Abstract
Anaplasma phagocytophilum is the causative agent of human granulocytic anaplasmosis, which is prevalent throughout China. In this study, we describe a rapid, simple, and sensitive loop-mediated isothermal amplification (LAMP) assay targeting the msp2 gene of A. phagocytophilum that is ideal for application in rural areas in China. This assay has the potential to detect A. phagocytophilum early in infection as an alternative to existing methods. A total of 42 suspected cases of infection with A. phagocytophilum, 15 serologically confirmed and 27 probable cases, were analyzed by the msp2 LAMP assay. To validate the accuracy of LAMP, previously established nested-PCR and real-time PCR assays were utilized. The sensitivity of LAMP was 25 copies per reaction (approximately 1,250 copies per ml blood) for A. phagocytophilum, and the assay did not detect false positives among 27 members of the order Rickettsiales and 17 common clinical pathogens. To evaluate the clinical applicability of the LAMP assay, a total of 42 clinical samples were examined. A positive LAMP result was obtained for 12 of the confirmed cases and for 14 of 27 suspected cases, while only 1 confirmed case and 3 cases (2 confirmed cases and 1 suspected case) were detected by nested PCR and real-time PCR, respectively. The LAMP assay described in this study demonstrated a high level of sensitivity comparable with that of nested PCR and real-time PCR for the detection of A. phagocytophilum. This LAMP assay is a valuable method for rapid, cost-effective, and simple detection of A. phagocytophilum in the rural areas of China.
INTRODUCTION
Human granulocytic anaplasmosis (HGA) is an emerging tick-borne disease caused by Anaplasma phagocytophilum, which was reported in the United States in 1990 and in Europe in 1997 (3, 6). Seroepidemiological data suggest that HGA infection rates in the United States are as high as 15% to 36% in some locations (7). An unusual case of nosocomial human-to-human A. phagocytophilum transmission was reported in 2006 in a regional hospital in Anhui Province, China (19). Subsequently, nationwide seroepidemiological surveys of A. phagocytophilum among high-risk agricultural groups in China demonstrated that the total seroprevalence rate of A. phagocytophilum was 13.94% (unpublished data). However, the greatest challenge to clinicians is the lack of a rapid, early, and accurate diagnostic method for the detection of anaplasmosis as an emerging infectious disease in China. The indirect immunofluorescence assay (IFA), the gold standard method proposed by the World Health Organization Collaborating Center for Rickettsial Reference and Research, depends on a specific, expensive fluorescence microscope and requires a pair of serum samples from both the acute and convalescent stages of the illness, which is not applicable as an early test for A. phagocytophilum infection. The utility of insensitive conventional PCR, time-consuming and easily contaminated nested PCR, and highly sensitive real-time PCR is also limited in rural areas because they require specific, expensive instruments. Because the incidence of A. phagocytophilum infection in China is high in rural areas, rapid and simple diagnostic methods are urgently needed.
Loop-mediated isothermal amplification (LAMP) is a novel nucleic acid amplification method that was developed in 2000 (12). The LAMP technique is highly sensitive, specific, and simple and can generate up to 109-fold amplification in less than an hour under isothermal conditions (60 to 65°C), making LAMP a potentially rapid and simple diagnostic tool for A. phagocytophilum infection. In this study, we aimed to develop a rapid, sensitive, and specific assay to detect A. phagocytophilum in rural areas.
MATERIALS AND METHODS
Laboratory strains.
A total of 44 strains were used to determine the specificity of the LAMP assay. Members of the order Rickettsiales, including Rickettsia prowazekii; Rickettsia typhi; Orientia tsutsugamushi types Karp, Kato, and Gilliam; Rickettsia sibirica; Rickettsia conorii; Rickettsia marmionii; Rickettsia akari; Rickettsia rickettsii; Rickettsia africa; Rickettsia parkeri; Rickettsia japonica; Rickettsia slovaca; Rickettsia aeschlimannii; Rickettsia montanensis; Rickettsia helvetica; Rickettsia felis; Rickettsia australis; Rickettsia canadensis; Rickettsia bellii; Rickettsia heilongjiangensis; Anaplasma phagocytophilum types Webster, MRK, Slovienie, and MD; Ehrlichia chaffeensis; and the related species Bartonella henselae and Bartonella quintana, were provided by the WHO Collaborating Center for Rickettsial Reference and Research (Marseille, France) and maintained in our laboratory. Genomic DNA was extracted from cultured cells using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). The genomic DNAs of other common clinical pathogenic bacteria, including Coxiella burnetii, Borrelia burgdorferi, Escherichia coli, Vibrio cholerae, Bacillus anthracis, Haemophilus influenzae, Listeria spp., Legionella spp., Yersinia pestis, Shigella dysenteriae, Neisseria meningitides, Leptospira spp., Mycobacterium tuberculosis, and Klebsiella pneumoniae, were obtained from relevant departments of the China National Institute of Communicable Disease Control and Prevention (ICDC). In addition, total blood DNAs from healthy humans, cattle, horses, goats, and mice were extracted as negative controls. DNA extracted from the above-mentioned bacterial strains was first amplified by PCR using prokaryotic bacterial 16S rRNA universal primers (15) before the specificity of the LAMP assay was tested in this study.
Clinical samples.
We obtained 42 samples from probable cases of infection with A. phagocytophilum from different hospitals (Beijing University First Hospital, Laizhou First People's Hospital, Beijing Friendship Hospital, and 302 Military Hospital of China) between 2007 and 2009. This study was approved by the Human Research Ethics Committee of the China Center for Disease Control and Prevention (China CDC). The cases were confirmed by the serological diagnostic criteria of anaplasmosis (4-fold increase of IgG antibody titer), by nested-PCR amplification of the 16S rRNA gene, or by real-time PCR amplification of the msp2 gene of A. phagocytophilum, which excluded other febrile illnesses in clinics (2, 17). Two milliliters of EDTA-anticoagulated blood and 2 ml of nonanticoagulated blood were collected during the acute phase of illness for culture and for detecting IgM and IgG antibodies, respectively, and DNA was extracted from the remaining clots for nested PCR (9), real-time PCR (18), and the LAMP assay described in this study. A second serum sample was collected from the convalescent stage (2 to 4 weeks after the acute phase) of the illness for detecting IgG antibodies.
LAMP primer design.
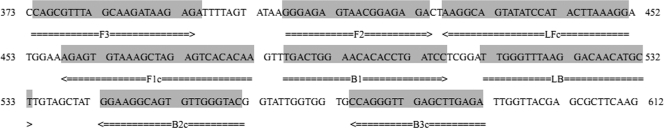
A set of universal primers targeted toward the expression site of the msp2 gene of the species A. phagocytophilum isolate EQ6 D22 (AY763471) were designed using PrimerExplorer V4 software (http://primerexplorer.jp) (Eiken Chemical Co. Ltd., Tokyo, Japan) based on conserved sequences determined by aligning 82 of the msp2 gene sequences obtained from GenBank: AY137510, AY164490 to AY164494, AY319262 to AY319265, AY642115, AY763468, AY763470 to AY763488, AY763490 to AY763499, DQ097228, DQ097231, DQ105669 to DQ105673, DQ519565 to DQ519570, EF143796 to EF143812, EU921902 to EU921906, FJ600592, FJ600595, FJ600600, FJ600601, and FJ600611. The primers were synthesized by Sangon Biotech (Shanghai, China). The primers were as follows: FIP(F1c-F2) (5′-TTGTIIGACTCIAGCTTTACACTCTGGGAGAGTAACGGAGAGA-3′) (43 nucleotides [nt]), BIP(B1c-B2) (5′-TGACTGGAACACICCTGATCCITIACCAACACTICCTTCC-3′) (40 nt), F3 (5′-CAGCGTTTAGCAAGATAAGAGA-3′) (22 nt), B3 (5′-TCTCAAGCTCAACCCTGG-3′) (18 nt), LF (5′-CCTTTAAGTATGGATATACTGCCTT-3′) (25 nt), and LB (5′-TTGGGTTTAAGGACAACATGCT-3′) (22 nt). The primer sequences and their positions in the expression site of the msp2 gene are shown in Fig. 1.
Fig. 1.
Names and locations of target sequences used as primers for the expression site of msp2 LAMP.
Reference plasmid.
To determine the sensitivity of the LAMP assay, a recombinant plasmid containing the target sequence of the A. phagocytophilum msp2 gene from the isolate EQ6 D22 (AY763471) was constructed as follows. A pair of primers was designed to span the sequences between the F3 and B3 primers: forward primer MP1 (5′-GAAGCGTAATGATGTCTA-3′) and reverse primer MP2 (5′-AGTAACAACATCATAAGC-3′). The PCR products (452 bp) were cloned into the pEASY-T1 vector using the pEASY-T1 Cloning Kit (Transgen, China). The recombinant plasmid was quantified with a NanoPhotometer (Implen, Germany) and was serially diluted (to concentrations of 55, 54, 53, 52, 51, and 50 copies/μl) to evaluate the limit of detection and the reproducibility of the msp2 LAMP assay.
LAMP reaction.
All LAMP reactions were performed with the Loopamp Kit (Eiken Chemical Co. Ltd., Tokyo, Japan) in a 25-μl mixture containing 1.6 μM (each) FIP and BIP primers, 0.8 μM LF and LB primers, 0.2 μM F3 and B3 primers, 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 8 mM MgSO4, 10 mM (NH4)2SO4, 0.1% Tween 20, 0.8 M betaine, 1.4 mM (each) deoxynucleoside triphosphates (dNTPs), and 1 μl Bst DNA polymerase (8 U/μl). The reaction mixture was incubated in a real-time turbidimeter LA200 (Teramecs, Tokyo, Japan) at 63°C for 60 min and then at 80°C for 5 min to terminate the reaction. The positive and negative samples were distinguished by a turbidity cutoff value of 0.1. After amplification, the LAMP products were detected by electrophoresis on 2% agarose gels with ethidium bromide staining or were determined by visual inspection after adding 1 μl of 1,000× SYBR green I.
Evaluation of the sensitivity, specificity, and reproducibility of the LAMP assay.
To compare the sensitivities of the msp2 LAMP assay and general PCR, the serially diluted reference plasmid (at concentrations of 55, 54, 53, 52, 51, and 50 copies/μl) containing the target DNA was used to define the limit of detection. A general PCR was performed with the primers MP1 and MP2. PCR amplification was performed in a 25-μl volume containing 0.2 μM each primer, 0.4 mM each dNTP, and 1 U of Taq DNA polymerase. The PCR was cycled 35 times using a denaturation step of 94°C for 50 s followed by annealing at 50°C for 30 s and extension at 72°C for 45 s. The PCR products were electrophoresed on a 1% TBE agarose gel and stained with ethidium bromide (1 μg/ml).
Genomic DNAs from the 44 strains, including 27 members of the order Rickettsiales and 17 common clinical pathogens, were detected by LAMP to determine the specificity of the msp2 LAMP assay. All detection assays were performed in triplicate.
Serially diluted reference plasmids were amplified in two ways (five times on 1 day and once on each of 5 days) to evaluate the reproducibility of the LAMP assay. The intra-assay coefficient of variation (CVi) and interassay coefficient of variation (CVo) were analyzed at the time of peak precipitation, as measured by turbidity on a real-time turbidimeter. Statistical analysis was conducted using SAS software version 9.1.
Clinical validation.
A total of 42 clinical samples were also assessed by 16S rRNA nested PCR (9) and msp2 real-time PCR (18), as described above, and 8 μl of extracted DNA was used as a template in a 25-μl reaction mixture. Positive results from the three methods were counted to analyze the difference between the LAMP assay and nested and real-time PCR using SPSS software.
RESULTS
Specificity of LAMP.
To determine the specificity of the LAMP assay for A. phagocytophilum, 27 members of the order Rickettsiales were tested for amplification; R. prowazekii; R. typhi; O. tsutsugamushi types Karp, Kato, and Gilliam; R. sibirica; R. conorii; R. marmionii; R. akari; R. rickettsii; R. africa; R. parkeri; R. japonica; R. slovaca; R. aeschlimannii; R. montanensis; R. helvetica; R. felis; R. australis; R. canadensis; R. bellii; R. heilongjiangensis; E. chaffeensis; and A. phagocytophilum types Webster, MRK, Slovienie, and MD. Only A. phagocytophilum tested positive, while the other species did not. Moreover, other common pathogens, including C. burnetii, B. henselae, B. quintana, B. burgdorferi, E. coli, V. cholerae, B. anthracis, H. influenzae, Listeria, Legionella, Y. pestis, S. dysenteriae, N. meningitides, Leptospira, M. tuberculosis, and K. pneumoniae, also did not test positive by LAMP assay. This indicates no false-positive amplification with these heterologous species in the LAMP assay.
Sensitivity of LAMP.
The limits of detection of LAMP and PCR for the msp2 gene were 25 (approximately 1,250 copies of A. phagocytophilum per ml blood) and 625 copies per reaction, respectively. This indicates that the LAMP assay is more sensitive than conventional PCR for detecting A. phagocytophilum DNA.
Reproducibility of LAMP.
The reproducibility of the minimal detection level of the LAMP assay was 1.17% of CVi and 3.47% of CVo, respectively (Table 1).
Table 1.
Summary of CVi and CVo of the time of peak precipitation of serially diluted plasmids
Intra-assay coefficient of variation.
Interassay coefficient of variation.
Examination of clinical samples.
A total of 42 samples were assayed, and 15 samples were confirmed as indicative of A. phagocytophilum infection in our laboratory: 2 samples by bacterial culture and 13 samples by measuring a 4-fold increase in the IgG titer in acute- and convalescent-phase serum samples. To evaluate the utility of the LAMP assay, we compared it to nested-PCR and real-time PCR assays previously developed in our laboratory (Table 2). Of these samples, 26 positive results were obtained by LAMP assay, including 12 of 15 confirmed cases and 14 of 27 suspected cases. This level of detection is significantly higher than the one positive result obtained by nested PCR (P < 0.001) and the three positive results obtained by real-time PCR (P < 0.001).
Table 2.
Comparison of the LAMP assay with nested PCR and real-time PCR for the detection of A. phagocytophilium in clinical samplesa
| Case no. | Diagnostic type | Resultb |
||
|---|---|---|---|---|
| LAMP | Nested PCR | Real-time PCR | ||
| 1 | 4-fold IgG titer increase | Neg. | Neg. | Neg. |
| 2 | Suspected case | Pos. | Neg. | Neg. |
| 3 | Suspected case | Neg. | Neg. | Neg. |
| 4 | Suspected case | Pos. | Neg. | Neg. |
| 5 | Suspected case | Pos. | Neg. | Neg. |
| 6 | Suspected case | Neg. | Neg. | Neg. |
| 7 | 4-fold IgG titer increase | Pos. | Neg. | Neg. |
| 8 | Suspected case | Neg. | Neg. | Neg. |
| 9 | Suspected case | Neg. | Neg. | Neg. |
| 10 | Suspected case | Neg. | Neg. | Neg. |
| 11 | Suspected case | Pos. | Neg. | Neg. |
| 12 | 4-fold IgG titer increase | Neg. | Neg. | Neg. |
| 13 | Suspected case | Neg. | Neg. | Pos. |
| 14 | Suspected case | Neg. | Neg. | Neg. |
| 15 | Suspected case | Pos. | Neg. | Neg. |
| 16 | Suspected case | Neg. | Neg. | Neg. |
| 17 | 4-fold IgG titer increase | Pos. | Pos. | Neg. |
| 18 | Suspected case | Neg. | Neg. | Neg. |
| 19 | Suspected case | Neg. | Neg. | Neg. |
| 20 | 4-fold IgG titer increase | Neg. | Neg. | Pos. |
| 21 | 4-fold IgG titer increase | Pos. | Neg. | Neg. |
| 22 | Suspected case | Pos. | Neg. | Neg. |
| 23 | Culture confirmed | Pos. | Neg. | Pos. |
| 24 | Suspected case | Pos. | Neg. | Neg. |
| 25 | Suspected case | Neg. | Neg. | Neg. |
| 26 | Suspected case | Pos. | Neg. | Neg. |
| 27 | Suspected case | Pos. | Neg. | Neg. |
| 28 | 4-fold IgG titer increase | Pos. | Neg. | Neg. |
| 29 | 4-fold IgG titer increase | Pos. | Neg. | Neg. |
| 30 | Suspected case | Pos. | Neg. | Neg. |
| 31 | Suspected case | Pos. | Neg. | Neg. |
| 32 | Suspected case | Pos. | Neg. | Neg. |
| 33 | 4-fold IgG titer increase | Pos. | Neg. | Neg. |
| 34 | Suspected case | Neg. | Neg. | Neg. |
| 35 | Suspected case | Neg. | Neg. | Neg. |
| 36 | 4-fold IgG titer increase | Pos. | Neg. | Neg. |
| 37 | 4-fold IgG titer increase | Pos. | Neg. | Neg. |
| 38 | Suspected case | Pos. | Neg. | Neg. |
| 39 | Suspected case | Pos. | Neg. | Neg. |
| 40 | Culture confirmed | Pos. | Neg. | Neg. |
| 41 | 4-fold IgG titer increase | Pos. | Neg. | Neg. |
| 42 | 4-fold IgG titer increase | Pos. | Neg. | Neg. |
All runs were performed in triplicate.
Pos., positive; Neg., negative.
DISCUSSION
HGA, caused by A. phagocytophilum, is a worldwide tick-borne zoonosis that has been described in recent years (17). The greatest challenge to clinicians is not therapy but the difficult diagnostic dilemma in the early phase of infection. It should be noted that rural areas are at the highest risk for A. phagocytophilum infection in China. This phenomenon, combined with the lack of a rapid and sensitive diagnostic method for A. phagocytophilum, may cause delays in treatment or misdiagnoses that could lead to severe disease and fatal outcomes (2). Conventional serodiagnosis and cultivation of bacteria in cell culture are not useful techniques for clinical diagnosis during the acute phase of A. phagocytophilum infection because they are relatively time-consuming. Modified PCR techniques, such as nested PCR and real-time PCR, are complicated and require a high-precision thermal cycler; therefore, these methods are not practical for diagnosing A. phagocytophilum in rural areas. In contrast, LAMP has the advantages of a rapid reaction, simple operation, and cost-effectiveness. The LAMP assay does not require sophisticated and expensive equipment; keeping a constant temperature of 60 to 65°C for 1 to 1.5 h is sufficient for the reaction (12). These features demonstrate that the LAMP assay is suitable for the detection of A. phagocytophilum in rural areas.
In this study, we developed a sensitive LAMP assay based on a conserved region in the msp2 gene, and the limit of detection for the assay was 25 copies per reaction. To evaluate the LAMP assay in clinical diagnoses, we assayed 42 clinical samples and compared the results to those of nested PCR and real-time PCR. Of the 42 samples, 26 tested positive by LAMP assay, while only 1 and 3 samples tested positive by nested PCR and real-time PCR, respectively. Some researchers have reported that the Bst polymerase in LAMP is less sensitive to the presence of inhibitors than is the Taq polymerase used in PCR (8, 16, 13). Thus, although nested PCR (1, 9) and real-time PCR (5, 18) have been reported to be more sensitive than the LAMP assay for the detection of A. phagocytophilum, in this study, the LAMP assay was shown to be more accurate and more sensitive than PCR methods on clinical samples. However, it is remarkable that there were three positive results of seroconversion that were not detected by the msp2 LAMP assay. Thus, we suggest that A. phagocytophilum infection should be confirmed by more than one method, if possible, to prevent misdiagnosis.
When evaluating the specificity of the msp2 LAMP assay, we tested 27 members of the order Rickettsiales and other pathogenic bacteria, and the results of this study showed that the specificity of the LAMP assay is 100%. The high specificity of LAMP methods has also been reported for other pathogens (4, 11, 13, 14, 16). Compared to conventional PCR techniques, 6 LAMP primers were designed to target 8 regions, while PCR methods used primers targeting 2 to 4 regions. Studies on other pathogens have also reported the high specificity of the LAMP assay, in keeping with the results of our work. Although confirmation of LAMP results by visual inspection is important for applying the technique in low-technology settings and could reduce amplicon carryover that may lead to false-positive results, it was shown to be less sensitive than agarose gel electrophoresis in our study (data not shown) and in the results obtained by Thekisoe et al. (16). In contrast, Mao et al. (10) reported consistent results, as detected by electrophoresis and the addition of SYBR green after amplification was completed; however, opening the tubes to add the dye introduces a high risk of contamination. This possibility should be considered for reducing false-positive results during field testing.
Extracting DNA using a kit would increase the cost per test and would require more time to complete the test. However, in this study, we detected DNA only in samples extracted from whole blood by the LAMP assay, which uses a kit and appears to be more sensitive than heat-freeze procedures. However, Paris et al. (14) detected O. tsutsugamushi, order Rickettsiales, in DNA samples extracted using a kit and by freeze-heat procedures, obtaining, respectively, 7/7 and 5/7 positive results. This finding suggests that more attention should be paid to testing boiled samples by the LAMP assay for greater suitability in rural areas.
In conclusion, we developed a rapid, simple, sensitive, and cost-effective LAMP assay for detecting A. phagocytophilum in clinical samples. Because the LAMP assay is fast and sensitive for testing clinical samples without sophisticated equipment and complicated operation, it could be a valuable alternative method of A. phagocytophilum diagnosis in the field and in the rural areas of China.
ACKNOWLEDGMENTS
This study was funded by the National Basic Research Program of China (973 Program) (2010CB530200) (2010CB530206) and the National Key Science and Technology Projects of China (no. 2009ZX10004-203 and 2008ZX10004-008).
Footnotes
Published ahead of print on 5 October 2011.
REFERENCES
- 1. Alberti A., Sparagano O. A. 2006. Molecular diagnosis of granulocytic anaplasmosis and infectious cyclic thrombocytopenia by PCR-RFLP. Ann. N. Y. Acad. Sci. 1081:371–378 [DOI] [PubMed] [Google Scholar]
- 2. Chapman A. S., et al. 2006. Diagnosis and management of tickborne rickettsial diseases: Rocky Mountain spotted fever, ehrlichioses, and anaplasmosis—United States: a practical guide for physicians and other health-care and public health professionals. MMWR Morb. Mortal. Wkly. Rep. 55:1–27 [PubMed] [Google Scholar]
- 3. Chen S. M., Dumler J. S., Bakken J. S., Walker D. H. 1994. Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. J. Clin. Microbiol. 32:589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Curtis K. A., Rudolph D. L., Owen S. M. 2008. Rapid detection of HIV-1 by reverse-transcription, loop-mediated isothermal amplification (RT-LAMP). J. Virol. Methods 151:264–270 [DOI] [PubMed] [Google Scholar]
- 5. Drazenovich N., Foley J., Brown R. N. 2006. Use of real-time quantitative PCR targeting the msp2 protein gene to identify cryptic Anaplasma phagocytophilum infections in wildlife and domestic animals. Vector Borne Zoonotic Dis. 6:83–90 [DOI] [PubMed] [Google Scholar]
- 6. Dumler J. S., et al. 2005. Human granulocytic anaplasmosis and Anaplasma phagocytophilum. Emerg. Infect. Dis. 11:1828–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. IJdo J. W., et al. 2000. The emergence of another tickborne infection in the 12-town area around Lyme, Connecticut: human granulocytic ehrlichiosis. J. Infect. Dis. 181:1388–1393 [DOI] [PubMed] [Google Scholar]
- 8. Kaneko H., Kawana T., Fukushima E., Suzutani T. 2007. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J. Biochem. Biophys. Methods 70:499–501 [DOI] [PubMed] [Google Scholar]
- 9. Luan M. C., Yu D. Z., Tang L., Zhang L. J. 2008. Identification of Orientia tsutsugamushi, spotted fever group and typhus group rickettsia by duplex and nested PCR methods. Asian Pac. J. Trop. Med. 1:1–8 [Google Scholar]
- 10. Mao X. L., et al. 2008. Rapid and sensitive detection of Singapore grouper iridovirus by loop-mediated isothermal amplification. J. Appl. Microbiol. 105:389–397 [DOI] [PubMed] [Google Scholar]
- 11. Nkouawa A., Sako Y., Nakao M., Nakaya K., Ito A. 2009. Loop-mediated isothermal amplification method for differentiation and rapid detection of Taenia species. J. Clin. Microbiol. 47:168–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Notomi T., et al. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Okada K., et al. 2010. A rapid, simple, and sensitive loop-mediated isothermal amplification method to detect toxigenic Vibrio cholerae in rectal swab samples. Diagn. Microbiol. Infect. Dis. 66:135–139 [DOI] [PubMed] [Google Scholar]
- 14. Paris D. H., Blacksell S. D., Newton P. N., Day N. P. 2008. Simple, rapid and sensitive detection of Orientia tsutsugamushi by loop-isothermal DNA amplification. Trans. R. Soc. Trop. Med. Hyg. 102:1239–1246 [DOI] [PubMed] [Google Scholar]
- 15. Roux V., Raoult D. 1995. Phylogenetic analysis of members of the genus Rickettisae by 16S rDNA sequencing. Res. Microbiol. 146:385–396 [DOI] [PubMed] [Google Scholar]
- 16. Thekisoe O. M., et al. 2007. Species-specific loop-mediated isothermal amplification (LAMP) for diagnosis of trypanosomosis. Acta Trop. 102:182–189 [DOI] [PubMed] [Google Scholar]
- 17. Thomas R. J., Dumler J. S., Carlyon J. A. 2009. Current management of human granulocytic anaplasmosis, human monocytic ehrlichiosis and Ehrlichia ewingii ehrlichiosis. Expert Rev. Anti Infect. Ther. 7:709–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang S. W., et al. 2011. Establishment and comparison of real-time PCR assays to detect Anaplasma msp2 gene with general TaqMan probes and TaqMan-MGB probe. Dis. Surveill. 1:12–14 [Google Scholar]
- 19. Zhang L., et al. 2008. Nosocomial transmission of human granulocytic anaplasmosis in China. JAMA 300:2263–2270 [DOI] [PubMed] [Google Scholar]