentachloronitrobenzene (33), and the plates were sealed using paraffin tape. Plates were examined for the presence of fungal colonies under a dissection microscope 10 to 15 days after collection, and putative fusaria were identified for subculturing. Growing hyphal tips of Fusarium-like colonies were transferred to carnation leaf agar (CLA) plates (28) for further identification. Rarely, two discrete colonies growing on the same Nash plate were treated as potentially distinct fusaria and independently subcultured from hyphal tips. Fusarium colonies mixed with bacteria and/or other fungi were transferred to another plate of Nash until it was possible to establish a pure culture from a growing hyphal tip or single spore. Because other fungi, including Penicillium, Aspergillus, Mucor, and unidentified dematiaceous hyphomycetes (darkly pigmented fungi producing mitotic spores), were rarely encountered, they were not included in the present study. For long-term storage, cultures growing on CLA were transferred to vials of sterile milk and lyophilized, vacuum sealed, given accession numbers (e.g., FRC S-2543), and stored in the culture collection of the Fusarium Research Center, University Park, PA, where they are available to all authorized users.
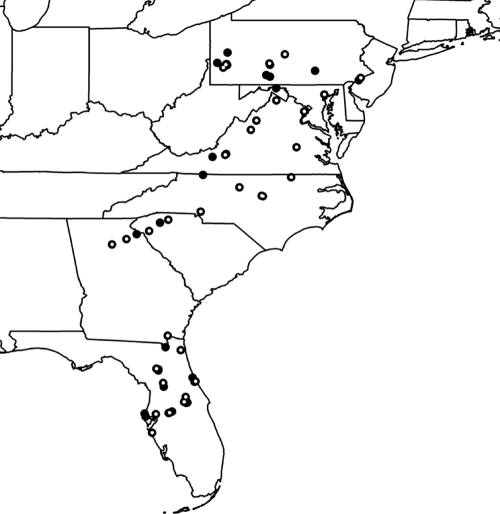
Fig. 7.
Generalized schematic map showing 57 collection areas (black dots) containing buildings yielding Fusarium. White points within black dots highlight sites that yielded the widespread clonal lineage FOSC ST 33. Buildings <10 miles apart were grouped into 57 “sites” for depiction using ArcMap 9.2 (Esri, Redlands, CA) and layers downloaded from the U.S. Census Bureau (http://www.census.gov/geo/www/cob/st2000.html).
DNA manipulation.
Total genomic DNA was isolated from mycelium using a modified DNeasy plant minikit protocol (Qiagen, Valencia, CA) and suspended in a 100-μl volume. Portions of six gene fragments were used for MLST based on published phylogenetic analyses, with different combinations of loci used for different species complexes as previously described: partial translation elongation factor 1α (TEF), the internal transcribed spacer region of the rRNA gene and domains D1 and D2 of the LSU (ribosomal DNA [rDNA]), the nuclear rDNA intergenic spacer region (IGS rDNA), two contiguous regions of the RNA polymerase II second largest subunit (RPB2), partial benA β-tubulin (β-TUB), and calmodulin (CAM) (Table 1). PCR was performed for TEF, rDNA, CAM, and RPB2 using GoTaq PCR kits (Promega, Madison, WI). Because of its longer amplicon length and secondary structure, PCR was performed for IGS using Platinum Taq kits (Invitrogen, Carlsbad, CA) as previously described (19, 44). PCR products were purified for DNA sequencing by incubating 5 μl of product with 0.33 μl of shrimp alkaline phosphatase (1 U/μl; USB, Cleveland, OH), 0.67 μl of exonuclease I (10 U/μl; Affymetrix, Santa Clara, CA), and 1 μl of shrimp alkaline phosphatase 10× reaction buffer (USB), prepared in a master mix for 30 min at 37°C, followed by 15 min at 80°C. Sanger sequencing was performed at the Penn State Genomics Core Facility, University Park, PA, on an ABI 3730 XL automated DNA sequencer (Applied Biosystems, Carlsbad, CA). Sequencher version 4.8 (Gene Codes, Ann Arbor, MI) was used to edit the raw sequence data, which was generated in both directions. Sequences were then aligned by using CLUSTAL W (http://www.align.genome.jp) and improved manually prior to the addition to existing MLST alignments from previous studies.
Table 1.
MLST markers used in this study for each species complex, together with primers used for PCR and DNA sequencing
| MLST locusa | Species complexb | Primer(s)c (reference) |
|---|---|---|
| TEF | FSSC, FOSC, FDSC, FIESC | EF1-EF2 (39) |
| rDNA | FSSC, FDSC, FIESC | ITS5 (57), NL4 (37) |
| β-TUB | FDSC | T1-T22 (37) |
| IGS | FOSC | CNS1, NL11, iNL11, NLa, CNSa, iCNS1 (38) |
| RPB2 | FSSC, FIESC | 5F2-7CR/7CF-11AR (30) |
| CAM | FIESC | CL1-CL2A (43) |
Characteristics of the various loci: TEF, 5′ end of the translation elongation factor 1α gene, including three introns; rDNA, portion of the nuclear rRNA gene operon that includes the ITS regions, 5.8S rRNA gene, and the D1-D2 region of the large (28S) rRNA gene; β-TUB, 5′ end of a beta-tubulin gene, including three introns; IGS, intergenic spacer region of the nuclear rRNA gene operon; RPB2, portion of the second largest RNA polymerase B-subunit gene; CAM, portion of the calmodulin gene.
FDSC, Fusarium dimerum species complex; FIESC, Fusarium incarnatum-F. equiseti species complex; FOSC, Fusarium oxysporum species complex; FSSC, Fusarium solani species complex.
All primers were used for both PCR and sequencing reactions, with the exception of those for used locus IGS, for which CNS1 and NL11 were used exclusively for the PCR.
MLST.
A BLAST search using TEF sequences against the FUSARIUM-ID (19) and GenBank databases (http://www.ncbi.nlm.nih.gov/GenBank/index.html) was used as an initial step to identify isolates to species and/or species complex. The results of these searches revealed that all isolates belonged to one of four species complexes: the FOSC, FSSC, FDSC, and the F. incarnatum-equiseti species complex (FIESC). Additional sequences were then generated for each isolate, using species complex-specific MLST schemes (Table 1). Once edited, sequences were manually added to existing sequence alignments for the respective species complex. Missing or ambiguous DNA sequence characters were assigned Ns in the alignments, and these characters were treated as missing data in the phylogenetic software PAUP* (Phylogenetic Analysis Using Parsimony and other Methods) v.4.0.b9 (53). To assign isolates to known STs, a parsimony tree was first generated for isolates from each species complex using PAUPRat (34, 50) implemented in PAUP* v.4.0.b9 with the following settings: set seed = 0, nreps = 200, pct = 15, set wtmode = uniform, set terse, with simple sequence addition and heuristic searches. Isolates were assigned to existing STs when they shared a common terminal node in the parsimony tree with a previously known ST, and their exact identity was confirmed by visual scrutiny of sequence alignments and chromatograms. Isolates were assigned to new STs when they formed a unique branch in the parsimony tree, confirmed by scrutiny of the sequence quality and alignments. Both indels and nucleotide substitutions were taken into account when assigning STs. (Note that, for the FIESC and FSSC, ST designations consist of a number indicating the phylogenetic species, followed by a letter or series of letters indicating the unique ST within the species. For the FOSC, only a number is provided to indicate the ST, since phylogenetic species boundaries have not been elucidated within this complex. ST nomenclature for the FDSC has not been developed.) To compare the diversity of drain fusaria with the diversity of clinical fusaria, the frequencies of human pathogenic STs belonging to different Fusarium species complexes were compiled from published phylogenetic analyses (8, 41, 42, 44, 49, 58) using MLST data.
RESULTS
Frequency and phylogenetic distribution of fusaria.
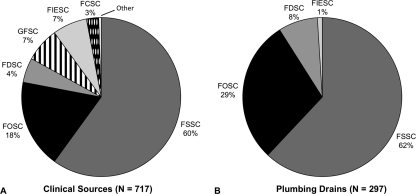
A total of 312 (66%) of 471 sinks sampled and 107 (82%) of 131 buildings in Pennsylvania, Virginia, Maryland, North Carolina, South Carolina, Georgia, Florida, and California yielded at least one Fusarium isolate, determined by the production of diagnostic conidia on Nash agar. Of these, 297 were successfully obtained as pure cultures and subjected to full MLST analysis, yielding 59 unique STs (multilocus DNA sequence data are available at GenBank). A total of 99% of STs were nested within one of three species complexes: 185 from the FSSC (62%), 85 from the FOSC (28%), and 25 from the FDSC (8.5%) (Fig. 1). Among clinical isolates, a review of STs from previous analyses (n = 717) indicated that ca. 60% came from the FSSC, 18% came from the FOSC, 5% came from the FDSC, 7% came from the Gibberella fujikuroi species complex (GFSC), 7% came from the FIESC, 3% came from the F. chlamydosporum species complex (FCSC), and <1% came from other species complexes (Fig. 1). In the present study, only two isolates were from the FIESC, and no members of the GFSC or FCSC were obtained from sink drains.
Fig. 1.
Frequencies of Fusarium species complexes characterized by MLST from clinical sources based on previous studies (2, 8, 38, 41, 44, 49, 58) (A) and from drains based on the present study (B). FDSC, Fusarium dimerum species complex; FIESC, Fusarium incarnatum-equiseti species complex; FOSC, Fusarium oxysporum species complex; FSSC, Fusarium solani species complex. GFSC, Gibberella fujikuroi species complex; FCSC, Fusarium chlamydosporum species complex.
MLST diversity of sink fusaria.
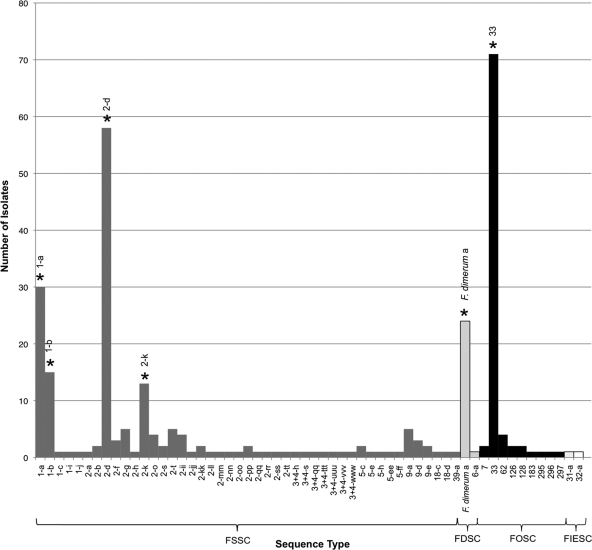
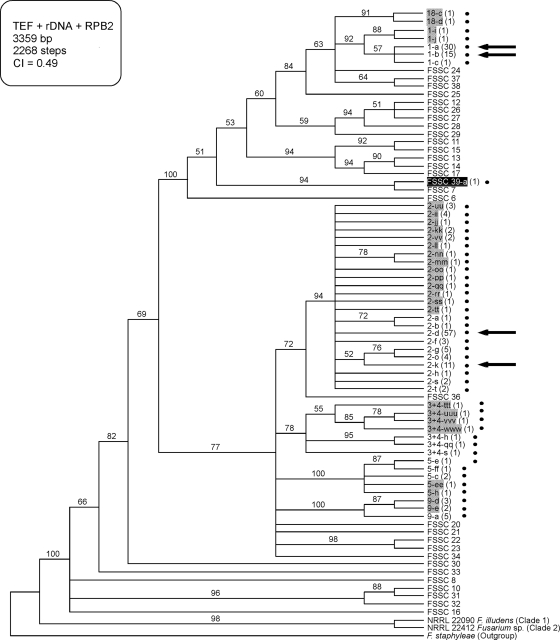
A total of 70% (209/297) of sink isolates belonged to six STs from four species in three species complexes. Nucleotide sequence data has been deposited in GenBank with accession numbers (JN235143 to JN235946). From most to least common, the STs were FOSC ST 33 (n = 71), FSSC 2-d (n = 58), FSSC 1-a (n = 30), F. dimerum ST a (n = 24), FSSC 1-b (n = 15), and FSSC 2-k (n = 11) (Fig. 2).
Fig. 2.
Frequencies of 59 STs which represent 297 fusaria isolated from drains in the present study. The six most common clinical STs are highlighted with asterisks: FSSC 1-a, FSSC 1-b, FSSC 2-d, FSSC 2-k, FOSC ST 33, and Fusarium dimerum a.
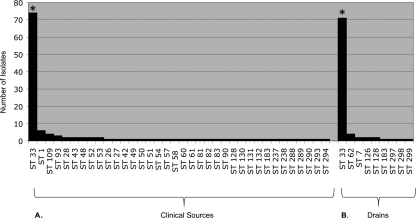
Based on our census of isolates from clinical sources in published MLST studies, these same six STs were the six most common in human infections (FOSC ST 33 = 74, FSSC 2-d = 44, FSSC 1-b = 29, F. dimerum a = 17, FSSC 1-a = 15, and FSSC 2-k = 7). FOSC ST 33 was the most common ST isolated from drain environments and clinical sources (Fig. 3).
Fig. 3.
Frequencies of FOSC STs isolated from clinical sources from previous studies (A) and drains in the present study (B). Asterisks highlight FOSC ST 33, the widespread clonal lineage known from human infections.
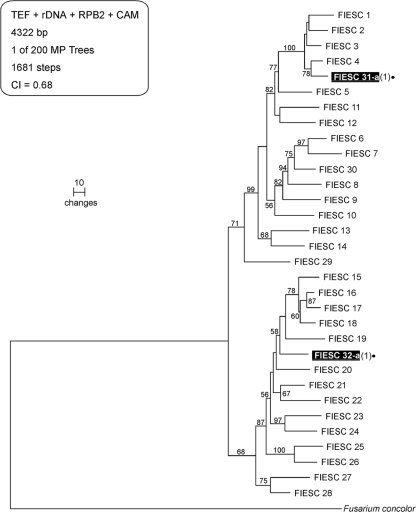
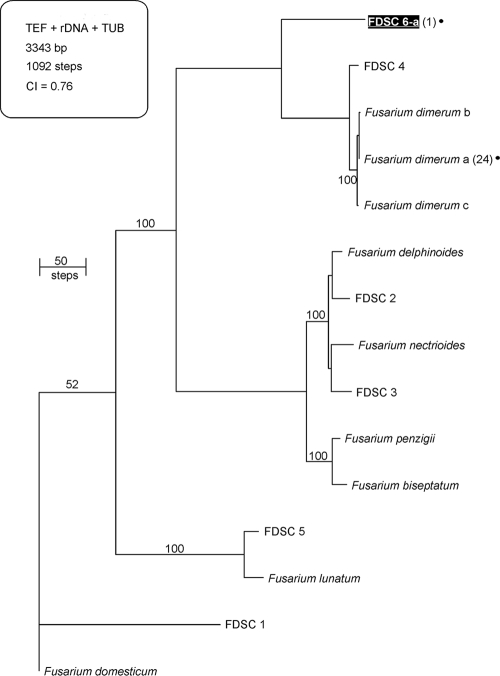
Of the 59 STs identified from sinks, 32 had not been observed in previous MLST studies of Fusarium (Fig. 4 to 6). These novel STs included members of four putatively new Fusarium species based on their clear sequence divergence, two in the FIESC (FIESC 29 and 30; Fig. 6), one in the FSSC (FSSC 39; Fig. 4), and one in the FDSC (FDSC 6; Fig. 5). In addition, putative interspecific hybrids were detected representing two novel STs from phylogenetic species FSSC 2, which possessed ribosomal DNA (rDNA) alleles that were an exact match for those from a different phylogenetic species, FSSC 9 (data not shown). The remaining 27 STs were known from previous MLST studies of Fusarium that mostly targeted clinical isolates. All 27 of these STs have been previously associated with at least one human infection.
Fig. 4.
Cladogram of the FSSC based on TEF, rDNA, and RPB2 highlighting the spectrum of FSSC diversity found in drains. The tree is rooted at F. staphyleae (NRRL 22316). All drain isolates (indicated by black dots) are members of FSSC clade 3 (36). Gray and black shading identify, respectively, novel FSSC STs and species discovered in the present study. Black arrows indicate the four most common FSSC STs recovered from sinks. Numbers in parentheses indicate the number of isolates recovered from drains in the present study. Numbers above nodes represent MP bootstrap support based on 1,000 replicates. MP, most-parsimonious; CI, consistency index.
Fig. 6.
Phylogram of the FIESC based on TEF, rDNA, RPB2, and CAM highlighting the spectrum of diversity found in drains. The tree is rooted at F. concolor (NRRL 13459). Black dots and shading identify two putatively novel FIESC species and STs, designated FIESC 31-a and FIESC 32-a, which were discovered in the present study. Numbers above nodes represent MP bootstrap support based on 1,000 replicates. MP, most-parsimonious; CI, consistency index.
Fig. 5.
Phylogram of the FDSC based on TEF, rDNA, and TUB highlighting the spectrum of FDSC diversity found in drains. The tree is rooted at F. domesticum (NRRL 29976). A black dot is used to identify two phylogenetically distinct STs within the FDSC isolated from sinks; black shading indicates the putatively novel species FDSC 6. Numbers of sink isolates from the present study are indicated in parentheses. Numbers above nodes represent MP bootstrap support based on 1,000 replicates. MP, most-parsimonious; CI, consistency index.
Geographic distribution.
Of the 107 buildings that yielded a Fusarium culture, 86 (79%) yielded one of the six common STs known from human infections. FOSC ST 33 was the most common ST recovered; it was found 71 times in 42 buildings (see Table S1 in the supplemental material) and has been previously described as a widespread clonal lineage associated with human infections (44). Moreover, FOSC ST 33 was found in every state sampled (Fig. 7 [the data for California are not shown]); it has been reported from human infections in five countries on two continents, as well as from a wide variety of environmental sources such as hospital water systems (see Table S2 in the supplemental material). FSSC 2-d, the next most commonly isolated ST, was found 58 times in 27 different buildings, 20 of which were in Florida. We found that 21/23 (91%) of residences and 86/108 (80%) of public buildings yielded Fusarium. Multiple STs were found in two of the eight sinks that were swabbed twice: one sink yielded STs FSSC 2-d and FOSC ST 128, and another yielded FSSC 2-d and FSSC 2-ii.
DISCUSSION
Widespread occurrence of pathogenic fusaria.
With 66% of sinks and 82% of buildings found to harbor Fusarium, it is clear that their inhabitants are exposed to these fungi on a regular basis. Because our swabbing method was far from exhaustive, these frequencies should be considered a minimum estimate. The fact that the six most common STs found in drains, representing 70% of the fusaria recovered, are the same six most common STs responsible for human infections strongly supports the hypothesis that plumbing surface biofilms serve as reservoirs for human pathogenic fusaria (2, 3, 8, 32). However, while we hypothesize that STs of opportunistic fusaria are very common in our indoor environment, we are quick to point out that Fusarium infections are relatively rare, even among the severely immunocompromised and immunosuppressed (31, 35, 48). Victims of the 2005–2006 outbreak of Fusarium keratitis did not show any tendency toward immune deficiency, but the frequency of infections was also very low, with approximately 255 confirmed cases worldwide among the many millions of users of the contact lens solution associated with the infections (13, 15, 41). It is noteworthy that five of the six most common STs we isolated from sinks represented 54% of the 67 isolates analyzed from infected corneas (i.e., FSSC 1-a, n = 13; FSSC 1-b, n = 3; FSSC 2-d, n = 9; FOSC 33, n = 10; and F. dimerum a, n = 24). This striking concordance supports the hypothesis that the water systems in the patients' communities and/or homes served as the environmental reservoir in the keratitis outbreaks (8).
Diversity of sink fusaria.
The six most common STs reflect a great deal of phylogenetic diversity within the genus, covering a minimum of four phylogenetic species in three divergent species complexes. In addition, based on their phylogenetic divergence at multiple loci, four isolates appear to represent putative novel phylogenetic species in three different species complexes, designated FIESC 29, FIESC 30, FSSC 39, and FDSC 6. Because evolutionarily defined fungal species are often morphologically cryptic (54), it is not surprising that putatively novel species were discovered in this survey, particularly in the species-rich FIESC, which has not been studied exhaustively (42). In addition to the four putatively new species, 28 novel STs were found within known phylogenetic species in the FSSC and the FDSC (Fig. 4 and 5), as well as within the FOSC (Fig. 3) where phylogenetic species delimitation has been confounded by clone-like reproduction and genealogical discordance (38). The 32 novel STs were observed most frequently as single isolates, with only 6 of them occurring more than once (Fig. 2). Of the 27 previously known STs, all had been associated with at least one human infection. Previous MLST surveys of clinical isolates indicate that such “singleton” STs are commonly associated with human infections (43–45, 58).
Connections to clinical fusaria.
Fusarium has been shown to comprise at least 10 phylogenetic groups referred to as species complexes, 7 of which are known to harbor human pathogens (45). Four of the seven Fusarium species complexes known to harbor human pathogens were isolated from drains in our study, with the GFSC, F. sambucinum species complex (FSamSC) and FCSC not observed and the FIESC only isolated twice. Although the FIESC, GFSC, and FCSC are less commonly associated with human infections, together they represent ∼17% of the fusaria known from human infections; however, they represented <1% of the sink isolates recovered in our survey. In contrast, more than 99% of the fusaria isolated from drains in the present study were members of the FOSC, FSSC, and FDSC, which were associated with ∼85% of human clinical isolates genotyped previously (Fig. 1). The six dominant STs represented a great deal of diversity, four coming from two species of the FSSC (FSSC 1-a, FSSC 1-b, FSSC 2-d, and FSSC 2-k), one from a species in the FDSC (F. dimerum a), and one from the FOSC (FOSC ST 33) (Fig. 2). These same species and STs are known to be the most common in Fusarium infections across different continents (see Table S2 in the supplemental material). The application of molecular markers with higher resolution at the population level, as has been previously applied to FOSC ST 33 (44), may reveal clonal and/or recombinant patterns of propagation within these common STs. We hypothesize that these cosmopolitan STs may thrive in manufactured water systems and may have been distributed around the world anthropogenically.
Biofilm adaptation and its role in infection.
Biofilms on plumbing surfaces are known to be communities comprising a diverse spectrum of fungi and other microbes (11). The widespread distribution of certain Fusarium species and STs in drains suggests that they are particularly well adapted to this environmental niche and/or highly fecund. Even though it is a semiselective medium containing the fungicide pentachloronitrobenzene (PCNB), we observed some other fungi and bacteria in the drain samples streaked onto Nash agar, including unidentified dematiaceous hyphomycetes (darkly pigmented, putatively asexual fungi), Aspergillus (which is known to occur in some hospital water systems) (4), and Penicillium. However, based on its very high frequency, it is clear that Fusarium is a ubiquitous component of biofilm microbial communities in plumbing systems.
The adaptations that make Fusarium biofilm growth possible may also facilitate infection of humans. For example, in the 2005–2006 mycotic keratitis outbreak, it was hypothesized that noncompliant use of a contact lens solution led to reduced efficacy of its antimicrobial properties, which allowed fusaria to establish biofilms on contact lens surfaces and in lens cases (8, 29). Likewise, Fusarium can form biofilms on surfaces of indwelling catheters (56). The biofilm may also play an important role in established infections in the human host by protecting the fungus from antifungal drug treatments, as biofilm-phase fusaria tend to be more resistant to antifungal drugs than those growing planktonically (14).
Fusarium taxa not sampled.
Additional studies of biofilms are needed to assess the extent to which sampling biases may have affected the observed frequencies of taxa due to the use of a semiselective medium. Although it is difficult to draw definitive conclusions about species that may have been expected based on published surveys of water systems (10, 17), it is important to note that the majority of the fusaria previously reported from water are known to grow competitively on the semiselective medium we used. Although members of the GFSC (reported as F. moniliforme and F. dlaminii), FIESC (reported as F. pallidoroseum and F. equiseti), and FSamSC (reported as F. graminearum, F. sporotrichoides, and F. culmorum) have been reported from water systems, these need to be verified molecularly because most of these species cannot be identified reliably using phenotypic data. Thus, our working hypothesis is that a large portion of the species previously reported from water systems were probably misidentified because the majority of these morpho-species harbor multiple phylogenetically distinct species (37, 40, 43, 45).
The near absence of representatives of the GFSC, FIESC, FCSC, and FSamSC in sinks may be explained in part by the way these fungi produce asexual spores. Members of the these species complexes typically produce dry, small asexual spores (microconidia), whereas members of the FSSC, FOSC, and FDSC produce microconidia in a wet or adhesive material, either in small masses (false heads) that adhere to the spore-forming cells (phialides) or in larger masses (pionnotes). Although biofilms in plumbing systems may prove to represent the most important environmental reservoir of human pathogenic fusaria, other sources, including dry indoor surfaces, soil, and plant material may be significant sources for infections caused by taxa within species complexes that do not form biofilms.
Biofilms may play a role in dispersal and metabolic diversity.
Adaptation and spread through indoor environments is likely enhanced by water dispersal, as a number of mycological surveys have recovered Fusarium in activated sludge (7), from tap water (26, 27), and specifically from tap water in hospitals (5, 6, 23, 24, 47, 55). In addition to their wide distribution in plumbing systems and in human infections, the common biofilm-forming Fusarium species are known to colonize other unusual manufactured substrates, as contaminants of soap, paint, ointments, pasteurized beverages in manufacturing plants, lubrication oils in heavy machinery, oily water, and coolant fluids (20, 45, 49, 58). Biofilms also may facilitate interactions between Fusarium and other metabolically diverse, plumbing-associated microbes (21). Biofilm-associated fusaria and their coresident microbes may function as polyextremophiles in plumbing systems by virtue of novel metabolic capabilities that allow them to exploit a variety of challenging environmental niches, including the ability degrade keratin (9, 18) and other refractory compounds (22). Metagenomic approaches for dissecting the biofilm microbiome are needed to fully characterize the microbial communities and to determine what impact they have on the diversity of fusaria and vice versa.
Supplementary Material
ACKNOWLEDGMENTS
We thank Stacy Sink for excellent technical assistance and Gregory F. Short and Nicolas A. Warren for their assistance with the surveys of Fusarium in plumbing systems.
This study was supported in part by financial support from Bausch & Lomb, Inc., as well as by a special Graduate Research grant provided by the Pennsylvania State College of Agricultural Sciences awarded to D.P.G.S. D.P.G.S. is supported by a USDA-AFRI grant in Microbial Genomics and a USDA National Needs Fellowship.
The mention of firm names or trade products does not imply that they are endorsed or recommended by the U.S. Department of Agriculture over other firms or similar products not mentioned.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 5 October 2011.
REFERENCES
- 1. Alfonso E. C., Rosa R. H. 1997. Fungal keratitis, p. 1253–1266. In Krachmer J. H., Mannis M. J., Holland E. J. (ed.), Cornea and external diseases: clinical diagnosis and management. Mosby, St. Louis, MO [Google Scholar]
- 2. Anaissie E. J., et al. 2001. Fusariosis associated with pathogenic Fusarium species colonization of a hospital water system: a new paradigm for the epidemiology of opportunistic mold infections. Clin. Infect. Dis. 33:1871–1878 [DOI] [PubMed] [Google Scholar]
- 3. Anaissie E. J., Penzak S. R., Dignani M. C. 2002. The hospital water supply as a source of nosocomial infections: a plea for action. Arch. Intern. Med. 162:1483–1492 [DOI] [PubMed] [Google Scholar]
- 4. Anaissie E. J., et al. 2002. Pathogenic Aspergillus species recovered from a hospital water system: a 3-year prospective study. Clin. Infect. Dis. 34:780–789 [DOI] [PubMed] [Google Scholar]
- 5. Arvanitidou M., Kanellou K., Constantinides T. C., Katsouyannopoulos V. 1999. The occurrence of fungi in hospital and community potable waters. Lett. Appl. Microbiol. 29:81–84 [DOI] [PubMed] [Google Scholar]
- 6. Arvanitidou M., et al. 2000. High level of recovery of fungi from water and dialysate in hemodialysis units. J. Hosp. Infect. 45:225–230 [DOI] [PubMed] [Google Scholar]
- 7. Awad M. F., Kraume M. 2011. Mycological survey of activated sludge in MBRs. Mycoses doi:10.1111/j.1439-0507.2010.01959 [DOI] [PubMed] [Google Scholar]
- 8. Chang D. C., et al. 2006. Multistate outbreak of Fusarium keratitis associated with use of a contact lens solution. JAMA 296:953–963 [DOI] [PubMed] [Google Scholar]
- 9. Colombo J. C., Cabello M., Arambarri A. M. 1996. Biodegradation of aliphatic and aromatic hydrocarbons by natural soil microflora and pure cultures of imperfect and lignolitic fungi. Environ. Pollut. 94:355–362 [DOI] [PubMed] [Google Scholar]
- 10. Cooke W. B., Kabler P. W. 1957. Plant disease fungi in sewage polluted water. Public Health Rep. 72:651–654 [PMC free article] [PubMed] [Google Scholar]
- 11. Doggett M. S. 2000. Characterization of fungal biofilms within a municipal water distribution system. Appl. Environ. Microbiol. 66:1249–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Domsch K. H., Gams W., Anderson T. H. 2007. Compendium of soil fungi, 2nd taxonomically revised edition by W. Gams. IHW-Verlag, Eching, Germany [Google Scholar]
- 13. Donnio A., et al. 2007. Outbreak of keratomycosis attributable to Fusarium solani in the French West Indies. Am. J. Ophthalmol. 143:356–358 [DOI] [PubMed] [Google Scholar]
- 14. Dyavaiah M., et al. 2007. Molecular characterization, biofilm analysis and experimental biofouling study of Fusarium isolates from recent cases of fungal keratitis in New York State. BMC Ophthalmol. 7:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Epstein A. B. 2007. In the aftermath of the Fusarium keratitis outbreak: what have we learned? Clin. Ophthalmol. 1:355–366 [PMC free article] [PubMed] [Google Scholar]
- 16. Farr D. F., Bills G. F., Chamuris G. P., Rossman A. Y. 1989. Fungi on plants and plant products in the United States. American Phytopathological Society, St. Paul, MN [Google Scholar]
- 17. Feldman A. E. 1957. Biota associated with sewage filtration. Sewage Indust. Wast. 29:538–540 [Google Scholar]
- 18. Friedrich J., Gradiöar H., Mandin D., Chaumont J. P. 1999. Screening fungi for synthesis of keratinolytic enzymes. Lett. Appl. Microbiol. 28:127–130 [Google Scholar]
- 19. Geiser D. M., et al. 2004. FUSARIUM-ID v.1.0: a DNA sequence database for identifying Fusarium. Eur. J. Plant Pathol. 110:473–479 [Google Scholar]
- 20. Gopinathan U., et al. 2002. The epidemiological features and laboratory results of fungal keratitis: a 10-year review at a referral eye care center in South India. Cornea 21:555–559 [DOI] [PubMed] [Google Scholar]
- 21. Gostincar C., Grube M., Gunde-Cimerman N. 2011. Evolution of fungal pathogens in domestic environments? Fungal Biol. doi:10.1016/j.funbio.2011.03.004 [DOI] [PubMed] [Google Scholar]
- 22. Hamada N., Abe N. 2009. Physiological characteristics of 13 common fungal species in bathrooms. Mycoscience 50:421–429 [Google Scholar]
- 23. Hayette M. P., et al. 2010. Filamentous fungi recovered from the water distribution system of a Belgian university hospital. Med. Mycol. 48:969–974 [DOI] [PubMed] [Google Scholar]
- 24. Hedayati M. T., Mayahi S., Movahedi M., Shokohi T. 2011. Study on fungal flora of tap water as a potential reservoir of fungi in hospitals from Sari city, Iran. J. Med. Mycol. 21:10–14 [DOI] [PubMed] [Google Scholar]
- 25. Imamura Y., et al. 2008. Fusarium and Candida albicans biofilms on soft contact lenses: model development, influence of lens type, and susceptibility to lens care solutions. Antimicrob. Agents Chemother. 57:171–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kanzler D., et al. 2008. Occurrence and hygienic relevance of fungi in drinking water. Mycoses 51:165–169 [DOI] [PubMed] [Google Scholar]
- 27. Kinsey G., Paterson R., Kelley J. 2003. Filamentous fungi in water systems, p. 77–98. In Horan N. J. (ed.), Handbook of water and wastewater microbiology. Academic Press, London, England [Google Scholar]
- 28. Leslie J. F., Summerell B. A., Bullock S. 2006. The Fusarium laboratory manual. Blackwell Publishing, Ames, IA [Google Scholar]
- 29. Levy B., Heiler D., Norton S. 2006. Report on testing from an investigation of Fusarium keratitis in contact lens wearers. Eye Contact Lens 32:1–7 [DOI] [PubMed] [Google Scholar]
- 30. Liu Y. J., Whelen S., Hall B. D. 1999. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 16:1799–1808 [DOI] [PubMed] [Google Scholar]
- 31. Martino P., Gastaldi R., Raccah R., Girmenia C. 1994. Clinical patterns of Fusarium infections in immunocompromised patients. J. Infect. 28:7–15 [DOI] [PubMed] [Google Scholar]
- 32. Mehl H. L., Epstein L. 2008. Sewage and community shower drains are environmental reservoirs of Fusarium solani species complex group 1, a human and plant pathogen. Environ. Microbiol. 10:219–227 [DOI] [PubMed] [Google Scholar]
- 33. Nash S. M., Snyder W. C. 1962. Quantitative estimations by plate counts of propagules of the bean root rot Fusarium in field soils. Phytopathology 52:567–572 [Google Scholar]
- 34. Nixon K. C. 1999. The parsimony ratchet, a new method for rapid parsimony analysis. Cladistics 15:407–414 [DOI] [PubMed] [Google Scholar]
- 35. Nucci M., Anaissie E. 2007. Fusarium infections in immunocompromised patients. Clin. Microbiol. Rev. 20:695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. O'Donnell K. 2000. Molecular phylogeny of the Nectria haematococca-Fusarium solani species complex. Mycologia 92:919–938 [Google Scholar]
- 37. O'Donnell K., Cigelnik E., Nirenberg H. I. 1998. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia 90:465–493 [Google Scholar]
- 38. O'Donnell K., et al. 2009. A two-locus DNA sequence database for typing plant and human pathogens within the Fusarium oxysporum species complex. Fungal Genet. Biol. 46:936–948 [DOI] [PubMed] [Google Scholar]
- 39. O'Donnell K., Kistler H. C., Cigelnik E., Ploetz R. C. 1998. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. U. S. A. 95:2044–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. O'Donnell K., Kistler H. C., Tacke B. K., Casper H. H. 2000. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc. Natl. Acad. Sci. U. S. A. 97:7905–7910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. O'Donnell K., et al. 2007. Phylogenetic diversity and microsphere array-based genotyping of human pathogenic fusaria, including isolates from the multistate contact lens-associated U.S. keratitis outbreaks of 2005 and 2006. J. Clin. Microbiol. 45:2235–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. O'Donnell K., et al. 2008. Molecular phylogenetic diversity, multilocus haplotype nomenclature, and in vitro antifungal resistance within the Fusarium solani species complex. J. Clin. Microbiol. 46:2477–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. O'Donnell K., et al. 2009. Novel multilocus sequence typing scheme reveals high genetic diversity of human pathogenic members of the Fusarium incarnatum-F. equiseti and F. chlamydosporum species complexes within the United States. J. Clin. Microbiol. 47:3851–3861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. O'Donnell K., et al. 2004. Genetic diversity of human pathogenic members of the Fusarium oxysporum complex inferred from multilocus DNA sequence data and amplified fragment length polymorphism analyses: evidence for the recent dispersion of a geographically widespread clonal lineage and nosocomial origin. J. Clin. Microbiol. 42:5109–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. O'Donnell K., et al. 2010. Internet-accessible DNA sequence database for identifying fusaria from human and animal infections. J. Clin. Microbiol. 48:3708–3718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Palmero D., et al. 2009. Species of Fusarium isolated from river and sea water of southeastern Spain and pathogenicity on four plant species. Plant Dis. 93:377–385 [DOI] [PubMed] [Google Scholar]
- 47. Panagopoulou P., et al. 2002. Environmental surveillance of filamentous fungi in three tertiary care hospitals in Greece. J. Hosp. Infect. 52:185–191 [DOI] [PubMed] [Google Scholar]
- 48. Raad I., et al. 2002. Epidemiology, molecular mycology, and environmental sources of Fusarium infection in patients with cancer. Infect. Control. Hosp. Epidemiol. 23:532–537 [DOI] [PubMed] [Google Scholar]
- 49. Schroers H. J., et al. 2009. Taxonomy and phylogeny of the Fusarium dimerum species group. Mycologia 101:44–70 [DOI] [PubMed] [Google Scholar]
- 50. Sikes D. S., Lewis P. O. 2001. PAUPRat:PAUP* implementation of the parsimony ratchet, beta software, version 1. Department of Ecology and Evolutionary Biology, University of Connecticut, Storrs, CT [Google Scholar]
- 51. Simmons R. B., Buffington J. R., Ward M., Wilson L. A., Ahearn D. G. 1986. Morphology and ultrastructure of fungi in extended-wear soft contact lenses. J. Clin. Microbiol. 24:21–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sutton D. A., Brandt M. E. 2011. Fusarium and other opportunistic hyaline fungi, p. 1853–1879. In Versalovic J., Carroll K., Funke G., Jorgensen J. H., Landry M. L. (ed.), Manual of clinical microbiology, 10th ed ASM Press, Washington, DC [Google Scholar]
- 53. Swofford D. L. 2002. PAUP* Phylogenetic analysis using parsimony (*and other methods), version 4.10 b. Sinauer Associates, Sunderland, MA [Google Scholar]
- 54. Taylor J. W., et al. 2000. Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 31:21–32 [DOI] [PubMed] [Google Scholar]
- 55. Warris A., et al. 2001. Recovery of filamentous fungi from water in a pediatric bone marrow transplantation unit. J. Hosp. Infect. 47:143–148 [DOI] [PubMed] [Google Scholar]
- 56. Wey S. B., Colombo A. 1997. Fungal infections of catheters, p. 139–156. In Seifert H., Jansen B., Farr B. (ed.), Catheter-related infections. Marcel Decker, New York, NY [Google Scholar]
- 57. White T. J., Bruns T., Lee S., Taylor J. 1990. Amplification and direct sequencing of fungal rRNA genes for phylogenetics, p. 315–322. In Innis M., Gelfand D., Sninsky J., White T. (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., San Diego, CA [Google Scholar]
- 58. Zhang N., et al. 2006. Members of the Fusarium solani species complex that cause infections in both humans and plants are common in the environment. J. Clin. Microbiol. 44:2186–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.