Abstract
The prevalence of heterogeneous intermediate-level resistance to vancomycin (hVISA) in Staphylococcus aureus was assessed by screening a large collection of recent isolates. Susceptibility testing by the Clinical and Laboratory Standards Institute broth microdilution method and the Etest GRD (glycopeptide resistance detection) method (bioMérieux) was performed on 4,210 clinically significant S. aureus isolates obtained in 2009 from 43 U.S. centers. Isolates with Etest GRD-positive results for hVISA were evaluated further by repeat GRD testing and population analysis profiling–area under the curve (PAP-AUC) analysis. No VISA (vancomycin MIC, 4 to 8 μg/ml) or vancomycin-resistant (MIC ≥ 16 μg/ml) strains were detected. The Etest GRD screen for hVISA was initially positive for 68 isolates (1.6%; all by teicoplanin MIC ≥ 8 μg/ml at 24 or 48 h). Among those 68 isolates, 45 were reproducibly GRD positive. PAP-AUC testing confirmed only 11 isolates as hVISA (all had reproducible GRD-positive results). The 11 hVISA isolates were from nine medical centers and appeared genetically diverse (ten different PFGE types). The rates of resistance (including intermediate) for hVISA were as follows: oxacillin, 82%; erythromycin, 82%; clindamycin, 73%; levofloxacin, 73%; trimethoprim-sulfamethoxazole, 9%; and daptomycin, 9%. All hVISA isolates were susceptible to linezolid, tigecycline, and ceftaroline. Our data suggest that the overall prevalence of hVISA in the United States is low (0.3%). The hVISA isolates represented 10.5% of isolates with vancomycin MICs of 2 μg/ml and 0.1% of isolates with vancomycin MICs of 1 μg/ml. The positive predictive value of GRD Etest for hVISA was 16.2% for initial screen positive and 24.4% for reproducibly positive results.
INTRODUCTION
Vancomycin has been the primary agent used to treat serious methicillin-resistant Staphylococcus aureus (MRSA) infections for many years, but the emergence of strains with reduced susceptibility to vancomycin has led to questions regarding its efficacy (12, 16). The first strain of S. aureus with heterogeneous intermediate resistance to vancomycin (hVISA) was reported in 1997 in a Japanese patient with MRSA pneumonia who did not respond to vancomycin therapy (7). Because of the difficulty in detection, the prevalence and clinical impact of hVISA have not been adequately studied (9). Retrospective reports have associated hVISA with treatment failure (2, 19), but a prospective study found no significant difference in cure rates for patients infected with hVISA compared to MRSA fully susceptible to vancomycin (8).
Strains defined as hVISA harbor a subpopulation of cells that are vancomycin intermediate yet appear susceptible by the routine Clinical and Laboratory Standards Institute (CLSI) broth microdilution method (6). The gold standard for detection of hVISA, population analysis profiling with area under the curve analysis (PAP-AUC), is too labor-intensive for routine use by clinical laboratories (27). The Etest GRD (glycopeptide resistance detection, for research use only) is a screening method for hVISA with reported sensitivity of 93 to 94% and specificity of 82 to 95% (13, 28). The objective of the present study was to assess the prevalence of hVISA in the United States by screening a large collection of recent clinical isolates with the Etest GRD. The positive predictive value (PPV) of the Etest GRD for hVISA was determined by testing GRD-positive isolates with the PAP-AUC method.
(Presented in part at the 48th Infectious Diseases Society of America Annual Meeting, Vancouver, British Columbia, Canada, 23 October 2010 [abstr. 1013].)
MATERIALS AND METHODS
Bacterial isolates.
A collection of 4,210 clinically significant S. aureus isolates obtained from 43 U.S. medical centers from June to December 2009 were screened for hVISA. Each center was asked to provide 100 consecutive S. aureus isolates (minimum of 25% from blood cultures) that did not represent colonization. Susceptibility by the CLSI broth microdilution method (3, 4) and genotyping by pulsed-field gel electrophoresis (PFGE) with comparison to U.S. type strains (17, 21) had been previously performed on all isolates. The mecA PCR-positive isolates (53.4% of collection) had been analyzed by SCCmec typing and Panton-Valentine leukocidin (PVL) gene analysis using modifications of previously published methods (14, 18, 20). Details of the epidemiologic trends for the entire collection were recently published (23).
Screen for hVISA.
Screening of all isolates for hVISA was performed by the GRD Etest method (bioMérieux, Marcy l'´Etoile, France) according to the manufacturer's instructions. The Etest GRD strip is double sided with 0.5- to 32-μg/ml gradients of vancomycin and teicoplanin on opposite ends. A standard 0.5 McFarland suspension was prepared from overnight growth on a blood agar plate and used to inoculate Mueller-Hinton with 5% blood agar prior to GRD strip placement. The standard vancomycin Etest was also set up on Mueller-Hinton agar. Plates were incubated in ambient air at 35°C. GRD tests were read after 24 and 48 h of incubation. The standard vancomycin Etest was read after 24 h of incubation. A positive GRD result for hVISA was defined as vancomycin or teicoplanin MIC of ≥8 μg/ml (GRD strip on Mueller-Hinton with 5% blood agar at 24 or 48 h) and a standard vancomycin Etest MIC of <4 μg/ml. Isolates with positive GRD results were confirmed with repeat GRD testing (as needed to generate a majority result) and evaluated by the PAP-AUC method. The source of organisms for repeat testing was stock cultures (stored at −70°C on beads) prepared when isolates were received from participating centers.
Confirmation of hVISA by PAP-AUC method.
PAP-AUC testing was performed with modifications (26) of a microdilution technique previously described (22, 25). Briefly, serial dilutions (10−1 to 10−7) of a 0.5 McFarland suspension were prepared in sterile saline. Spiral plating of 100 μl of 10−6 and 10−7 dilutions onto Difco brain heart infusion (BHI) agar (BD Diagnostics) was performed to determine viable counts. For each of the five dilutions (10−1 to 10−5) one aliquot (100 μl) was spiral plated, and four 10-μl droplets were dropped onto 11 different BHI agar plates with different concentrations of vancomycin (0, 0.25, 0.5, 0.75, 1.0, 1.5, 2.0, 2.5, 3.0, 4.0, 6.0, and 8.0 μg/ml). After air drying, the plates were incubated at 35°C. After 24 and 48 h of incubation, colony counts (log10 CFU/ml) were determined and plotted against the vancomycin concentration. The AUC was calculated using GraphPad Prism 5 (San Diego, CA). Isolates with an AUC ratio of ≥0.90 (AUC of test isolate divided by AUC of hVISA control strain Mu3) were defined as hVISA (27).
RESULTS
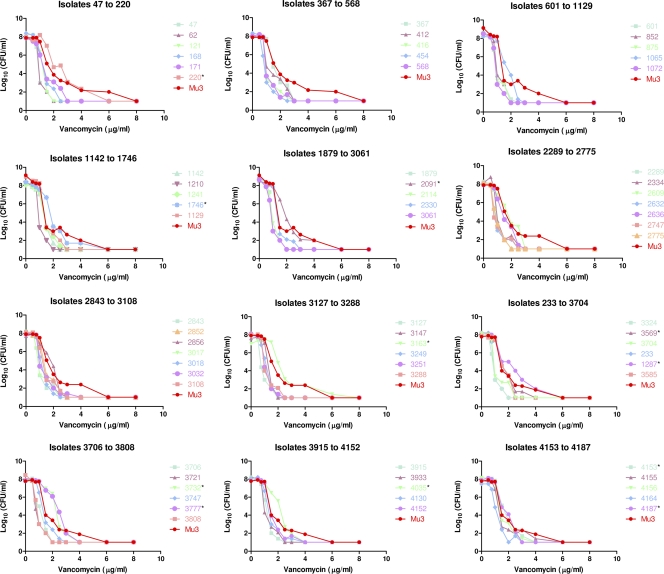
No VISA (vancomycin MIC = 4 to 8 μg/ml) or vancomycin-resistant (MIC ≥ 16 μg/ml) S. aureus strains were detected by broth microdilution MIC or Etest. The Etest GRD screen for hVISA was initially positive for 68 isolates (1.6%; all by teicoplanin MIC ≥ 8 μg/ml at 24 or 48 h). Among those 68 isolates, 45 (66.2%) were GRD positive with repeat testing. PAP-AUC testing confirmed only 11 isolates as hVISA, representing 16.2% of the 68 initial screen-positive and 24.4% of the 45 reproducible GRD-positive results (Table 1 and Fig. 1). Eight isolates were “near hVISA,” with PAP-AUC ratios of 0.85-0.89.
Table 1.
PAP-AUC data for 11 hVISA isolatesa
| Isolate | AUC | Mu3 AUC | AUC ratio |
|---|---|---|---|
| 220 | 27.01 | 26.31 | 1.03 |
| 1287 | 25.23 | 22.19 | 1.14 |
| 1746 | 23.38 | 23.43 | 0.998 |
| 2091 | 24.62 | 23.43 | 1.05 |
| 3163 | 25.91 | 23.71 | 1.09 |
| 3569 | 20.16 | 22.19 | 0.91 |
| 3732 | 23.54 | 22.19 | 1.06 |
| 3777 | 24.39 | 22.19 | 1.1 |
| 4035 | 23.09 | 22.19 | 1.04 |
| 4153 | 21.27 | 22.19 | 0.96 |
| 4187 | 20.71 | 22.19 | 0.93 |
Isolates with an AUC of ≥0.90 compared to the AUC of Mu3 were defined as hVISA.
Fig. 1.
Log10 viable counts plotted against vancomycin concentrations for 68 GRD-positive S. aureus isolates. The 11 isolates confirmed as hVISA by PAP-AUC are indicated by an asterisk.
The vancomycin MIC distribution of hVISA strains by identification method with a comparison to the GRD screen-negative S. aureus population is shown in Table 2. Nine of the eleven hVISA strains were MRSA. The vancomycin MICs of the two methicillin-susceptible hVISA isolates were 1 and 2 μg/ml. The rates of resistance (including intermediate-level resistance) for the 11 hVISA isolates compared to the 4,142 GRD screen-negative isolates were, respectively, as follows: oxacillin, 81.8% versus 53.3%; erythromycin, 81.8% versus 65.3%; clindamycin (including inducible), 72.7% versus 29.9%; levofloxacin, 72.7% versus 39.3%; trimethoprim-sulfamethoxazole, 9.1% versus 1.5%; daptomycin, 9.1% versus 0.2%; linezolid, 0% versus 0.02%; and ceftaroline, 0% versus 0.9%. No isolates in the collection were resistant to tigecycline.
Table 2.
Vancomycin MIC frequency distribution of hVISA and other S. aureus
| Vancomycin MIC (μg/ml)a | No. of isolates (column %) |
||||
|---|---|---|---|---|---|
| PAP-AUC positive |
Etest GRD positive |
Other (S. aureus)d | |||
| hVISAb | Near hVISAc | Repeat | Initial | ||
| ≤0.25 | 1 (2.2) | 1 (1.5) | 4 (0.1) | ||
| 0.5 | 217 (5.2) | ||||
| 1 | 5 (45) | 5 (63) | 35 (78) | 57 (84) | 3,874 (94) |
| 2 | 6 (55) | 3 (37) | 9 (20) | 10 (15) | 47 (1.1) |
| Total | 11 | 8 | 45 | 68 | 4,142 |
Determined by the CLSI broth microdilution method.
PAP-AUC ratio of ≥0.90. These 11 hVISA isolates were obtained from nine medical centers.
PAP-AUC ratio of 0.85 to 0.89.
Etest GRD screen negative.
Six of the eleven hVISA isolates (55%) were from blood cultures compared to only 26% of the GRD screen-negative isolates. Two hVISA isolates were from a wound or abscess. The three other hVISA isolates were from tissue, lower respiratory tract, and normally sterile body fluid (not cerebrospinal fluid) sources, respectively. Eight of the eleven hVISA isolates (73%) were from inpatients compared to 54% of GRD screen negative isolates. A higher portion of GRD screen negative isolates were nosocomial (recovered >48 h after admission; 14% versus 9% of the 11 hVISA).
The 11 hVISA isolates were received from nine medical centers and PFGE analysis revealed 10 different PFGE types. Only three isolates had PFGE patterns representative of a major U.S. type strain: USA300 (SCCmec type IV, PVL positive), USA100 (SCCmec type II, PVL negative), and USA200 (SCCmec type II, PVL negative). The other eight hVISA isolates were not closely related to common PFGE types.
DISCUSSION
A unique aspect of our study is the large population of recent isolates (n = 4,210) screened for hVISA without preselection criteria based on vancomycin MIC or presence of mecA. Most (82%) of the eleven hVISA detected were MRSA, representing 0.4% of the MRSA population in the present study. The vancomycin MIC was also predictive of hVISA. The prevalence of hVISA isolates among isolates with broth microdilution vancomycin MICs of 2 μg/ml was 10.5% (6 of 57 isolates) compared to only 0.1% (5 of 3,931) of isolates with vancomycin MICs of 1 μg/ml.
Global variation in hVISA prevalence is likely, but interpretation of published data is difficult due to variation in study design (i.e., selection of isolates tested and lab methodologies) (9). The higher prevalence of hVISA that has been reported by investigators looking at only one institution may reflect clonal spread within the facility rather than true prevalence for the larger region (5, 24). The prevalence of hVISA among MRSA tested at one Turkish hospital over a 4-year period increased from 1.6% (in 1998) to 36% (in 2001) in a pattern suggestive of clonal spread, but strain typing was not performed (24). A French study demonstrated 11% of 2,300 S. aureus isolates from one hospital were hVISA and PFGE analysis revealed 93% of the 255 hVISA isolates belonged to a single clone (5). Hiramatsu reported hVISA among 20% of MRSA in one Japanese hospital (7), but a subsequent nationwide study screening 6,625 MRSA isolates from 278 hospitals found no hVISA (10).
A Canadian surveillance program screened 475 MRSA isolates from hospitalized patients in 1995 to 2006 (all isolates with a vancomycin MIC of 2 μg/ml and ∼5% of isolates with MICs of 1 μg/ml) using GRD Etest and Etest macromethod and found only 25 Etest screen-positive isolates confirmed as hVISA by PAP-AUC (1). On the basis of those rates, the authors estimated the prevalence of hVISA among this MRSA population in Canada to be 1.3% (1). Besides geographical differences, our lower rate of hVISA among MRSA in the present study (0.4%) may be attributed to the inclusion of outpatient isolates (44% of the entire population) since 73% of our hVISA were from inpatients. Compared to the present study results for MRSA by vancomycin MIC, the Canadian study (1) reported a lower prevalence of hVISA among MRSA isolates with vancomycin MICs of 2 μg/ml (8.1% versus 14.3%) and a higher prevalence among MICs of 1 μg/ml (1.5% versus 0.2%).
Testing of all MRSA isolates from patients with persistent or recurrent bacteremia (n = 22) during a 1-year period in a Detroit hospital (4-μg/ml vancomycin screen, followed by PAP-AUC confirmation) yielded only three isolates of hVISA (13.6%) (11). The authors of that study concluded that hVISA was not a common reason for poor therapeutic response (11).
Our results suggest that the Etest GRD is a useful tool for rapid screening of hVISA, but that it tends to overestimate the hVISA rates compared to PAP-AUC confirmation (1.6% initial and 1.1% reproducibly GRD positive versus 0.3% PAP-AUC positive). The PPV of GRD Etest results for hVISA were 16.2% for initial screen positive and 24.4% for reproducibly positive results. Focusing only on the population of S. aureus isolates with vancomycin MICs of 2 μg/ml (n = 57), the PPV was 60% for initial screen positive results and 66.7% for reproducibly positive results. A weakness of our study was the use of PAP-AUC to test GRD positive isolates only. Our approach to hVISA screening is supported by the Canadian study that used both Etest GRD and Etest macromethod with reported sensitivities of 100 and 44%, respectively, with lower specificity of the Etest GRD (36% versus 98%) in comparison to PAP-AUC as the gold standard (1). However, this study was also limited by the performance of PAP-AUC on only 18 randomly selected isolates not screen positive by Etest GRD or Etest macromethod (1).
A separate study that tested 100 hVISA and 50 vancomycin susceptible strains by Etest GRD and Etest macromethod in comparison to PAP-AUC showed Etest GRD to be more sensitive than Etest macromethod at 48 h (93% versus 83%), but less specific (82% versus 94%) (13). Positive and negative predictive values were not calculated due to the large number of hVISA not being reflective of a normal population of isolates (13). In an earlier evaluation the prototype Etest GRD detected 98% of 60 hVISA strains, and the reported specificity for hVISA/VISA was 95% (collection included 15 VISA and 75 vancomycin-susceptible isolates) (28). In contrast to these previous studies (1, 13, 28), a recent evaluation of 140 MRSA blood isolates reported lower sensitivities for Etest GRD and Etest macromethod with both detecting only 57% of 21 hVISA (26).
Our data suggest the overall prevalence of hVISA in the United States is low (0.3% of all S. aureus; 0.4% of MRSA). The prevalence of hVISA among S. aureus isolates with a vancomycin MIC of 2 μg/ml was 10.5% (14.3% of MRSA, 4.5% of MSSA). Recently, published Infectious Diseases Society of America practice guidelines for treatment of MRSA infections suggest that for vancomycin susceptible MRSA (MIC ≤ 2 μg/ml), clinical response, independent of MIC, should determine whether vancomycin therapy is continued (15). When S. aureus isolates are screened for hVISA using the Etest GRD system, the low PPV demonstrated by the present study (16% overall, 60% for a vancomycin MIC of 2 μg/ml, and 9% for a vancomycin MIC of 1 μg/ml) suggests confirmatory testing with PAP-AUC is necessary.
ACKNOWLEDGMENTS
We thank the following individuals for providing the isolates of S. aureus characterized in this study: Joseph Schwartzman, Dartmouth-Hitchcock Medical Center, Lebanon, NH; Andrew Onderdonk, Brigham and Women's Hospital, Boston, MA; Laura Ovittore, Danbury, Hospital, Danbury, CT; Daniel Shapiro, Lahey Clinic, Burlington, MA; Phyllis Della-Latta, Columbia Presbyterian Hospital, New York, NY; Allan Truant, Temple University Hospital, Philadelphia, PA; Deanna Kiska, SUNY Upstate Medical Center, Syracuse, NY; Paul Bourbeau, Danville, PA; Dwight Hardy, University of Rochester Medical Center, Rochester, NY; and Christine Ginocchio, North Shore–LIJ Health System, Lake Success, NY (Northeast); Betty Forbes, VA Commonwealth University School of Medicine, Richmond, VA; Peter Gilligan, University of North Carolina Hospital, Chapel Hill, NC; Lisa Steed, Medical University of South Carolina, Charleston, SC; Robert Jerris, Children's Healthcare of Atlanta, Atlanta, GA; James Snyder, University of Louisville Hospital, Louisville, KY; Kenneth Rand, Shands Hospital–University of Florida, Gainesville, FL; Diane Halstead, Baptist Medical Center, Jacksonville, FL; Teresa Barnett, University of South Alabama, Mobile, AL; and Yi-Wei Tang, Vanderbilt University Medical Center, Nashville, TN (Southeast); Gerri Hall, Cleveland Clinic, Cleveland, OH; Wanita Howard, University of Iowa Health Care, Iowa City, IA; Mary Beth Perri, Henry Ford Hospital, Detroit, MI; Gerald Denys, Clarian Pathology Laboratory, Indianapolis, IN; Mary Hayden, Rush University Medical Center, Chicago, IL; Richard Thomson, Jr., Evanston Northwestern Healthcare, Evanston, IL; Joan Hoppe-Bauer, Barnes Jewish Hospital, St. Louis, MO; Rebecca Horvath, University of Kansas Medical Center, Kansas City, KS; Steven Cavalieri, Creighton University, Omaha, NE; Glenn Hansen, Hennepin County Hospital, Minneapolis, MN (Midwest); James Versalovic, Houston, TX; Paul Southern, Jr., Dallas, TX; James Jorgenson, San Antonio, TX; Sara Hobbie, Tulsa, OK; Michael Wilson, Denver, CO; Ann Croft, Salt Lake City, UT; and Michael Saubolle, Phoenix, AZ (Southwest); and Ann Robinson, Pathology Associates Medical Lab, Spokane, WA; Janet Hindler, UCLA Medical Center, Los Angeles, CA; Rohan Nadarajah, UCSF Medical Center, San Francisco, CA; Susan Sharp, Northwest Kaiser Permanente, Portland, OR; Brad Cookson, University of Washington Medical Center, Seattle, WA; and Matt Bankowski, Diagnostic Laboratory Services, Inc., Honolulu, HI (West).
G.V.D. has received research funding from Abbott Laboratories, Schering-Plough, Bayer Pharmaceutical, Merck, Shionogi, Cubist, and Astra-Zeneca. He has been on the speakers' bureaus of Abbott Laboratories, Aventis, Astra-Zeneca, Pfizer, Astellas, and Schering-Plough. D.J.D. has received research funding from Forest Laboratories, Merck, Pfizer, Schering-Plough, Astellas, and bioMérieux. S.S.R. has received research funding from Abbott Laboratories, BD Diagnostics, bioMérieux, Forest Laboratories, and Schering-Plough. She has received an honorarium from bioMérieux for an educational presentation.
Financial support for this project was provided by Forest Laboratories, Inc. (New York, NY). Scientific Therapeutics Information, Inc. (Springfield, NJ), provided editorial assistance funded by Forest Laboratories for the manuscript.
Footnotes
Published ahead of print on 5 October 2011.
REFERENCES
- 1. Adam H. J., et al. 2010. Detection and characterization of heterogeneous vancomycin-intermediate Staphylococcus aureus isolates in Canada: results from the Canadian nosocomial infection surveillance program, 1995–2006. Antimicrob. Agents Chemother. 54:945–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Charles P. G., Ward P. B., Johnson P. D., Howden B. P., Grayson M. L. 2004. Clinical features associated with bacteremia due to heterogeneous vancomycin-intermediate Staphylococcus aureus. . Clin. Infect. Dis. 38:448–451 [DOI] [PubMed] [Google Scholar]
- 3. Clinical and Laboratory Standards Institute. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 8th ed. CLSI document M7–A8. CLSI, Wayne, PA. [Google Scholar]
- 4. Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial susceptibility testing; 19th informational supplement. CLSI document M100–S19. CLSI, Wayne, PA. [Google Scholar]
- 5. Garnier F., et al. 2006. A 1 year surveillance study of glycopeptide-intermediate Staphylococcus aureus strains in a French hospital. J. Antimicrob. Agents Chemother. 57:146–149 [DOI] [PubMed] [Google Scholar]
- 6. Hiramatsu K. 2001. Vancomycin-resistant Staphylococcus aureus: a new model of antibiotic resistance. Lancet Infect. Dis. 1:147–155 [DOI] [PubMed] [Google Scholar]
- 7. Hiramatsu K., et al. 1997. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 350:1670–1673 [DOI] [PubMed] [Google Scholar]
- 8. Horne K. C., et al. 2009. Prospective comparison of the clinical impacts of heterogeneous vancomycin-intermediate methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-susceptible MRSA. Antimicrob. Agents Chemother. 53:3447–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Howden B. P., Davies J. K., Johnson P. D., Stinear T. P., Grayson M. L. 2010. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin. Microbiol. Rev. 23:99–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ike Y., et al. 2001. Nationwide survey shows that methicillin-resistant Staphylococcus aureus strains heterogeneously and intermediately resistant to vancomycin are not disseminated throughout Japanese hospitals. J. Clin. Microbiol. 39:4445–4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Khosrovaneh A., et al. 2004. Frequency of reduced vancomycin susceptibility and heterogeneous subpopulation in persistent or recurrent methicillin-resistant Staphylococcus aureus bacteremia. Clin. Infect. Dis. 38:1328–1330 [DOI] [PubMed] [Google Scholar]
- 12. Kollef M. H. 2007. Limitations of vancomycin in the management of resistant staphylococcal infections. Clin. Infect. Dis. 45:S191–S195 [DOI] [PubMed] [Google Scholar]
- 13. Leonard S. N., Rossi K. L., Newton K. L., Rybak M. J. 2009. Evaluation of the Etest GRD for the detection of Staphylococcus aureus with reduced susceptibility to glycopeptides. J. Antimicrob. Chemother. 63:489–492 [DOI] [PubMed] [Google Scholar]
- 14. Lina G., et al. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128–1132 [DOI] [PubMed] [Google Scholar]
- 15. Liu C., et al. 2011. Clinical practice guidelines by the Infectious Diseases Society of America for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children. Clin. Infect. Dis. 52:e18–e55 [DOI] [PubMed] [Google Scholar]
- 16. Lodise T. P., et al. 2008. Relationship between vancomycin MIC and failure among patients with methicillin-resistant Staphylococcus aureus bacteremia treated with vancomycin. Antimicrob. Agents Chemother. 52:3315–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McDougal L. K., et al. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mendes R. E., et al. 2007. Rapid detection and identification of metallo-β-lactamase-encoding genes by multiplex real-time PCR assay and melt curve analysis. J. Clin. Microbiol. 45:544–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moore M. R., Perdreau-Remington F., Chambers H. F. 2003. Vancomycin treatment failure associated with heterogeneous vancomycin-intermediate Staphylococcus aureus in a patient with endocarditis and in the rabbit model of endocarditis. Antimicrob. Agents Chemother. 47:1262–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Oliveira D. C., de Lencastré H. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pfaller M. A., Caliendo A. M., Versalovic J. 2010. Chromosomal restriction fragment analysis by pulsed-field gel electrophoresis: application to molecular epidemiology, p. 12.4.5.1–12.4.5.7.In Garcia L. S. (ed.), Clinical microbiology procedures handbook, 3rd ed. ASM Press, Washington, DC. [Google Scholar]
- 22. Pfeltz R. F., Schmidt J. L., Wilkinson B. J. 2001. A microdilution plating method for population analysis of antibiotic-resistant staphylococci. Microb. Drug Resist. 7:289–295 [DOI] [PubMed] [Google Scholar]
- 23. Richter S. S., et al. 2011. Activity of ceftaroline and epidemiologic trends in Staphylococcus aureus isolates collected from 43 medical centers in the United States in 2009. Antimicrob. Agents Chemother. 55:4154–4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sancak B., Ercis S., Menemenlioglu D., Colakoglu S., Hascelik G. 2005. Methicillin-resistant Staphylococcus aureus heterogeneously resistant to vancomycin in a Turkish university hospital. J. Antimicrob. Chemother. 56:519–523 [DOI] [PubMed] [Google Scholar]
- 25. Satola S. W., Caliendo A. M., Farley M. M., Patel J. B., Burd E. M. 2009. Lack of heteroresistance among Staphylococcus aureus isolates with vancomycin MICs of 2 micrograms per milliliter by automated testing. J. Clin. Microbiol. 47:2680–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Satola S. W., Farley M. M., Anderson K. F., Patel J. B. 2011. Comparison of detection methods for heteroresistant vancomycin-intermediate Staphylococcus aureus with the population analysis profile method as the reference method. J. Clin. Microbiol. 49:177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wootton M., et al. 2001. A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J. Antimicrob. Chemother. 47:399–404 [DOI] [PubMed] [Google Scholar]
- 28. Yusof A., et al. 2008. Evaluation of a new Etest vancomycin-teicoplanin strip for detection of glycopeptide-intermediate Staphylococcus aureus (GISA), in particular, heterogeneous GISA. J. Clin. Microbiol. 46:3042–3047 [DOI] [PMC free article] [PubMed] [Google Scholar]