Abstract
The performance of the Bruker Biotyper matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometer (MS) for the identification of dermatophytes from clinical cultures was compared to that of dermatophyte identification using 28S rRNA gene sequencing. The MALDI Biotyper library (MBL; version 3.0) was used alone and in combination with a supplemented library containing an additional 20 dermatophyte spectra (S-MBL). Acquired spectra were interpreted using both the manufacturer-recommended scores (genus, ≥1.7; species, ≥2.0) and adjusted cutoff values established by this study (genus, ≥1.5; species, ≥1.7); identifications required a minimum 10% difference in scores between the top two different organisms to be considered correct. One hundred well-characterized, archived dermatophyte isolates and 71 fresh dermatophyte cultures were evaluated using both libraries and both sets of cutoff criteria. Collectively, the S-MBL significantly outperformed the MBL at both the genus (93% versus 37.4%; P < 0,0001) and species (59.6% versus 20.5%; P < 0.0001) levels when using the adjusted score criteria. Importantly, application of the lowered cutoff values significantly improved genus (P = 0.005)- and species (P < 0.0001)-level identification for the S-MBL, without leading to an increase in misidentifications. MALDI-TOF MS is a cost-effective and rapid alternative to traditional or molecular methods for dermatophyte identification, provided that the reference library is supplemented to sufficiently encompass clinically relevant, intraspecies strain diversity.
INTRODUCTION
Dermatophytes, comprising species within the genera Epidermophyton, Microsporum, and Trichophyton, exclusively invade and infect keratinized tissues (i.e., hair, skin, and nails). These organisms are among the most common fungal infections in humans, accounting for over 5 million physician visits annually in the United States (7, 26). Dermatophytes primarily cause superficial dermal mycoses and rarely progress to more invasive disease (8). In the United States, Trichophyton rubrum remains the most common cause of tinea infections and the most frequently isolated dermatophytic pathogen (7, 24).
Laboratory identification of dermatophytes is typically based on macroscopic observation of colony morphology (i.e., pigmentation, growth rate, texture, etc.) on selective media, followed by microscopic examination of conidia. In addition, specialized Trichophyton agars may be used for differentiation of species within the genera (14). These phenotypic techniques require experienced technologists and are often labor-intensive with a prolonged turnaround time. With the advancement of modern molecular techniques, some laboratories have turned to dermatophyte identification schemes involving sequencing of specific ribosomal DNA regions or real-time PCR amplification of discrete genes (2, 20). While these methods are highly accurate and rapid, they are costly and can be complex to implement.
Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (MS) may present an alternative to conventional dermatophyte identification. This technology is based on the generation of an isolate-specific spectral profile, which is largely determined by the ribosomal protein content. The acquired protein spectra are subsequently compared to an extensive library of reference spectra using specific analysis software, which yields a list of top-matching identifications.
Recent studies have validated the MALDI-TOF technology as a rapid and cost-effective technique for bacterial (19, 23, 25) and yeast (5, 18, 27) identification. The assessment of MALDI-TOF for species-level identification of filamentous fungi, however, is not as extensive. Many initial fungal MALDI-TOF MS studies focused on isolate identification using purified fungal spores (3, 13, 15, 28). Only more recently has this technology been applied directly to unfractionated fungal colonies (10, 11, 17). Studies examining the application of MADLI-TOF MS specifically for dermatophyte identification are limited (6), and to our knowledge, there have been no published studies to date on the performance of the Bruker Biotyper system (Bruker Daltonics, Billerica, MA) for the identification of dermatophytes.
In this study, we compared the Bruker Biotyper MALDI-TOF MS system to sequencing of the D2 region of the 28S fungal ribosomal gene for the identification of clinical dermatophyte isolates. The MALDI Biotyper library (MBL; version 3.0) was used unmodified from the manufacturer and in combination with a laboratory-developed, spectral database containing supplemental dermatophyte spectra (S-MBL). Subsequently, we assessed the ability of the Biotyper to identify both unusual and commonly encountered clinical isolates.
(This study was presented at the 51st Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, IL, 17 to 20 September 2011.)
MATERIALS AND METHODS
Dermatophyte culture isolates. (i) Archived isolates.
One hundred previously identified clinical dermatophyte isolates were grown on Mycosel agar (BD Diagnostics, NJ) at 30°C for 3 days prior to analysis by the Bruker Biotyper MALDI-TOF MS system (Bruker Daltonics, Bremen, Germany). Both common (identified at least weekly in the clinical laboratory) and uncommon (identified monthly to yearly in the clinical laboratory) dermatophyte isolates were analyzed.
(ii) Fresh isolates.
During a 1-month period between March and April of 2011, 92 clinical dermatophyte isolates (29 from in-house patient specimens and 63 cultured isolates received from other laboratories) were simultaneously tested by 28S ribosomal DNA sequencing and MALDI-TOF MS. These specimens were at various stages of growth, collected from multiple sources (i.e., nail, skin specimens), and cultured on a variety of media, including chocolate agar (Thermo Fisher Scientific, MA) (n = 1), inhibitory mold agar (IMA; Thermo Fisher Scientific) (n = 2), modified dermatophyte test medium (BD Diagnostics, NJ) (n = 3), Mycosel agar (BD Diagnostics) (n = 37), nutrient agar (BD Diagnostics) (n = 11), and Sabouraud's agar (Thermo Fisher Scientific) (n = 38).
D2 28S rRNA sequencing.
Filamentous fungi macroscopically and microscopically resembling dermatophytes were submitted for sequencing of the D2 region of the 28S rRNA gene as previously described (9). Isolates submitted for sequencing were also plated on IMA to confirm purity. Resultant sequences are compared against those in a laboratory-developed, previously validated sequence library (9) or the MicroSeq fungal library (Applied Biosystems Fungal Library, version 2.0; Foster City, CA).
Specimen extraction and preparation for MALDI-TOF MS.
Fungal analysis by the Biotyper MALDI-TOF MS system requires chemical extraction of the isolated organism. Briefly, an approximately 2-mm by 2-mm piece of the colony was selected using a sterile loop and submerged in 1 ml of 70% ethanol (Sigma-Aldrich, St. Louis, MO), pelleted, decanted, and dried for 10 min using a speed vacuum (Eppendorf, Hamburg, Germany). The pellet was resuspended in 50 μl of 70% formic acid (Fluka [Sigma-Aldrich], St. Louis, MO) and briefly vortexed, and 50 μl of acetonitrile (Fluka) was added. After a brief vortexing, 1 μl of the supernatant was spotted on a steel plate (BigAnchor 24 well; Bruker) and allowed to air dry. Two microliters of matrix (α-cyano-4-hydroxycinnamic acid [HCCA; Bruker Daltonics, Billerica, MA] dissolved in 50% acetonitrile and 2.5% trifluoroacetic acid) was added and allowed to dry completely. For each run, positive (Candida krusei) and negative (70% alcohol blank) controls were also processed. A bacterial test standard (Bruker Daltonics) was used for instrument calibration.
MALDI-TOF MS analysis was performed on the MicroFlex LT system (Bruker) using the default settings. Briefly, protein ions (mass range, 2 to 20 kDa) generated with a 337-nm nitrogen laser were detected in the positive linear mode using the MALDI Biotyper automation control program. Captured spectra were analyzed using the MALDI Biotyper library (version 3.0), which at the time of this study contained 3,995 unique reference spectra.
Supplemented spectrum library entries.
The standard Bruker library (MBL) was supplemented with 20 additional dermatophyte spectra obtained from clinical isolates (identified by D2 28S rRNA sequencing) and 4 American Type Cell Culture Collection (ATCC; Manassas, VA) strains. These encompassed nine dermatophyte species (Table 1), four of which (Microsporum audouinii, M. persicolor, Trichophyton soudanense, T. verrucosum) were not represented by the MBL. Isolates used for library entry were grown on Mycosel medium for 2 to 5 days (Table 1) prior to extraction as described above. Each extract was spotted onto eight individual wells, and spectra were collected in triplicate, yielding a total of 24 spectra per isolate, according to the manufacturer's recommendations. A minimum of 22 quality spectra were required for library entry. The supplemented library is referred to as the S-MBL and consists of the MBL plus the 20 additional dermatophyte spectra.
Table 1.
Dermatophytes represented in MALDI Biotyper and supplemented MALDI Biotyper libraries
| Library | Dermatophyte isolate | Age(s) of isolate used for library entry (days) |
|---|---|---|
| MALDI Biotyper, version 3.0 (MBL) | Arthroderma benhamiae 24 VML (teleomorph of T. mentagrophytes) | Not available |
| Epidermophyton floccosum 27 VML | Not available | |
| Microsporum canis 30 VML | Not available | |
| M. gypseum RV491 | Not available | |
| M. gypseum 26 VML | Not available | |
| M. gypseum VML | Not available | |
| Trichophyton interdigitale RV06 32 VML | Not available | |
| T. mentagrophytes 31 VML | Not available | |
| T. rubrum 25 VML | Not available | |
| T. rubrum VML | Not available | |
| T. tonsurans RV491 | Not available | |
| T. tonsurans 28 VML | Not available | |
| T. tonsurans VML | Not available | |
| Supplemental spectra (S-MBL) | E. flocossuma | 4 |
| M. audouiniia | 2, 5 | |
| M. canisa | 2, 5 | |
| M. persicolor (ATCC 22439) | 2 | |
| T. mentagrophytes (ATCC MYA-4439) | 2, 5 | |
| T. mentagrophytes (ATCC 16781) | 2 | |
| T. mentagrophytesa | 4 | |
| T. rubruma | 2, 5 | |
| T. rubrum (ATCC MYA-4438) | 2, 5 | |
| T. soudanensea | 2, 5 | |
| T. tonsuransa | 2 | |
| T. tonsuransa | 3 | |
| T. verrucosuma | 2, 5 | |
| 2 |
Clinical isolate.
Analysis.
For each phase of the study, spectral results were analyzed using the MBL alone and the S-MBL. Possible score outputs for the Biotyper range from 0 (no spectrum match) to 3.0 (perfect match). Data analysis was performed using both the manufacturer-recommended cutoff scores of ≥1.7 for genus-level identification and ≥2.0 for species-level identification and adjusted, lowered score values of ≥1.5 and ≥1.7 for genus- and species-level identification, respectively. These altered criteria were chosen on the basis of previously described studies which analyzed the effect of decreased scores on bacterial and yeast identification efficiency (1, 5). In addition, for the genus- or species-level identification to be considered correct, at least a 10% difference in scores between the top score and the score of the next different organism was required (4, 23).
When using the previously archived isolates, where there was always abundant growth on the culture plate, in instances where the initial spectral score was 0, the isolate was reextracted on the same day and reanalyzed using automated analysis. However, in the study using fresh clinical isolates where a colony was tested as soon as it was visible on the plate, in instances where the initial spectral score was 0, spectra were gathered manually due to a lack of sufficient growth on the culture plate for reextraction. Manual acquisition allows the operator to choose a specific spot in the well for ionization and does not rely on automated direction of the laser. If this method did not provide better spectral scores, the isolate was reextracted from the purity plate 2 to 4 days later and analyzed as described above.
Statistics.
The McNemar test of paired proportions was used to analyze the MBL and S-MBL comparison at the genus and species levels, as well as S-MBL performance, using the manufacturer's recommended and the lowered score cutoff criterion. P values of less than 0.05 were considered statistically significant. All analyses were performed using SAS software (version 9.1; SAS Institute, Cary, NC).
RESULTS
For the first phase of this study, 100 common and uncommon, archived dermatophyte isolates were processed using MALDI-TOF and analyzed using the MBL and the S-MBL. Since T. rubrum is encountered frequently in the clinical laboratory relative to other genera and species, analysis of this species was limited to 27 isolates in order to increase the diversity of dermatophytes examined. Of the 100 isolates, 12 failed to generate spectra (score, 0) during the initial MALDI-TOF run and required reextraction and reprocessing prior to analysis. During the second phase of this study, of the 92 fresh clinical isolates grown in or submitted to our laboratory for dermatophyte identification, the following were excluded from analysis: 19 were not dermatophytes (1 Streptomyces albus isolate with a score of 2.03, 1 Chaetomium sp. isolate with a score of 1.516, 17 isolates identified as bacteria by the laboratory with scores of <1.5 by MALDI-TOF), and 2 isolates were identified by sequencing as Trichophyton violaceum (a species not represented by either the MBL or the S-MBL). Of the remaining 71 samples, 20 failed to produce spectra (score, 0) during the initial MALDI-TOF run. Improved spectra were acquired for 18 of these isolates by either manual generation of spectra (n = 4) or reextraction of the isolate from the IMA purity plate (n = 14). The performance of MALDI-TOF for archived and fresh isolates was similar, and therefore, the results are presented in a combined format.
As seen in Table 2, using the MBL alone and the recommended manufacturer score cutoff values, MALDI-TOF correctly identified 22.2% and 4.7% of isolates to the genus and species levels, respectively. In comparison, analysis of these spectra using the S-MBL improved isolate identification to 86.0% for genus-level recognition and 36.8% for species-level recognition. Notably, lowering the preset score values to ≥1.5 for genus-level confidence and ≥1.7 for species-level confidence significantly improved the MALDI-TOF performance, with the S-MBL significantly outperforming the MBL at both the genus (93% versus 37.4%; P < 0.0001) and species (59.6% versus 20.5%; P < 0.0001) levels. With respect to the S-MBL, lowering the score cutoff values increased both genus- and species-level identification with statistical significance (P < 0.005 in both cases).
Table 2.
Combined results for archived and fresh isolates: comparison of MBL and S-MBL using manufacturer-recommended and decreased score criteria
| Dermatophytes tested (no. of isolates) | No. (%) of isolates |
|||||||
|---|---|---|---|---|---|---|---|---|
| Genus-level score ≥ 1.5 |
Genus-level score ≥ 1.7 |
Species-level score ≥ 1.7 |
Species-level score ≥ 2.0 |
|||||
| MBL | S-MBL | MBL | S-MBL | MBL | S-MBL | MBL | S-MBL | |
| E. floccosum (n = 1) | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 |
| M. canis (n = 23) | 6 | 16 | 3 | 16 | 3 | 14 | 2 | 11 |
| M. gypseum (n = 2) | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 |
| T. mentagrophytes (n = 29) | 15 | 26 | 3 | 23 | 1 | 19 | 0 | 12 |
| T. rubrum (n = 72) | 11 | 60 | 6 | 57 | 6 | 26 | 0 | 17 |
| T. tonsurans (n = 40) | 20 | 27 | 16 | 23 | 16 | 20 | 2 | 14 |
| T. verrucosum (n = 4) | 0 | 4 | 0 | 4 | 0 | 4 | 0 | 3 |
| Correcta | 53 (38.1) | 135 (97.1) | 29 (20.9) | 125 (89.9) | 27 (19.4) | 85 (61.1) | 4 (2.9) | 58 (41.7) |
| Reanalyzed isolates (n = 32)b | 11 | 24 | 9 | 22 | 8 | 17 | 4 | 5 |
| Total correct, including repeatsc | 64 (37.4) | 159 (93.0) | 38 (22.2) | 147 (86.0) | 35 (20.5) | 102 (59.6) | 8 (4.7) | 63 (36.8) |
The number correct does not include isolates requiring reanalysis. Percent correct is based on n = 139 (total number of isolates tested [n = 171] minus the number of isolates requiring reanalysis [n = 32]).
M. canis (n = 9), M. gypseum (n = 1), T. mentagrophytes (n = 2), T. rubrum (n = 8), and T. tonsurans (n = 12); 2 were not available for reanalysis due to insufficient growth.
Percent correct is based on n = 171.
Misidentifications.
Genus-level misidentification occurred once and only when using the MBL. A single Epidermophyton floccosum isolate, which is not represented in the MBL, was identified as a Trichophyton sp. (score, 1.628) and met the 10% score difference criterion required for a confident identification.
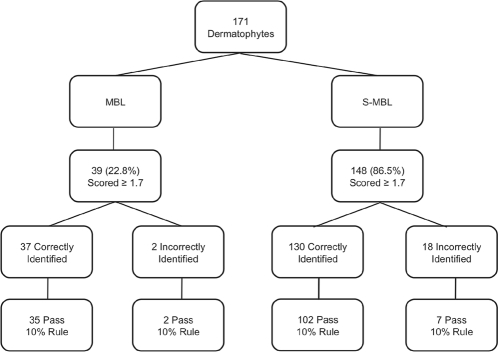
With regard to species-level identification, upon analysis of all isolates (archived and fresh), the MBL alone correctly identified the species of 37 out of the 39 (92.3%) isolates scoring ≥1.7; 2 isolates were incorrectly identified (Trichophyton mentagrophytes called T. tonsurans) and passed the 10% criterion for correct species identification (Fig. 1). In contrast, using the S-MBL, the species of 130 of 148 (87.8%) isolates scoring ≥1.7 were correctly identified. However, of those 130 isolates, 27 did not meet the 10% difference criterion (Fig. 1) and would require a method other than MALDI-TOF for identification. These were considered incorrect identifications for the purposes of data analysis. Eighteen of the 148 isolates were misidentified: 1 Microsporum canis isolate identified as M. audouinii, 1 T. mentagrophytes isolate identified as T. tonsurans, and 16 T. rubrum isolates identified as T. soudanense. Of these 18 isolates, 11 did not pass the 10% rule and would require a method other than MALDI-TOF for identification. The remaining 7 isolates passed the 10% difference criterion and would be incorrectly identified using MALDI-TOF. These 7 isolates, determined to be T. rubrum by the laboratory, were incorrectly identified as T. soudanense by the Biotyper, matching to a single entry in the S-MBL. Reanalysis of these isolates following omission of the T. soudanense reference spectrum led to the correct and confident identification of all T. rubrum isolates to the species level.
Fig. 1.
Comparison of the MBL and S-MBL species-level identifications.
DISCUSSION
Dermatophyte species identification by the Bruker Biotyper MALDI-TOF MS system was evaluated in comparison to that by D2 sequencing. The MALDI-TOF system is an excellent alternative to dermatophyte identification by traditional microscopy or molecular techniques due to its speed and cost-effectiveness. Also, because each acquired spectrum is compared against the entire Biotyper database, preliminary technician identification of a colony as a dermatophyte is unnecessary. However, we show that supplementation of the version 3.0 MALDI Biotyper library is required for optimal dermatophyte species identification.
Species identification by the Biotyper is largely limited by the spectra in the reference library. In the current version, microorganisms commonly seen in the clinical laboratory have more reference spectra than infrequently isolated organisms. In theory, a higher number of library entries per organism would encompass more intraspecies diversity occurring due to variable protein expression, growth conditions, or age (1, 25). While all three dermatophyte genera are represented in the current MBL version, the maximum number of spectra present for any species is three, and single reference spectra are present for Epidermophyton floccosum, Microsporum canis, and Trichophyton interdigitale (Table 1); many dermatophyte species are not included. In order to increase database coverage of strain variability, we created a supplemental library with 20 additional dermatophyte spectra.
Combining all identifications using both archived and fresh isolates and adhering to the manufacturer's recommended score cutoff values, the MBL correctly identified only 22.2% of isolates to the genus level, compared to 86.0% when using the S-MBL. Species-level identification was likewise improved from 4.7% to 36.8% with use of the S-MBL. Database supplementation is therefore necessary for accurate and efficient dermatophyte identification by the Bruker Biotyper. Further supplementation of the S-MBL with additional dermatophyte spectra, such as spectra for T. violaceum, would likely increase percent identification.
The increased percent identification elicited with the S-MBL is likely due to a number of factors. As mentioned above, the added dermatophyte spectra likely enhanced the species-specific diversity covered by the library, which allowed an increased capacity of the database to differentiate between subtle intraspecies protein variability. Alternatively, as dermatophyte conidia germinate and age, the expressed protein profiles change (16), making isolate age an important factor to consider with regard to MALDI-TOF analysis, both when making spectral library entries and when testing clinical samples. All manually supplemented reference spectra in the S-MBL were collected when the isolates were 2 to 5 days old, but the ages of the dermatophyte strains used by the manufacturer to build the original MBL are not available. In our study, archived strains were tested after 3 days of growth, while fresh isolates were processed when an adequate colony size was present to sample for both MALDI-TOF and D2 sequencing. Therefore, if the isolates used for MBL development were cultured for prolonged periods of time prior to library entry, the associated protein spectra may be significantly different from those found in younger cultures. This may explain the poor level of identification obtained with the MBL alone. Finally, the type of medium used for culture has also been suggested to play a role in altering microorganism protein profiles (21). We found no evidence of a medium effect on dermatophyte identification during our clinical study, which analyzed a range of isolates (5 to 30) of 5 different dermatophyte species grown on 6 different types of media (see Materials and Methods; data not shown).
In an effort in further enhance the percentage of acceptable identifications, we reanalyzed our data using lowered genus and species score cutoff values: ≥1.5 for genus-level confidence and ≥1.7 for species-level confidence. Combining our results from archived and fresh isolates and using these criteria with the S-MBL, MALDI-TOF correctly identified 93% (previously 86.0% at a score cutoff value of ≥1.7; P = 0.0005) and 59.6% (previously 36.8% at a score cutoff value of ≥2.0; P < 0.0001) of isolates to the genus and species levels, respectively (Table 2). The increase in accurate species identification obtained with the lower score values was not met with an increase in misidentifications. On the basis of these data, using the S-MBL with the lowered score criterion is preferred for dermatophyte identification.
Few studies have focused on MALDI-TOF-based identification of dermatophytes. Using the Voyager DE Pro MS system (Applied Biosystems, Darmstadt, Germany) and the Saramis spectral database (AnagnosTec, Potsdam, Germany), Erhard and colleagues achieved high-confidence-level (99.9%) identifications for all but one of the dermatophyte isolates tested (6). However, comparison of our study and theirs is difficult due to the utilization of two different reference spectrum databases and because the majority of isolates tested in the study of Erhard et al. (6) were T. rubrum strains, while our study assayed a wider range of dermatophytes, including multiple strains of representative species from each genera except Epidermophyton.
Our criterion for dermatophyte identification to both the genus and species levels required a 10% difference in scores between the top-scoring organism and the next genus and species identified. There are two types of errors that can occur when using the 10% difference criterion. First, in situations where the isolate is correctly identified by the top score, if that score does not differ by more than 10% from the next-highest score, the identification is considered doubtful and the isolate should be reanalyzed or identified using another method. Importantly, this 10% score difference criterion was met for all genus-level identifications when using the S-MBL, regardless of which score cutoff value was used. We are therefore confident that MALDI-TOF and the S-MBL will accurately determine the genus of any pure dermatophyte specimen analyzed. Doubtful species identifications were encountered when using the S-MBL, where 38 of 148 (27.7%) isolates scoring ≥1.7 would have required reprocessing or alternative methods for confident identification to the species level. While this represents over one-quarter of the specimens analyzed, the process for extraction is straightforward and the cost per sample is minimal (∼$0.50) (5).
The second type of error that may occur is a true misidentification, in which case the species of the isolate is incorrectly identified but the associated score meets the 10% confidence criterion. Of the 148 isolates scoring ≥1.7, 7 (4.7%) were misidentified to the species level with the S-MBL. Interestingly, these were all T. rubrum isolates incorrectly identified as T. soudanense on the basis of the same manually added T. soudanense reference spectra. This strain of T. soudanense appears to be extremely similar to the tested T. rubrum isolates, and the Biotyper using the S-MBL was unable to adequately differentiate these two organisms. Since T. soudanense is a rare clinical isolate, deletion of this spectral entry is an option to prevent future misidentifications of the more frequently encountered organism T. rubrum. Alternatively, pigment production on potato dextrose agar can be used as a simple method to differentiate these two organisms (http://www.mycology.adelaide.edu.au/, accessed 8 June 2011) (14).
The question arises, however, whether clinical management of dermatophyte infections requires species identification beyond genus-level identification. Depending on the extent of pathology, tinea infections are typically treated with either oral or topical antifungals, including griseofulvin, terbinafine, or select azoles (12). Little is mentioned in the literature regarding species-specific treatment guidelines, and while some resistance has been documented (primarily in T. rubrum against terbinafine), active alternatives remain available (22). Each laboratory performing MALDI-TOF MS identification of dermatophytes should determine whether species-level identification is warranted or whether genus-level identification is sufficient for patient care.
In summary, the ability to use MALDI-TOF to rapidly and simply identify ∼93% of the dermatophyte isolates encountered in the laboratory to the genus level and ∼60% to the species level dramatically reduces the turnaround time associated with traditional phenotypic methods, as well as the cost and labor associated with molecular methods. MALDI-TOF MS is an excellent alternative to microscopy and sequencing for dermatophyte identification, provided the spectral database sufficiently encompasses intraspecies strain diversity. While the current Biotyper library requires user supplementation for optimal performance in dermatophyte identification, the process of reference spectrum addition is relatively simple. Furthermore, spectral analysis using decreased score cutoff values increases the percentage of dermatophytes whose species are correctly identified, without significantly compromising identification accuracy. Future studies will be aimed at expanding MALDI-TOF-based identification to other filamentous fungi and further increasing library robustness.
ACKNOWLEDGMENTS
We thank the technologists in the Mayo Clinic Mycology Laboratory for their help in collecting and preparing the isolates used in this study.
Footnotes
Published ahead of print on 28 September 2011.
REFERENCES
- 1. Alatoom A. A., Cunningham S. A., Ihde S. M., Mandrekar J., Patel R. 2011. Comparison of direct colony method versus extraction method for identification of gram-positive cocci by use of Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 49:2868–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bergmans A. M., et al. 2010. Evaluation of a single-tube real-time PCR for detection and identification of 11 dermatophyte species in clinical material. Clin. Microbiol. Infect. 16:704–710 [DOI] [PubMed] [Google Scholar]
- 3. Chen H. Y., Chen Y. C. 2005. Characterization of intact Penicillium spores by matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 19:3564–3568 [DOI] [PubMed] [Google Scholar]
- 4. Degand N., et al. 2008. Matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of nonfermenting gram-negative bacilli isolated from cystic fibrosis patients. J. Clin. Microbiol. 46:3361–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dhiman N., Hall L., Wohlfiel S. L., Buckwalter S. P., Wengenack N. L. 2011. Performance and cost analysis of matrix-assisted laser desorption ionization–time of flight mass spectrometry for routine identification of yeast. J. Clin. Microbiol. 49:1614–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Erhard M., Hipler U. C., Burmester A., Brakhage A. A., Wostemeyer J. 2007. Identification of dermatophyte species causing onychomycosis and tinea pedis by MALDI-TOF mass spectrometry. Exp. Dermatol. 17:356–361 [DOI] [PubMed] [Google Scholar]
- 7. Foster K. W., Ghannoum M. A., Elewski B. E. 2004. Epidemiologic surveillance of cutaneous fungal infection in the United States from 1999 to 2002. J. Am. Acad. Dermatol. 50:748–752 [DOI] [PubMed] [Google Scholar]
- 8. Gong J. Q., et al. 2007. Deep dermatophytosis caused by Trichophyton rubrum: report of two cases. Mycoses 50:102–108 [DOI] [PubMed] [Google Scholar]
- 9. Hall L., Wohlfiel S., Roberts G. D. 2004. Experience with the MicroSeq D2 large-subunit ribosomal DNA sequencing kit for identification of filamentous fungi encountered in the clinical laboratory. J. Clin. Microbiol. 42:622–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hettick J. M., et al. 2008. Discrimination of Aspergillus isolates at the species and strain level by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry fingerprinting. Anal. Biochem. 380:276–281 [DOI] [PubMed] [Google Scholar]
- 11. Hettick J. M., et al. 2008. Discrimination of Penicillium isolates by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry fingerprinting. Rapid Commun. Mass Spectrom. 22:2555–2560 [DOI] [PubMed] [Google Scholar]
- 12. Kakourou T., Uksal U. 2010. Guidelines for the management of tinea capitis in children. Pediatr. Dermatol. 27:226–228 [DOI] [PubMed] [Google Scholar]
- 13. Kemptner J., et al. 2009. Evaluation of matrix-assisted laser desorption/ionization (MALDI) preparation techniques for surface characterization of intact Fusarium spores by MALDI linear time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 23:877–884 [DOI] [PubMed] [Google Scholar]
- 14. Larone D. H. 2002. Medically important fungi: a guide to identification, 4th ed American Society for Microbiology Press, Washington, DC [Google Scholar]
- 15. Li T. Y., Liu B. H., Chen Y. C. 2000. Characterization of Aspergillus spores by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 14:2393–2400 [DOI] [PubMed] [Google Scholar]
- 16. Liu T., et al. 2007. The use of global transcriptional analysis to reveal the biological and cellular events involved in distinct development phases of Trichophyton rubrum conidial germination. BMC Genomics 8:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marinach-Patrice C., et al. 2009. Use of mass spectrometry to identify clinical Fusarium isolates. Clin. Microbiol. Infect. 15:634–642 [DOI] [PubMed] [Google Scholar]
- 18. Marklein G., et al. 2009. Matrix-assisted laser desorption ionization–time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J. Clin. Microbiol. 47:2912–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mellmann A., et al. 2008. Evaluation of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry in comparison to 16S rRNA gene sequencing for species identification of nonfermenting bacteria. J. Clin. Microbiol. 46:1946–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ninet B., et al. 2003. Identification of dermatophyte species by 28S ribosomal DNA sequencing with a commercial kit. J. Clin. Microbiol. 41:826–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pennanec X., Dufour A., Haras D., Rehel K. 2010. A quick and easy method to identify bacteria by matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 24:384–392 [DOI] [PubMed] [Google Scholar]
- 22. Peres N. T., Maranhao F. C., Rossi A., Martinez-Rossi N. M. 2010. Dermatophytes: host-pathogen interaction and antifungal resistance. An. Bras. Dermatol. 85:657–667 [DOI] [PubMed] [Google Scholar]
- 23. Saffert R. T., et al. 2011. Comparison of Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometer to BD Phoenix automated microbiology system for identification of gram-negative bacilli. J. Clin. Microbiol. 49:887–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seebacher C., Bouchara J. P., Mignon B. 2008. Updates on the epidemiology of dermatophyte infections. Mycopathologia 166:335–352 [DOI] [PubMed] [Google Scholar]
- 25. Seng P., et al. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 49:543–551 [DOI] [PubMed] [Google Scholar]
- 26. Smith E. S., Fleischer A. B., Feldman S. R., Williford P. M. 2002. Characteristics of office-based physician visits for cutaneous fungal infections. An analysis of 1990 to 1994 National Ambulatory Medical Care Survey Data. Cutis 69:191–198, 201-202 [PubMed] [Google Scholar]
- 27. Stevenson L. G., Drake S. K., Shea Y. R., Zelazny A. M., Murray P. R. 2010. Evaluation of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of clinically important yeast species. J. Clin. Microbiol. 48:3482–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Welham K. J., Domin M. A., Johnson K., Jones L., Ashton D. S. 2000. Characterization of fungal spores by laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 14:307–310 [DOI] [PubMed] [Google Scholar]