Abstract
Hepatitis E is recognized as a zoonosis, and swine are known reservoirs, but how broadly enzootic its causative agent, hepatitis E virus (HEV), is remains controversial. To determine the prevalence of HEV infection in animals, a serological assay with capability to detect anti-HEV-antibody across a wide variety of animal species was devised. Recombinant antigens comprising truncated capsid proteins generated from HEV-subgenomic constructs that represent all four viral genotypes were used to capture anti-HEV in the test sample and as an analyte reporter. To facilitate development and validation of the assay, serum samples were assembled from blood donors (n = 372), acute hepatitis E patients (n = 94), five laboratory animals (rhesus monkey, pig, New Zealand rabbit, Wistar rat, and BALB/c mouse) immunized with HEV antigens, and four pigs experimentally infected with HEV. The assay was then applied to 4,936 sera collected from 35 genera of animals that were wild, feral, domesticated, or otherwise held captive in the United States. Test positivity was determined in 457 samples (9.3%). These originated from: bison (3/65, 4.6%), cattle (174/1,156, 15%), dogs (2/212, 0.9%), Norway rats (2/318, 0.6%), farmed swine (267/648, 41.2%), and feral swine (9/306, 2.9%). Only the porcine samples yielded the highest reactivities. HEV RNA was amplified from one farmed pig and two feral pigs and characterized by nucleotide sequencing to belong to genotype 3. HEV infected farmed swine primarily, and the role of other animals as reservoirs of its zoonotic spread appears to be limited.
INTRODUCTION
Hepatitis E virus (HEV), the causative agent of hepatitis E, is a nonenveloped, single-stranded, positive-sense RNA virus that belongs to the Hepeviridae family (11). Among the mammalian HEV, at least four genotypes have been recognized (47). In addition, two putative genotypes of HEV, one genotype from the Norway rat (Rattus norvegicus) (25) and the other from a wild boar (56), were recently reported. In addition to mammalian HEV strains, avian HEV (38) and the newly described cutthroat trout virus represent new genera (3). Among mammalian HEV, genotypes 1 and 2 are primarily associated with fecal-oral transmission among humans which in developing countries can lead to waterborne epidemics of jaundice. Genotypes 3 and 4 circulate in humans and several animal species, and are associated with sporadic infection among humans in industrialized countries (57). Hepatitis E is mostly self-limiting but can progress to fulminant liver failure especially in pregnant women, and chronic HEV infection resulting in cirrhosis has been observed among solid-organ-transplant recipients and other immunosuppressed people (32).
HEV infection and hepatitis E in industrialized countries, including the United States, are more common than previously recognized (33, 58). A study conducted in a large civilian, noninstitutionalized U.S. population found an average anti-HEV-IgG seroprevalence rate of 21%, with a strongly positive age-wise correlation (30). Although HEV may be imported from people who have traveled to regions where HEV is hyperendemic, it can also be acquired by those who have not so traveled or been in contact with travelers.
Non-travel-associated hepatitis E is thought to be a zoonosis, the vertebrate reservoirs identified being pigs and boars (Sus scrofa) and deer (Cervus nippon, Cervus elaphus, and Capreolus rufus). Worldwide, anti-HEV seroprevalence rates in farmed swine are consistently high and, especially among piglets, HEV RNA and infectious virions can be found in blood and stools (38). In Europe and Japan, anti-HEV IgG and HEV RNA are also detected in boars and deer, although not as extensively as in farmed pigs (4, 26, 38, 58). HEV genomic sequences amplified from these three varieties of ungulates are closely related to those from geographically proximate humans, and instances of direct HEV transmission from them to humans have been documented (38, 58). Implicated routes of transmission include (i) the ingestion of their meat and offal as food, (ii) occupational or recreational contact with their body fluids and excreta, (iii) contact with environmental water contaminated with sewage, wastewater, and sludge effluents, and (iv) ingestion of shellfish feeding from such contaminated water (6, 33, 50, 58).
An increasingly disparate variety of other animals has been implicated as playing host to HEV. These include rodents (1, 10, 12, 14, 25, 27, 28, 51, 60, 64), cats (31, 35, 41, 43), dogs (1, 35, 41, 60, 71), horses (14, 16, 49, 71), donkeys (14), goats (14, 16, 45, 52, 71), sheep (5, 45, 61, 66, 68), cattle (1, 5, 14, 21, 60, 63, 71), rabbits (16, 17), nonhuman primates (46, 67), mongooses (34, 42), chickens (18, 23, 52, 60, 71), ducks (16, 71), and an assortment of other birds (71, 72). Except for farmed Rex rabbits (Oryctolagus cuniculus) in China (15, 16, 36), the evidence for HEV as being enzootic in species other than pigs, boars, deer, and chickens is weak, for several reasons. First, HEV RNA or antigen is infrequently, if ever, detected in their blood, bile or feces (1, 14, 16, 25, 35, 51, 63, 69). Although HEV RNA sequences have occasionally been amplified from single animals (71, 72), such detection would not necessarily implicate HEV enzooticity in the species to which those animals belong; rather, they could be accidental hosts or their samples (especially if of fecal origin) might have been contaminated with HEV shed from true maintenance hosts sharing the same habitats (58). Second, the anti-HEV detection rates are widely variable and never as high as in pigs and chickens. Such variability may be due to geographical differences in genus- or species-specific HEV prevalence or to variation in performance characteristics of the assays used for antibody detection.
Many enzyme immunoassays (EIAs) developed for anti-HEV detection are of the indirect format, so require that the second-layer (reporter) antibody be genus specific. This requirement restricts their use in serosurveys across the animal kingdom and also necessitates the production of genus-specific positive controls via immunization of animals representing the genus or species under investigation (45). Moreover, in most assays, the antigens used for antibody capture are derived from HEV genotype 1 or genotypes 1 and 2. Because the HEV genotypes thus far identified to infect animals belong instead to genotype 3, genotype 4, and the newer putative genotypes, the possibility that low antibody detection rates reflect weak binding of antibody to heterotypic antigens rather than true antibody absence (2, 19, 24, 44, 45, 48, 62) cannot be excluded.
In the United States, no studies have investigated HEV infection in animals other than rodents (10, 12, 27, 53), pigs (38, 39), chickens (23), and sika deer (68). To evaluate the extent of HEV enzooticity more broadly, we devised a double-antigen sandwich assay (DASA) for anti-HEV detection that is trans-genus-wide and circumvents the obligatory requirement in indirect assays for the analyte reporter to be an antibody. The antibody capture reagent consists of a cocktail of recombinant HEV proteins derived from all four genotypes. The detector of the bound antibody comprises the same mixture but with the proteins conjugated to a reporter enzyme. After development and validation, DASA was applied to detect anti-HEV in 35 genera of animals that were wild, feral, owned, or otherwise held captive. All samples found DASA-reactive were tested for HEV RNA. The results suggest that HEV is not extensively enzootic in the United States.
MATERIALS AND METHODS
HEV antigen preparation.
HEV antigens (hereafter called p166 antigens) were generated from amino acid positions 452 to 617 of open reading frame 2 (ORF2) of the following strains: HEV-Morocco F86 (genotype 1), Mexico-14 (genotype 2), US-1 (genotype 3), and China-9829 (genotype 4). Each antigen is a 166-amino-acid, truncated capsid protein which in dimeric form contains an immunodominant, conformation-dependent epitope (37). The antigens were tagged with histidine (His) and mixed equimolarly. Another antigenic cocktail was prepared by tagging glutathione S-transferase (GST) to the same mixture of p166 antigens and conjugating them to horseradish peroxidase (HRP) by using a SureLINK HRP conjugation kit (KPL, Gaithersburg, MD) (37, 70).
Study samples.
For assay development and validation, the following serum samples were assembled: 372 sera obtained from U.S. blood donors (BBI Diagnostics, West Bridgewater, MA); 94 sera derived from patients identified with acute hepatitis E in whom anti-HEV IgM and HEV RNA were detected (8); one serum sample each from a rhesus monkey, a pig, a New Zealand White rabbit, a Wistar rat, and a BALB/c mouse derived from blood taken 8 weeks after immunization with recombinant HEV-ORF2 antigens; and four serum panels generated from pigs that were experimentally infected with HEV and followed up weekly for 8 weeks (13). For diagnostic sensitivity determinations, a World Health Organization (WHO) reference reagent for HEV antibody was purchased from the National Institute of Biological Standards and Controls (South Mimms, United Kingdom) (product code 95/584). To determine the anti-HEV detection rate in various animal species, 4,936 serum samples were collected. The diversity and origins of the source animals are summarized in Table 1.
Table 1.
Location, captivity status, and anti-HEV seropositivity rates of study animals
| Animal | Location of habitat (U.S. states) | Captivity status | No. of anti-HEV-positive samples/total no. of samples tested (%) |
|---|---|---|---|
| Alpaca (Lama pacos) | IN | Domesticated (in zoo) | 0/2 |
| Addax (Addax nasomaculatus) | IN | Captive, wildlife (in zoo) | 0/3 |
| Badger (Taxidea taxus) | CA | Wild | 0/2 |
| Bat (Eptesicus fuscus) | Eastern United States | Wild | 0/14 |
| Bear (Ursus americanus) | IN | Captive, wildlife (in zoo) | 0/1 |
| Bison (Bison bison) | WY | Domesticated (as livestock) | 3/65 (4.6) |
| Cat (Felis silvestris) | GA | Domesticated (as pet) | 0/163 |
| GA | Feral | 0/22 | |
| NY | Domesticated (as pet) | 0/14 | |
| Cattle (Bos taurus) | IW, KS, and WY | Domesticated (as livestock) | 174/1,156 (15) |
| Cockatoo (Eolophus sp. and Cacatua sp.) | AZ, CA, FL, IL, MD, and MN | Domesticated (as pet) | 0/10 |
| Colobus (Colubus guereza) | IN | Captive, wildlife (in zoo) | 0/2 |
| Coyotes (Canis latrans) | CA | Wild | 0/302 |
| Deer, white-tailed (Odocoileus virgineanus) | CT and IN | Wild | 0/788 |
| IN | Captive, wildlife (in zoo) | 0/1 | |
| Dog (Canis lupus familiaris) | AL, GA, IL, and NY | Domesticated (as pet) | 2/212 (0.9) |
| Ferret (Mustela putorius furo) | NY | Domesticated (as pet) | 0/1 |
| Fox, gray (Vulpes vulpes) | CA | Wild | 0/1 |
| Horse (Equus caballus) | IW | Domesticated (as pet or for sport) | 0/201 |
| Kangaroo (Macropus giganticus) | IN | Domesticated (in zoo) | 0/1 |
| Mountain lion (Felis concolor) | CA | Wild | 0/3 |
| Macaw (Ara sp.) | FL, IL, KY, MI, PA, TN, TX, and WA | Domesticated (as pet) | 0/10 |
| Muntjac (Muntiacus reevesi) | IN | Captive, wildlife (in zoo) | 0/1 |
| Opossum (Didelphis virginiana) | CT | Wild | 0/21 |
| Ostrich (Struthio camelus) | IN | Captive, wildlife (in zoo) | 0/1 |
| Otter, river (Lontra canadensis) | CA | Wild | 0/15 |
| Parrot, African gray (Psittacus erithacus) | AZ, CA, FL, KY, NH, and PA | Domesticated (as pet) | 0/10 |
| Parrot, Amazon (Amazona sp.) | FL, GA, IL, MD, and MI | Domesticated (as pet) | 0/10 |
| Rabbit, eastern cottontail (Sylvilagus floridanus) | TX | Wild | 0/14 |
| Rabbit, blacktailed (Lepus californicus) | TX | Wild | 0/156 |
| Raccoon (Procyon lotor) | CA | Wild | 0/313 |
| Rat, Norway (Rattus norvegicus) | MD | Wild | 2/318 (0.6) |
| Skunk (Mephitis mephitis) | Eastern United States | Wild | 0/92 |
| Squirrel (Sciurus carolinensis) | CT | Wild | 0/63 |
| Swine (Sus scrofa) | AK, FL, GA, NC, SC, and TX | Feral | 9/306 (2.9) |
| Swine (Sus scrofa domesticus) | IW | Domesticated (as livestock) | 267/648 (41.2) |
| Wallaby (Macropus rufogriseus) | IN | Captive, wildlife (in zoo) | 0/5 |
| Warthog (Phacochoerus aethiopicus) | IN | Captive, wildlife (in zoo) | 0/1 |
| Total | 457/4,936 (9.3) |
Anti-HEV detection.
Wells of flat-bottom, 96-well, Nunc Polysorp microplates were coated with His-tagged p166 antigens (0.5 μg/ml, 100 μl/well) and incubated at room temperature overnight. Unbound antigens were washed with 10 mM phosphate-buffered saline containing 0.05% Tween 20 (PBS-T). To each well, undiluted test serum (100 μl/well) was added. The plates were then incubated at 37°C for 1 h. After a washing step with PBS-T, 100 μl of the GST-tagged, HRP-conjugated p166 antigens was added, and the plates incubated at 37°C for 1 h. After washing, tetramethylbenzidine was added as substrate and, following color development, optical density (OD) readings were read spectrophotometrically. A signal/cutoff (s/co) value of ≥1 was considered a positive reaction. Human sera and dilutions of the WHO reagent were tested in duplicate. Samples from animals, which were often made available for the present study in small volumes, were tested once.
HEV genome detection and characterization.
Two sets of HEV universal primers were designed to amplify RNA from conserved regions of the viral genome. One set was used for amplification of a 578-bp segment of ORF1, and the other set used a 643-bp segment from ORF2. The sequences and positions of the primers are listed in Table 2. Briefly, total RNA was extracted from 100 μl of serum and eluted in 50 μl of elution buffer. Reverse transcription and first-round PCR were performed using a Qiagen OneStep RT-PCR kit. Reverse transcription was carried out at 50°C for 45 m and terminated by heating at 95°C for 15 m, after which PCR consisting of 35 cycles proceeded, with each cycle consisting of denaturation at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 80 s, with a final extension at 72°C for 10 min. Next, 3 μl of the first-round PCR product was submitted for nested PCR using Taq DNA polymerase (Roche) consisting of 35 cycles, with each cycle involving denaturation at 94°C for 30 min, annealing at 56°C for 30 s, extension at 72°C for 80 s, and a final extension at 72°C for 10 min. Amplicons were purified and sequenced by using a Applied Biosystems BigDye v3.1 sequencing kit and an Applied Biosystems 3130 genetic analyzer. Sequence analysis was conducted by using SeqMan and MegAlign programs from the Lasergene DNA and protein analysis software (version 7.0). Phylogenetic trees were constructed by using the neighbor-joining method (MEGA version 4.0).
Table 2.
Nucleotide sequences and positions of primers used for RT-PCR amplification and sequencing of HEV RNA
| Function | Location in HEV genome | Primera | Nucleotide sequence (5′–3′) | Positionsb |
|---|---|---|---|---|
| Reverse and first-round PCR | ORF1 | JM502(F) | AGGCCCAYCAGTTYATWAAGGCTC | 32–55 |
| JM505(R) | TASCCWGCACTAGWGTCMCCCTC | 713–691 | ||
| ORF2 | JM10(F) | GAYGGSACYAAYACYCATATWATGGC | 5648–5673 | |
| JM34(R) | TYGGCTCGCCATTGGCYGAGAC | 6371–6350 | ||
| Nested PCR and sequencing | ORF1 | JM503(F) | CTGGCRTYACWACTGCYATTGAGC | 56–79 |
| JM504(R) | TRCCRGGKGGKARCAGSACCTC | 631–610 | ||
| ORF2 | JM11(F) | GARGCWTCWAATTAYGCCCAGTAYCG | 5678–5703 | |
| JM33(R) | CAGCCGACGAAATCAATTCTGTCG | 6320–6297 |
F, forward; R, reverse.
Numbered according to the nucleotide sequence of HEV Bur82 (GenBank accession number M73218).
RESULTS
DASA development and validation.
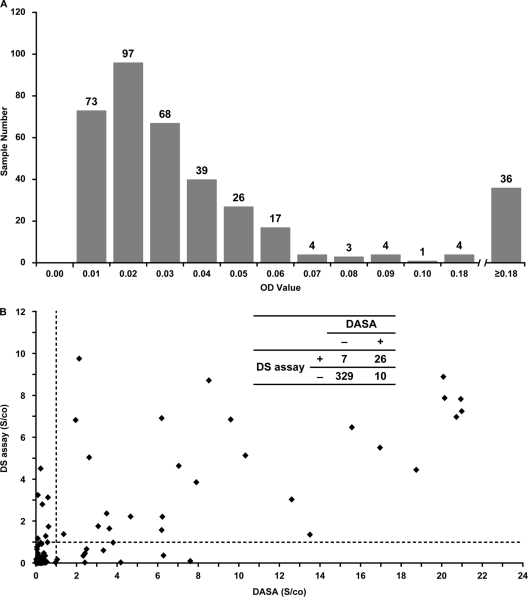
(i) Setting the assay cutoff. Initial testing by DASA of 372 blood donor samples showed that OD values ranged from 0 to 3.7 (Fig. 1A). For the purpose of calculating the DASA cutoff value, the same panel of samples was tested for reactivity in the Diagnostic Systems (DS) anti-HEV-IgG EIA (Saronno, Italy). The specificity and sensitivity of the HEV antigens incorporated in the DS assay for anti-HEV detection had been evaluated earlier (9). A total of 33 samples were found to be reactive in the DS assay, and 5 samples that exhibited OD values of >0.5 after testing by DASA were excluded. The cutoff was then set as the average OD of the remaining 334 samples plus three times the standard deviation, i.e., 0.03 + (3 × 0.049), to give a value of 0.18.
Fig. 1.
Comparison of DASA and DS EIA reactivities. (A) Distribution of signal/cutoff OD values. Numerals atop bars denote sample numbers. (B) Correlation between signal cutoff values. (Inset: 2 × 2 table showing concordance of detection).
(ii) Diagnostic specificity determination. Of 372 blood donor samples, 36 were determined to be anti-HEV-positive by DASA based on the established cutoff of 0.18 (Fig. 1B). The concordance between DASA and DS reactivities was 95.4%, and the diagnostic specificity of DASA determined to be 97.1%.
(iii) Diagnostic and analytic sensitivity determinations. A total of 94 serum samples collected from patients with hepatitis E were used to constitute an anti-HEV-positive panel. All of the samples in this panel were reactive by DASA, indicating a diagnostic sensitivity of 100% (confidence interval = 94 to 100%). To evaluate the analytic sensitivity, DASA was applied to serial, 2-fold dilutions of the WHO HEV-antibody reference reagent. The endpoint reactivity was determined as 0.062 U/ml.
(iv) Trans-genus detection and quantification of immune sera of laboratory animals. Immune sera, obtained from a rhesus monkey, pig, rabbit, rat, and mouse that had been immunized with HEV antigens, were tested by DASA. With reference to the WHO reference reagent dilution curve, the sera were determined to contain, respectively, 743, 206, 1,076, 1,154, and 1,063 U/ml of anti-HEV.
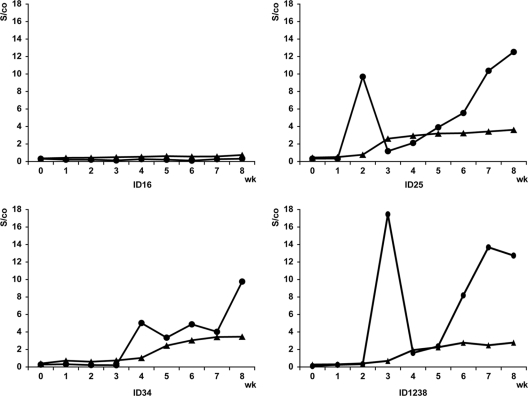
Seroconversion detection in swine.
Four seroconversion panels assembled from pigs experimentally infected with HEV that were sampled weekly postinoculation were tested by DASA, and the reactivities were compared to an indirect EIA that uses as an antigen the 55-kDa-capsid protein derived from Sar-55 HEV strain expressed from a recombinant baculovirus, hereafter referred to as the 55-kDa-EIA (13). As shown in Fig. 2, the control, ID16, remained anti-HEV negative at all of the time points tested, while ID25, ID34, and ID1238, which were inoculated with human genotype 3, human genotype 4, and swine genotype 3 HEV, respectively, showed evidence of seroconversion. In these three pigs, although anti-HEV was first detected at about the same time points by both DASA and the 55-kDa-EIA (between the second and fourth weeks postinoculation), the s/co values obtained from DASA were mostly higher than those from the 55k-Da-EIA. A more striking difference between the two assays is that for DASA the s/co values at the first time point for each of the three pigs (week 2 for ID25, week 4 for ID34, and week 3 for ID238) were higher than those generated by the 55-kDa-EIA. These initial DASA s/co values declined by the following week and thereafter rose again (Fig. 2).
Fig. 2.
Anti-HEV reactivities in swine seroconversion panels. Symbols: •, DASA; ▴, 55-kDa-EIA. ID16, uninoculated control; ID25, ID34, and ID1238, inoculated with human genotype 3, human genotype 4, and swine genotype 3 HEV, respectively.
DASA reactivity in diverse animal groups.
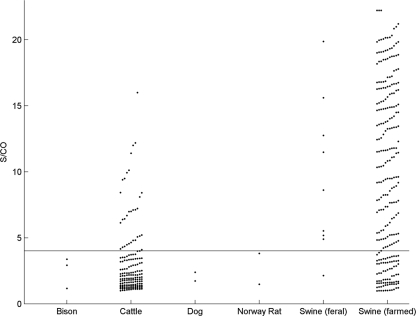
A total of 4,936 serum samples drawn from various animals were tested by DASA. Positivity was determined in 457 samples from five species (Table 1). The positivity rates among farmed swine, feral swine, bison, cattle, dogs, and Norway rats were, respectively, 41.2, 2.9, 4.6, 15, 0.9, and 0.6%. The highest s/co values were obtained from porcine and bovine samples, i.e., 70 and 20%, respectively, yielding s/co values of >4 (Fig. 3). The values from samples from bison, dogs, and rats were all <4.
Fig. 3.
Distribution of DASA reactivities in five animal species. Horizontal line indicates the s/co of 4.
HEV genomes in DASA-reactive animal samples.
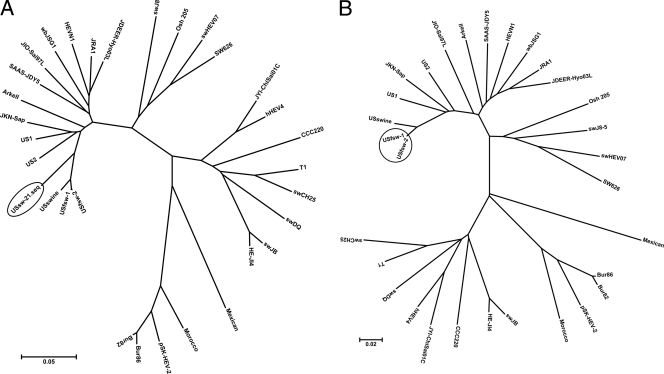
A total of 457 DASA-reactive samples were processed for reverse transcription-PCR (RT-PCR) to amplify HEV RNA from ORFs 1 and 2. Both fragments were amplified from two feral swine samples, designated USfsw-1 and USfsw-2. The ORF1 fragment was only amplified from one sample from a farmed pig, designated USsw-21. Phylogenetic analysis following nucleotide sequencing of the amplicons indicated that USfsw-1 and USfsw-2 belonged to genotype 3, sharing 99.2 and 100% identities with each other in the ORF1 and ORF2 regions, respectively (Fig. 4). The USsw-21 strain also belonged to genotype 3 and showed 91.9 and 91.7% ORF-1 sequence identities to USfsw-1 and USfsw-2, respectively. In the ORF1 region amplified (Fig. 4A), comparisons with GenBank HEV sequences indicated that the sequences from the three new HEV strains were more closely related to sequences from HEV strains known to circulate in the United States than to those elsewhere. Thus, USfsw-1 and USfsw-2 showed 89.8 and 90.5% identities with US1, 89.8 and 90.5% identities with US2, and 97 and 97.4% identities with US swine, respectively. USsw-21 shared 89.3, 88.9, and 92.7% identities with US1, US2, and US swine. For ORF2 (Fig. 4B), the USfsw-1/USfsw-2 sequence was 90.6, 90.1, and 97.5% identical to US1, US2, and US swine, respectively.
Fig. 4.
Phylogenetic trees comparing sequence similarities of USfsw-1 (GenBank accession numbers JN 701455 and JN 701453), USfsw-2 (GenBank accession numbers JN 701456 and JN 701454), and USsw-21 (GenBank accession number JN 701457) to representative GenBank sequences. (A) ORF1; (B) ORF2.
DISCUSSION
Cocirculation of genotype 3 and 4 HEV strains among humans and animals provides opportunities for cross-species transmission and may underlie the occurrences of non-travel-associated hepatitis E in industrialized countries, including the United States (38, 58). To effect programs for the surveillance of HEV infection in humans and animals, several molecular and serological approaches have been developed and applied. Molecular techniques such as RT-PCR generate data that identify active HEV infection by revealing the presence of the HEV genome in the tissues, body fluid, and excreta of the infected hosts. Viremia and fecal HEV shedding are mostly transient, however, and persistent HEV infection is rare, so the likelihood of identifying active infection in a person or an animal in cross-sectional investigations is small. Thus, in the present study, only 3 of 457 animals determined to be DASA-reactive were found to carry HEV RNA in their blood.
Serological EIAs are widely used for detection of anti-HEV, a marker of prevalent HEV infection (29). Most of these EIAs employ for antibody capture antigens that are derived from HEV genotypes 1 and 2 and so may not be sufficiently sensitive to detect heterotypic antibodies generated in human and animal hosts infected by genotype 3 (19, 24, 45, 48) or genotype 4 (2, 23, 62). Moreover, some indirect EIAs tend to generate false-positive reactivities (52, 65) and therefore require, in order to validate the specificity of their reactivities, the extra steps of neutralization, immunoblotting, or prior production of genus-specific positive controls (1, 12, 35, 45). To circumvent such problems inherent to indirect EIAs, DASAs have been developed. A principal advantage of DASAs is that they enable transgenus antibody detection. Another advantage is that sensitivity is potentiated by the detection of total rather than class-specific antibodies. DASAs have particular capability to detect IgM. Being decavalent, IgM substantially amplifies DASA OD readings, because for each pentameric immunoglobulin complex from which one Fab end has bound to the capture antigen, nine other Fab ends are potentially free to bind to the reporter antigen. Thus, in the course of testing the porcine seroconversion panels, we observed peaks of DASA s/co values in the immediate few weeks after experimental infection (Fig. 2), reflecting the possible detection of anti-HEV-IgM that were being transiently generated during acute infection.
The DASA we developed has several unique features. First, it incorporates antigens representing all four HEV genotypes, so allowing for the detection of antibodies generated in the host regardless of the genotype of the infecting HEV. Other DASAs developed for anti-HEV detection (14, 22, 55, 63) mostly utilize antigenic preparations derived from one genotype only. Second, the highly immunodominant p166 antigens (37) are used to capture antibody, as well as to report its solid-phase capture, thereby conferring both sensitivity and specificity to anti-HEV detection. The fine specificity is exemplified in the low background OD readings generated from the blood-donor samples (Fig. 1), thereby lending DASA the capability to test samples without predilution. Testing samples undiluted preserves sensitivity by conserving the original concentration of the analyte and facilitates ease of use for field studies. Third, the antigen mixture utilized for antibody capture was His tagged, whereas the mixture for antibody detection was GST tagged. Such a design was conceived to enhance the specificity of the DASA by minimizing solid-phase capture of antibody against His or GST that might be carried in the test sample.
Owing to the absence of gold standards for HEV serology, evaluating the performance characteristics of the DASA described here required various assemblages of test samples and reagents. Considering that as much as a fifth of the U.S. general population may be anti-HEV positive (30), it would be inappropriate, for the purpose of establishing the DASA cutoff, to include OD reactivities generated from every U.S. blood donor serum sample acquired. Therefore, only those of blood donor samples that were unreactive in the DS assay and unreactive or weakly reactive in DASA were included. The established cutoff value was then used as basis to determine the diagnostic specificity of DASA. Next, to determine diagnostic sensitivity, we incorporated another panel, constituted from sera derived from people who were undergoing acute hepatitis E. Although such a panel would not represent prevalent HEV infection, the presence of HEV RNA assured that its constituents originated from truly HEV-infected hosts. Nonetheless, such a panel preferentially favors the detection of IgM, so the ability of DASA to identify monomeric immunoglobulins may not have been fully assessed. An evaluation using sera obtained from five species of laboratory animals that had been immunized with HEV proteins provided proof of concept that DASA can detect anti-HEV generated across different genera and species. Lastly, the added usage of the WHO reference reagent permitted the analytic sensitivity of our DASA to be determined; this reagent was also used as a standard against which the content of anti-HEV in animal sera could be measured.
HEV-infected animals potentially serve as reservoirs of infection to other animals and to humans. Since the discovery of the first porcine HEV strain (39), evidence has mounted to indicate that HEV is epizootic in swine (38). The present study reveals a DASA reactivity rate of 41% among farmed pigs, which is consistent with anti-HEV seropositivity rates determined in other studies (ranging between 15 and 90%). Because our study samples were collected from adult pigs, it is not unexpected to find just one sample to carry HEV RNA, since the majority of pigs in the United States are infected between 2 and 4 months of age (39). The HEV seroprevalence in feral pigs, in contrast to farmed ones, has not been reported in the United States. We found that 3% of the feral pigs sampled were DASA reactive, and these seropositive pigs were collected in the vicinity of domestic swine farms (data not published). The seropositivity proportion is substantially lower than in farmed pigs, which not only suggests that HEV is less prevalent among feral than farmed pigs but implicates the pig-farming environment as potentially fostering HEV spread among swine. Such disparity might be due to geographical differences in the endemicity of HEV in farmed pigs and the proximity of farmed to feral pigs, which would facilitate cross-transmission of microbial agents (7). Phylogenetic analyses indicated that all of the three newly identified porcine HEV strains belonged to genotype 3 and were most closely related to human and porcine HEV strains in the United States than to those known to circulate elsewhere. These findings suggest the potential in the United States for HEV transmission between farmed and feral swine, as well as cross-species transmission from them to humans (and vice versa).
Subclinical human infection with attenuated HEV strains such as porcine HEV might explain the high anti-HEV seropositivity rates among people living in developed countries (33, 38, 58). Nonetheless, much of the U.S. population, being predominantly urban (59), is neither likely to come into frequent contact with swine or their excreta nor disposed to adopt eating habits that would entail ingestion of raw or inadequately cooked pork, other pig meat, or offal including liver (6, 20, 54, 58). These considerations suggest that even if swine are maintenance hosts for HEV, they may not necessarily serve as the only reservoir of HEV infection (47).
Accordingly, our studies extended to determine whether HEV can infect animals other than swine. Given the enormous diversity of the animal kingdom, exhaustive sampling was not possible. Emphasis was placed on investigating animals (pigs, cattle, deer, bison, horses, cats, coyotes, dogs, rats, psittacines, raccoons, skunks, and squirrels) which share habitats or are in frequent contact with humans or whose meat and offal may be eaten by humans. Testing samples from the more exotic animals (alpacas, addaxes, badgers, bats, bear, colobuses, ferret, fox, kangaroo, mountain lions, macaws, muntjac, ostrich, otters, wallabies, and warthog) was primarily to explore the possible broader tropism of HEV, but samples available from them were limited.
Among the nonporcine samples tested, anti-HEV positivity was found only among cattle, bison, dogs and Norway rats, the rates being 15, 4.6, 0.9, and 0.6%, respectively, with HEV RNA amplified from none. Since the samples from cattle, bison, dogs, and rats yielded weak OD readings (Fig. 3), the possibility that their DASA reactivities were not specific to anti-HEV cannot be excluded. It is also possible that the viruses infecting these species may be more distantly related, antigenically and genetically, to genotypes 1 to 4 of mammalian HEV, thus resulting in weaker reactivity.
The near absence of DASA reactivity among the rats sampled was unexpected. Previous HEV seroprevalence studies of rodents in the United States have yielded contradictory results. An early study of feral rats trapped in Louisiana, Maryland, and Hawaii found anti-HEV-seropositivity rates ranging between 44 and 90%, the seropositive species being Rattus norvegicus, R. rattus, and R. exulans (27). In a subsequent investigation of 26 species of wild U.S. rodents, the highest seropositivity rate (60%) was found in Rattus, and rodents caught in urban areas displayed a significantly higher rate than those in rural areas (12). Norway rats sampled in Los Angeles yielded a 14% seropositivity rate (53). Yet another study, conducted in North Carolina, found none of house mice (Mus musculus domesticus) and Norway rats trapped in pig farms to be seropositive (64). However, an investigation conducted in Baltimore found anti-HEV among 75% of Norway rats sampled (10). Our 0.2% seropositivity rate was obtained among Norway rats that also originated from Baltimore. The poor concordances in anti-HEV seropositivity rates obtained from these various studies likely reflect variability in the performance of the serological assays applied, although geographical and sampling variations may contribute. A recent publication from Germany reported the detection of HEV RNA in two Norway rats, but the nucleotide sequences generated were only 60% related to human HEV strains (25). The ability of such divergent HEV strains to effect cross-species transmission to humans seems remote. Nonetheless, the extensive sequence divergence between rat HEV and genotypes 1 to 4 of mammalian HEVs may also explain the lower anti-HEV-seropositivity rate among rats because of the limited antigenic cross-reactivity to rat HEV, since the DASA is based on mammalian HEV genotypes 1 to 4. Nonetheless, considering that peridomestic rodent infestation of human dwellings is rife (40), definitive evidence of HEV infection in rodents would need to be sought (57).
The development of a DASA for the detection of anti-HEV is described here. Its performance characteristics were validated to be sufficiently specific and sensitive for trans-genus HEV seroprevalence studies. When the assay was applied to nearly 5,000 sera collected from animals representing 35 genera sampled from the United States, the rate and extent of reactivity were substantial only among the porcine samples, in which HEV-specific nucleotide sequences also were detected. Owing to the limited numbers of samples available from the more exotic species of animals, the status of HEV infection in them would require further studies when more samples become available. The findings thus far obtained are consistent with limited enzooticity of HEV in the United States. Animals other than swine are unlikely maintenance hosts to HEV and so would not play appreciable roles as reservoirs of its transmission.
ACKNOWLEDGMENTS
We thank N. Rasuleva for technical assistance and G. Anderson, C. Rupprecht, and Furkids for assistance with the sourcing of test samples.
G.E.G. was supported by NSF grant EF0525751.
Footnotes
Published ahead of print on 12 October 2011.
REFERENCES
- 1. Arankalle V. A., et al. 2001. Prevalence of anti-hepatitis E virus antibodies in different Indian animal species. J. Viral Hepat. 8:223–227 [DOI] [PubMed] [Google Scholar]
- 2. Arankalle V. A., Lole K. S., Deshmukh T. M., Chobe L. P., Gandhe S. S. 2007. Evaluation of human (genotype 1) and swine (genotype 4)-ORF2-based ELISAs for anti-HEV IgM and IgG detection in an endemic country and search for type 4 human HEV infections. J. Viral Hepat. 14:435–445 [DOI] [PubMed] [Google Scholar]
- 3. Batts W., Yun S., Hedrick R., Winton J. 2011. A novel member of the family Hepeviridae from cutthroat trout (Oncorhynchus clarkii). Virus Res. 158:116–123 [DOI] [PubMed] [Google Scholar]
- 4. Boadella M., et al. 2010. Increasing contact with hepatitis E virus in red deer, Spain. Emerg. Infect. Dis. 16:1994–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chang Y., et al. 2009. Zoonotic risk of hepatitis E virus (HEV): a study of HEV infection in animals and humans in suburbs of Beijing. Hepatol. Res. 39:1153–1158 [DOI] [PubMed] [Google Scholar]
- 6. Colson P., et al. 2010. Pig liver sausage as a source of hepatitis E virus transmission to humans. J. Infect. Dis. 202:825–834 [DOI] [PubMed] [Google Scholar]
- 7. Corn J. L., Cumbee J. C., Barfoot R., Erickson G. A. 2009. Pathogen exposure in feral swine populations geographically associated with high densities of transitional swine premises and commercial swine production. J. Wildl. Dis. 45:713–721 [DOI] [PubMed] [Google Scholar]
- 8. Dong C., Dai X., Shao J. S., Hu K., Meng J. H. 2007. Identification of genetic diversity of hepatitis E virus (HEV) and determination of the seroprevalence of HEV in eastern China. Arch. Virol. 152:739–746 [DOI] [PubMed] [Google Scholar]
- 9. Drobeniuc J., et al. 2010. Serologic assays specific to immunoglobulin M antibodies against hepatitis E virus: pangenotypic evaluation of performances. Clin. Infect. Dis. 51:e24–e27 [DOI] [PubMed] [Google Scholar]
- 10. Easterbrook J. D., et al. 2007. A survey of zoonotic pathogens carried by Norway rats in Baltimore, Maryland, U.S.A. Epidemiol. Infect. 135:1192–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Emerson S. U., et al. 2004. Hepevirus, p. 853–857. In Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. (ed.), Virus Taxonomy. Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, London, England [Google Scholar]
- 12. Favorov M. O., Kosoy M. Y., Tsarev S. A., Childs J. E., Margolis H. S. 2000. Prevalence of antibody to hepatitis E virus among rodents in the United States. J. Infect. Dis. 181:449–455 [DOI] [PubMed] [Google Scholar]
- 13. Feagins A. R., Opriessnig T., Huang Y. W., Halbur P. G., Meng X. J. 2008. Cross-species infection of specific-pathogen-free pigs by a genotype 4 strain of human hepatitis E virus. J. Med. Virol. 80:1379–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fu H., et al. 2010. Hepatitis E virus infection among animals and humans in Xinjiang, China: possibility of swine to human transmission of sporadic hepatitis E in an endemic area. Am. J. Trop. Med. Hyg. 82:961–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Geng J., et al. 2011. Study on prevalence and genotype of hepatitis E virus isolated from Rex Rabbits in Beijing, China. J. Viral Hepat. 18:661–667 [DOI] [PubMed] [Google Scholar]
- 16. Geng J. B., et al. 2010. Hepatitis E virus (HEV) genotype and the prevalence of anti-HEV in 8 species of animals in the suburbs of Beijing. Zhonghua Liu Xing Bing Xue Za Zhi 31:47–50 (In Chinese.) [PubMed] [Google Scholar]
- 17. Geng Y., et al. 2011. The serological prevalence and genetic diversity of hepatitis E virus in farmed rabbits in China. Infect. Genet. Evol. 11:476–482 [DOI] [PubMed] [Google Scholar]
- 18. Haqshenas G., Shivaprasad H. L., Woolcock P. R., Read D. H., Meng X. J. 2001. Genetic identification and characterization of a novel virus related to human hepatitis E virus from chickens with hepatitis-splenomegaly syndrome in the United States. J. Gen. Virol. 82:2449–2462 [DOI] [PubMed] [Google Scholar]
- 19. Herremans M., Duizer E., Jusic E., Koopmans M. P. 2007. Detection of hepatitis E virus-specific immunoglobulin a in patients infected with hepatitis E virus genotype 1 or 3. Clin. Vaccine Immunol. 14:276–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Horowitz R. 2006. Putting meat on the American table: taste, technology, transformation. Johns Hopkins University Press, Baltimore, MD [Google Scholar]
- 21. Hu G. D., Ma X. 2010. Detection and sequences analysis of bovine hepatitis E virus RNA in Xinjiang Autonomous region. Bing Du Xue Bao. 26:27–32 (In Chinese.) [PubMed] [Google Scholar]
- 22. Hu W. P., et al. 2008. Double-antigen enzyme-linked immunosorbent assay for detection of hepatitis E virus-specific antibodies in human or swine sera. Clin. Vaccine Immunol. 15:1151–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang F. F., et al. 2002. Heterogeneity and seroprevalence of a newly identified avian hepatitis e virus from chickens in the United States. J. Clin. Microbiol. 40:4197–4202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jimenez de Oya N., et al. 2009. Serological immunoassay for detection of hepatitis E virus on the basis of genotype 3 open reading frame 2 recombinant proteins produced in Trichoplusia ni larvae. J. Clin. Microbiol. 47:3276–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johne R., et al. 2010. Detection of a novel hepatitis E-like virus in faeces of wild rats using a nested broad-spectrum RT-PCR. J. Gen. Virol. 91:750–758 [DOI] [PubMed] [Google Scholar]
- 26. Kaba M., Davoust B., Marie J. L., Colson P. 2010. Detection of hepatitis E virus in wild boar (Sus scrofa) livers. Vet. J. 186:259–261 [DOI] [PubMed] [Google Scholar]
- 27. Kabrane-Lazizi Y., et al. 1999. Evidence for widespread infection of wild rats with hepatitis E virus in the United States. Am. J. Trop. Med. Hyg. 61:331–335 [DOI] [PubMed] [Google Scholar]
- 28. Karetnyi Iu V., et al. 1993. The possible involvement of rodents in the spread of viral hepatitis E. Zh. Mikrobiol. Epidemiol. Immunobiol. 4:52–56 (In Russian.) [PubMed] [Google Scholar]
- 29. Khudyakov Y., Kamili S. 2011. Serological diagnostics of hepatitis E virus infection. Virus Res. 161:84–92 [DOI] [PubMed] [Google Scholar]
- 30. Kuniholm M. H., et al. 2009. Epidemiology of hepatitis E virus in the United States: results from the Third National Health and Nutrition Examination Survey, 1988–1994. J. Infect. Dis. 200:48–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kuno A., et al. 2003. Sporadic acute hepatitis E of a 47-year-old man whose pet cat was positive for antibody to hepatitis E virus. Hepatol. Res. 26:237–242 [DOI] [PubMed] [Google Scholar]
- 32. Labrique A., Kuniholm M. H., Nelson K. E. 2010. The global impact of hepatitis E: new horizons for an emerging virus, p. 54–92. In Grayson L. (ed.), Emerging infections 9. American Society for Microbiology, Washington, DC [Google Scholar]
- 33. Lewis H. C., Wichmann O., Duizer E. 2010. Transmission routes and risk factors for autochthonous hepatitis E virus infection in Europe: a systematic review. Epidemiol. Infect. 138:145–166 [DOI] [PubMed] [Google Scholar]
- 34. Li T. C., et al. 2006. Serologic evidence for hepatitis E virus infection in mongoose. Am. J. Trop. Med. Hyg. 74:932–936 [PubMed] [Google Scholar]
- 35. Liu J., et al. 2009. Prevalence of antibody to hepatitis E virus among pet dogs in the Jiang-Zhe area of China. Scand. J. Infect. Dis. 41:291–295 [DOI] [PubMed] [Google Scholar]
- 36. Ma H., et al. 2010. Experimental infection of rabbits with rabbit and genotypes 1 and 4 hepatitis E viruses. PLoS One 5:e9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meng J., et al. 2001. Identification and characterization of the neutralization epitope(s) of the hepatitis E virus. Virology 288:203–211 [DOI] [PubMed] [Google Scholar]
- 38. Meng X. J. 2010. Hepatitis E virus: animal reservoirs and zoonotic risk. Vet. Microbiol. 140:256–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Meng X. J., et al. 1997. A novel virus in swine is closely related to the human hepatitis E virus. Proc. Natl. Acad. Sci. U. S. A. 94:9860–9865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miller J. E., et al. 1994. Rodents, p. B1–B187. In Hygnstrom S. E., Timm R. M., Larson G. E. (ed.), Prevention and control of wildlife damage, vol. 1 University of Nebraska Press, Lincoln [Google Scholar]
- 41. Mochizuki M., et al. 2006. Epidemiological study of hepatitis E virus infection of dogs and cats in Japan. Vet. Rec. 159:853–854 [PubMed] [Google Scholar]
- 42. Nakamura M., et al. 2006. Hepatitis E virus infection in wild mongooses of Okinawa, Japan: demonstration of anti-HEV antibodies and a full-genome nucleotide sequence. Hepatol. Res. 34:137–140 [DOI] [PubMed] [Google Scholar]
- 43. Okamoto H., Takahashi M., Nishizawa T., Usui R., Kobayashi E. 2004. Presence of antibodies to hepatitis E virus in Japanese pet cats. Infection 32:57–58 [DOI] [PubMed] [Google Scholar]
- 44. Pan J. S., et al. 2010. Application of truncated immunodominant polypeptide from hepatitis E virus (HEV) ORF2 in an assay to exclude nonspecific binding in detecting anti-HEV immunoglobulin M. J. Clin. Microbiol. 48:779–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Peralta B., et al. 2009. Anti-HEV antibodies in domestic animal species and rodents from Spain using a genotype 3-based ELISA. Vet. Microbiol. 137:66–73 [DOI] [PubMed] [Google Scholar]
- 46. Purcell R. H., Emerson S. U. 2001. Animal models of hepatitis A and E. ILAR J. 42:161–177 [DOI] [PubMed] [Google Scholar]
- 47. Purdy M. A., Khudyakov Y. E. 2011. The molecular epidemiology of hepatitis E virus infection. Virus Res. 161:31–39 [DOI] [PubMed] [Google Scholar]
- 48. Rose N., et al. 2010. The use of Bayesian methods for evaluating the performance of a virus-like particles-based ELISA for serology of hepatitis E virus infection in swine. J. Virol. Methods 163:329–335 [DOI] [PubMed] [Google Scholar]
- 49. Saad M. D., et al. 2007. Hepatitis E virus infection in work horses in Egypt. Infect. Genet. Evol. 7:368–373 [DOI] [PubMed] [Google Scholar]
- 50. Said B., et al. 2009. Hepatitis E outbreak on cruise ship. Emerg. Infect. Dis. 15:1738–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shao Z. J., et al. 2009. Epidemiological screening for hepatitis E virus in bile specimens from livestock in northwest China. J. Clin. Microbiol. 47:814–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shukla P., Chauhan U. K., Naik S., Anderson D., Aggarwal R. 2007. Hepatitis E virus infection among animals in northern India: an unlikely source of human disease. J. Viral Hepat. 14:310–317 [DOI] [PubMed] [Google Scholar]
- 53. Smith H. M., et al. 2002. Prevalence study of antibody to ratborne pathogens and other agents among patients using a free clinic in downtown Los Angeles. J. Infect. Dis. 186:1673–1676 [DOI] [PubMed] [Google Scholar]
- 54. Smoler R. W. 1990. The useful pig: 150 succulent pork recipes. Harper-Collins, New York, NY [Google Scholar]
- 55. Takahashi K., et al. 2007. An easy ELISA for detecting antibodies to hepatitis E virus irrespective of animal species. Kanzo 48:338–340 [Google Scholar]
- 56. Takahashi M., Nishizawa T., Sato H., Sato Y., Jirintai, Nagashima S., Okamoto H. 2011. Analysis of the full-length genome of a hepatitis E virus isolate obtained from a wild boar in Japan that is classifiable into a novel genotype. J. Gen. Virol. 92:902–908 [DOI] [PubMed] [Google Scholar]
- 57. Teo C. G. 2007. The two clinico-epidemiological forms of hepatitis E. J. Viral Hepat. 14:295–297 [DOI] [PubMed] [Google Scholar]
- 58. Teo C. G. 2010. Much meat, much malady: changing perceptions of the epidemiology of hepatitis E. Clin. Microbiol. Infect. 16:24–32 [DOI] [PubMed] [Google Scholar]
- 59. Theobald D. M. 2001. Land-use dynamics beyond the American urban fringe. Geographical Rev. 91:544–564 [Google Scholar]
- 60. Vitral C. L., et al. 2005. Serological evidence of hepatitis E virus infection in different animal species from the Southeast of Brazil. Mem. Inst. Oswaldo Cruz 100:117–122 [DOI] [PubMed] [Google Scholar]
- 61. Wang Y., Ma X. 2010. Detection and sequences analysis of sheep hepatitis E virus RNA in Xinjiang Autonomous region. Wei Sheng Wu Xue Bao 50:937–941 (In Chinese.) [PubMed] [Google Scholar]
- 62. Wang Y., et al. 2001. Detection of sporadic cases of hepatitis E virus (HEV) infection in China using immunoassays based on recombinant open reading frame 2 and 3 polypeptides from HEV genotype 4. J. Clin. Microbiol. 39:4370–4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang Y. C., et al. 2002. Prevalence, isolation, and partial sequence analysis of hepatitis E virus from domestic animals in China. J. Med. Virol. 67:516–521 [DOI] [PubMed] [Google Scholar]
- 64. Withers M. R., et al. 2002. Antibody levels to hepatitis E virus in North Carolina swine workers, non-swine workers, swine, and murids. Am. J. Trop. Med. Hyg. 66:384–388 [DOI] [PubMed] [Google Scholar]
- 65. Wu F. B., Ouyan H. Q., Tang X. Y., Zhou Z. X. 2008. Double-antigen sandwich time-resolved immunofluorometric assay for the detection of anti-hepatitis C virus total antibodies with improved specificity and sensitivity. J. Med. Microbiol. 57:947–953 [DOI] [PubMed] [Google Scholar]
- 66. Wu J. Y., Kang Q., Bai W. S., Bai Z. H. 2010. Seroepidemiological survey of sheep hepatitis E virus infection in Aksu region of Xinjiang Autonomous. Bing Du Xue Bao. 26:234–237 (In Chinese.) [PubMed] [Google Scholar]
- 67. Yamamoto H., et al. 2008. Serological evidence for hepatitis e virus infection in laboratory monkeys and pigs in animal facilities in Japan. Exp. Anim. 57:367–376 [DOI] [PubMed] [Google Scholar]
- 68. Yu C., et al. 2007. Using improved technology for filter paper-based blood collection to survey wild Sika deer for antibodies to hepatitis E virus. J. Virol. Methods 142:143–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yu Y., et al. 2009. Seroepidemiology and genetic characterization of hepatitis E virus in the northeast of China. Infect. Genet. Evol. 9:554–561 [DOI] [PubMed] [Google Scholar]
- 70. Zhang H., Dai X., Shan X., Meng J. 2008. The Leu477 and Leu613 of ORF2-encoded protein are critical in forming neutralization antigenic epitope of hepatitis E virus genotype 4. Cell Mol. Immunol. 5:447–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhang W., et al. 2008. Hepatitis E virus infection among domestic animals in eastern China. Zoonoses Public Health 55:291–298 [DOI] [PubMed] [Google Scholar]
- 72. Zhang W., et al. 2008. Cross-species infection of hepatitis E virus in a zoo-like location, including birds. Epidemiol. Infect. 136:1020–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]