Abstract
We have developed a new research assay that combines sequence-specific sample preparation and isothermal amplification for the detection of Chlamydia trachomatis and Neisseria gonorrhoeae infections. The assay targets both the omp gene and the cryptic plasmid of C. trachomatis and the multicopy opa gene of N. gonorrhoeae, which are amplified and detected in a single reaction. We evaluated the ability of the assay to detect C. trachomatis and N. gonorrhoeae infections in first-catch urine, swab, and liquid-based cytology samples. Total agreement between the new assay and APTIMA Combo 2 varied between 95.3% and 100%, depending on the sample type and target detected. Total agreement between the new assay and BD ProbeTec varied between 96.7% and 100%, depending on the sample type and target detected. The assay has a simple work flow, and endpoint results can be achieved in 3 h, including sample preparation. The assay described here was evaluated for research use and was compared to commercially available assays.
INTRODUCTION
Chlamydia trachomatis and Neisseria gonorrhoeae are among the most common causes of sexually transmitted infections (3). Infection with these organisms is often asymptomatic, which hinders diagnosis or treatment. Persistent infection can have serious negative consequences, including pelvic inflammatory disease and infertility. Untreated and undiagnosed infections also contribute to the further spread of the disease. Screening programs are recommended to prevent these consequences (17). Of the various testing methods currently available, nucleic acid amplification tests are among the most sensitive and specific methods for detection of C. trachomatis and N. gonorrhoeae organisms in clinical specimens (7, 9). The high sensitivity of nucleic acid amplification tests allows them to be used with less-invasive clinical samples, such as first-catch urine, which facilitates establishment of screening programs.
The assay described here combines sequence-specific sample preparation and isothermal amplification for the detection of C. trachomatis and N. gonorrhoeae infections. Sequence-specific sample preparation is based on antibody capture of DNA-RNA hybrids. Ribo-oligonucleotide probes hybridize to specific target sequences, and the resulting hybrids are captured by antibodies specific for DNA-RNA hybrids. This process enriches the desired nucleic acid sequences in the sample. Antibodies are coupled to magnetic beads. Target amplification is accomplished by thermophilic helicase-dependent amplification (tHDA) (16). Amplification of captured strands of the DNA target occurs directly on magnetic beads through the combined action of primers, thermostable DNA polymerase, and helicase. Target DNA is detected by use of dually labeled probes in either endpoint or real-time detection modes. C. trachomatis and N. gonorrhoeae targets are amplified and detected simultaneously in a single closed reaction vessel.
We investigated the ability of the assay to detect C. trachomatis and N. gonorrhoeae infections in clinical specimens. Residual samples previously evaluated by commercially available FDA-cleared assays were tested in the new assay, and agreement with comparator assays was calculated. The clinical samples tested (n = 828) included urine (n = 244), swab (n = 60), and liquid-based cytology (LBC) (n = 524) samples. APTIMA Combo 2, BD ProbeTec, and HC2 CT/GC were used as comparator assays. The assay described here is under development and not available commercially. It is presented for research use only and not for diagnostic use.
MATERIALS AND METHODS
Assay design.
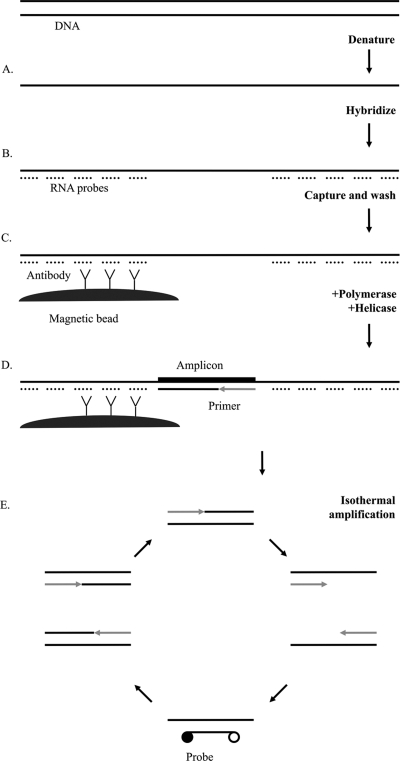
Figure 1 illustrates the principles of the assay. The assay targets included the omp gene in the C. trachomatis genome, a region of the C. trachomatis cryptic plasmid, and the multicopy opa gene in N. gonorrhoeae. Multiple targets were detected by using different fluorophores on target-specific probes. Both C. trachomatis genomic and plasmid targets were detected by a single fluorophore, while N. gonorrhoeae was detected by a separate fluorophore. Asymmetric primer concentrations generate an excess of the DNA strand containing the recognition site for the fluorescent probe. Fluorescence is evaluated in real-time and endpoint detection modes. The work flow of the assay for 96 samples includes 45 min for sample preparation and amplification setup and 120 min of amplification followed by endpoint detection.
Fig. 1.
Schematic illustration of the tHDA assay for detection of C. trachomatis and N. gonorrhoeae. (A) Samples are lysed, and DNA is denatured. (B) RNA probes are hybridized. (C) RNA-DNA hybrids are captured on a monoclonal hybrid capture antibody conjugated to magnetic beads. (D) tHDA reagents are added to washed beads. (E) DNA polymerase extends primers hybridized to single-stranded DNA. UvrD helicase unwinds the double-stranded DNA for the next round of amplification. Exponential amplification occurs through continuous isothermal (65°C) activity of the helicase and polymerase. Hybridization of fluorescent-labeled probes generates a signal.
Sample preparation.
One-milliliter portions of urine and PreservCyt samples were tested without dilution. Five-hundred-microliter portions of BD SurePath samples were diluted with 500 μl of molecular biology grade water. One hundred microliters of the transport medium from swab samples was diluted with 900 μl of molecular biology grade water. After dilution where necessary, 1 ml each of negative controls, positive controls, and clinical samples were added to 5-ml polypropylene tubes. Five hundred microliters of denaturation reagent (Qiagen, Gaithersburg, MD) and 250 μl of lysis buffer (7.5% Sarkosyl, 2.5% NP-40 alternative, 10 mM dithiothreitol [DTT]) were added, followed by heating at 68.5°C in an Eppendorf Thermomixer heated shaker (Eppendorf, Hauppauge, NY) for 7.5 min. Oligoribonucleotide probes (Integrated DNA Technologies, Coralville, IA) were diluted in a hybridization reagent (1 M N,N-bis(2-hydroxyethyl)taurine, 0.5% NP-40 alternative, 2.6% 90,000-molecular-weight polyacrylic acid [Polysciences, Warrington, PA]). Probes consisted of 20 25-mer oligonucleotides for each target. Probes were complementary to sequences immediately adjacent to the amplicon region. All probes hybridized to the same strand of DNA. Eight hundred microliters of this probe mix was added to each reaction mixture and shaken for 15 s at 900 rpm. Hybrid capture antibodies (Qiagen, MD) conjugated to magnetic beads (Thermo-Fisher Scientific Inc., Waltham, MA) were diluted in a blocking reagent (Qiagen, MD), and 25 μl of a 0.08% solids solution of magnetic beads was added to the reaction mixture. The reaction mixture was incubated at 50°C for 22.5 min in an Eppendorf Thermomixer heated shaker with shaking at 900 rpm. The reaction vessel was placed on a magnet stand, and beads were pelleted for 5 min. Supernatant was aspirated, and beads were resuspended in 500 μl of wash buffer (100 mM Tris [pH 7.5], 100 mM KCl, 40 mM NaCl, 3.5 mM MgSO4, 0.05% Tween 20). Beads were washed three times. After washing, magnetic beads were resuspended in 20 μl of Tris-EDTA (TE) buffer and transferred to a PCR plate.
tHDA.
The amplification mixture consists of 60 mM Tris-HCl, 20 mM KCl, 6 mM MgSO4, 0.14 M dimethyl sulfoxide (DMSO), 0.15 M sorbitol, 40 mM NaCl, 0.6 mM deoxynucleoside triphosphates (dNTPs), 6 mM dATP, 20 units Geobacillus stearothermophilus large-fragment polymerase (BioHelix Inc., Beverly, MA), 100 ng Tte-UvrD helicase (BioHelix Inc.), and 25 ng ET single-stranded binding protein (BioHelix Inc.). Primers (Integrated DNA Technologies) were used at a 1:3 forward/reverse asymmetric ratio (40 nM:120 nM for N. gonorrhoeae and C. trachomatis genomic target and 50 nM:150 nM for C. trachomatis plasmid). Dually labeled probes (60 nM) (Integrated DNA Technologies) specific to each amplicon provide the signal. Table 1 shows details of primer sequences and probe fluorophores. The plate holding target DNA was incubated in a thermal block for 5 min at 65°C. Following incubation, the sample plate was kept at 65°C while amplification reagents were added. The plate was sealed with optical PCR film and incubated at 65°C for 120 min, with fluorescence detection occurring every 60 s. Endpoint detection of fluorescence was performed at 25°C. Amplification and detection were performed on an iQ5 thermal cycler (Bio-Rad, Hercules, CA) using the Texas Red (excitation, 575 nm; emission, 625 nm) and Cy5 (excitation, 630 nm; emission, 685 nm) filters.
Table 1.
Sequences of primers and dually labeled fluorescent probes
| Target | Primer or probe | Primer or probe sequence (5′ → 3′) | Probe fluorophore/quencher | Amplicon size (bp) |
|---|---|---|---|---|
| N. gonorrhoeae genomic | Forward primer | ACCCGATATAATCCGTCCTTCA | 5′-TEX615/3′-IAbRQSp | 72 |
| Reverse primer | TTCGGCTCCTTATTCGGTTTAAC | |||
| Probe | CGTCCTTCAACATCAGTGAAAATCG | |||
| C. trachomatis genomic | Forward primer | ATTTGCCGCTTTGAGTTCTGCTTCCT | 5′-TYE665/3′-IAbRQSp | 75 |
| Reverse primer | ATCATAAGGCTTGGTTCAGCAGGATTC | |||
| Probe | CCTCCTTGCAAGCTCTGCCTGTGGGG | |||
| C. trachomatis plasmid | Forward primer | AGGCGATTTAAAAACCAAGGTCGATGT | 5′-TYE665/3′-IAbRQSp | 89 |
| Reverse primer | GAAGAATTGATCCAACACCCTTATCG | |||
| Probe | CCGTATGTGAATGTCGAACTCATCCG |
Determination of analytical performance.
A sample of C. trachomatis serovar D with enumerated inclusion-forming units (IFU) and a sample of N. gonorrhoeae with enumerated CFU were obtained from Zeptometrix (Buffalo, NY). Ten-fold serial dilutions from 5 × 105 IFU to 5 × 10−5 IFU of the C. trachomatis serovar were performed. Ten-fold serial dilutions of N. gonorrhoeae from 3 × 107 to 3 × 10−3 CFU were performed. Analytical specificity was tested by challenging the assay with 106 copies each of various microorganisms and viruses. The following 34 organisms were tested for cross-reactivity in the assay: Acinetobacter baumanii, adenovirus, Anaerococcus hydrogenalis, Bacteroides fragilis, Bifidiobacterium bifidum, Candida albicans, Clostridium difficile, Corynebacterium genitalium, Enterobacter cloacae, Enterococcus faecalis, Escherichia coli, Fusobacterium nucleatum, Gardnerella vaginalis, Haemophilus ducreyi, herpes simplex virus type 1, herpes simplex virus 2, human papillomavirus type 16 plasmid, human papillomavirus type 18 plasmid, Klebsiella pneumoniae, Lactobacillus acidophilus, Mycoplasma hominis, Neisseria flava, Neisseria meningitides, Neisseria perflava, Peptostreptococcus anaerobius, Proteus vulgaris, Pseudomonas aeruginosa, Serratia marcescens, Staphylococcus aureus, Staphylococcus epidermidis, Streptococcus agalactiae, Streptococcus pyogenes, Trichomonas vaginalis, and Ureaplasma urealyticum.
Determination of clinical performance.
Three independent studies using clinical samples were performed. The studies each investigated different sample types and comparator assays. The first study was performed on samples originally tested internally, while the other two studies were performed on samples originally tested at independent laboratories. Results from each study were used to optimize the criteria for calling positive samples in the next study.
(i) Study I.
Two hundred sixty-seven PreservCyt deidentified samples previously assayed by the HC2 CT/GC DNA assay and archived by the internal biobanking unit were tested in the new assay. HC2 CT/GC DNA results were not blinded to the user. HC2 CT/GC DNA testing was performed in-house as per the manufacturer's instructions. The new assay was performed as described above. Samples were considered positive by the new assay if they achieved a signal-to-noise ratio of 3 or greater and a signal greater than 3 standard deviations of noise above background. There was no indeterminate zone in this study. Discrepant samples were retested by quantitative PCR (qPCR) with the Quantitect virus PCR kit (Qiagen, Hilden, Germany). A Stratagene Mx 3005p thermocycler (Stratagene, La Jolla, CA) was used for amplification and real-time detection of PCR products. Sixty cycles of 95°C for 15 s to 55°C for 30 were performed, and the target was quantitated against a 10-fold serial dilution of C. trachomatis or N. gonorrhoeae DNA. Detection of any amount of target in the qPCR was counted as a positive result. Primers and probes used in the tHDA assay were used for the qPCR assay, but each target was tested for individually rather than in multiplex.
(ii) Study II.
Thirty urine, 29 PreservCyt, 30 SurePath, 30 cervical swab, and 30 vaginal swab deidentified samples previously assayed at an independent laboratory using the APTIMA Combo 2 assay or the BD ProbeTec assay were shipped to Qiagen. APTIMA Combo 2 and BD ProbeTec assays were performed as per the manufacturer's instructions. Comparator test results were blinded to the operator. The new assay was performed as described above. Samples were considered positive by the new assay if they had greater than 500 relative fluorescence units (RFU) in the endpoint reading. For samples in the range of RFU considered indeterminate (250 to 500), the presence or absence of a real-time curve was used to classify them as positive or negative.
(iii) Study III.
One hundred ninety-eight PreservCyt samples and 214 first-catch urine deidentified samples previously assayed at an independent laboratory by the APTIMA Combo 2 assay were tested in the new assay. The APTIMA Combo 2 assay was performed as per the manufacturer's instructions. After the results of APTIMA Combo 2 testing were known, an aliquot of stored neat urine was added to a conical tube containing 250 μl stability buffer. Comparator assay results were not blinded to the user. The new assay was performed as described above, except 1.25 ml of sample was used and lysis buffer was excluded. In this study, cutoffs of 300 RFU for C. trachomatis samples and 350 RFU for N. gonorrhoeae samples were used. Samples with RFU above these cutoffs were considered positive. There was no indeterminate zone in this study.
RESULTS
Assay characteristics.
The detailed technical performance and development of the assay are described in a separate publication (6a). Briefly, the assay has limits of detection of 25 C. trachomatis elementary bodies (EBs)/ml and 25 N. gonorrhoeae cells/ml. Either target could be detected with this sensitivity in the presence of a 6-log excess of the other. Sequence-specific sample preparation is necessary to overcome tHDA inhibition caused by extraneous DNA. The analytical sensitivity of the assay for pathogens was determined by analyzing serial dilutions of enumerated (CFU/IFU) C. trachomatis and N. gonorrhoeae strains in PreservCyt medium. For this experiment, the two C. trachomatis targets were detected separately to determine if the two amplicon regions had similar sensitivities. The assay detected a C. trachomatis target concentration of 0.05 IFU/assay in both amplicon regions. N. gonorrhoeae was detectable at concentrations of as low as 0.003 CFU/assay with some variability and was detectable consistently at a level of 3 CFU/assay. When measured by endpoint fluorescence, the assay has little to no dynamic range, as sufficient amplicon is generated by even very low copy numbers to saturate the detection. Signal-to-noise values generated for the tested dilutions ranged from 10.0 to 27.3 for C. trachomatis and from 5.6 to 24.6 for N. gonorrhoeae. Since 1 ml of sample was used, limits of detection were expressed as 0.05 IFU/ml and 3 CFU/ml, respectively. C. trachomatis trachoma serovars A to C, urogenital serovars D to K, and lymphogranuloma serovars L1 to L3 were all detected in the assay at similar limits of detection. The analytical specificity of the tHDA assay was demonstrated by challenging the assay with high titers of bacteria and viruses and demonstrating that no signal was generated in an endpoint reaction (the tested organisms are listed in “Determination of analytical performance” above). A wide variety of sample media commonly used for detection of C. trachomatis and N. gonorrhoeae were tested, including first-catch urine samples, culture media (e.g., M4 or M4RT), liquid-based cytology media(e.g., PreservCyt samples or BD SurePath samples), and HPV testing media (e.g., STM). The new assay was able to accommodate all of these medium types without modification. Detection of 25 C. trachomatis EBs and 25 N. gonorrhoeae cells spiked into clinical urine specimens, clinical PreservCyt, clinical SurePath, M4RT, and STM resulted in signal-to-noise ratios of greater than 5 for all targets in each medium. Additionally, the assay results were equivalent when used in combination with collection media used in other commercial assays, such as the APTIMA urine transport medium. Twenty-five C. trachomatis EBs or N. gonorrhoeae cells spiked into APTIMA urine transport medium gave the same fluorescent signal as EBs or cells spiked into a clean control.
The ability of the new assay to detect infection in clinical specimens was evaluated in several small studies. In each of these studies, clinical samples that were previously tested by FDA-cleared commercially available assays were tested in the new assay. Table 2 shows the agreement between the new assay and the FDA-cleared assays. The first study evaluated residual PreservCyt samples previously tested by the Digene HC2 CT/GC DNA test, a nucleic acid hybridization assay with signal amplification. The HC2 CT/GC DNA test puts out a composite result for both organisms, so a positive result for either target in the new assay was considered a positive for correlation to results of the HC2 CT/GC DNA test. The positive agreement of the new test with the HC2 test was 88.7% (110/124), and the negative agreement was 98.6% (141/143). Discrepant samples were tested by qPCR. Of 14 HC2-positive, new-assay-negative samples, 14 were negative by qPCR. Of two HC2 negative, new-assay-positive samples, one was positive and one was negative by qPCR.
Table 2.
Concordance of results of tHDA C. trachomatis/N. gonorrhoeae assay with those of other assays
| Study | Specimen | Comparator test | n |
C. trachomatis |
N. gonorrhoeae |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Positive |
Negative |
Total | Positive |
Negative |
Total | ||||||||
| % | No./total | % | No./total | % | No./total | % | No./total | ||||||
| 1a | PreservCyt | HC2 | 267 | ||||||||||
| 2 | Urine | APTIMA Combo 2 | 30 | 100.0 | 16/16 | 100.0 | 14/14 | 100.0 | 100.0 | 7/7 | 100.0 | 23/23 | 100.0 |
| PreservCyt | APTIMA Combo 2 | 29 | 93.8 | 15/16 | 100.0 | 13/13 | 96.6 | 100.0 | 7/7 | 100.0 | 22/22 | 100.0 | |
| SurePath | APTIMA Combo 2 | 30 | 100.0 | 20/20 | 100.0 | 10/10 | 100.0 | 100.0 | 4/4 | 100.0 | 26/26 | 100.0 | |
| Cervical swab | BD ProbeTec | 30 | 94.7 | 18/19 | 100.0 | 11/11 | 96.7 | 100.0 | 18/18 | 100.0 | 12/12 | 100.0 | |
| Vaginal swab | BD ProbeTec | 30 | 92.9 | 13/14 | 100.0 | 16/16 | 96.7 | 100.0 | 2/2 | 100.0 | 28/28 | 100.0 | |
| 3 | Urine | APTIMA Combo 2 | 214 | 92.9 | 92/99 | 98.3 | 113/115 | 95.3 | 75.0 | 18/24 | 97.9 | 186/190 | 95.3 |
| PreservCyt | APTIMA Combo 2 | 198 | 93.3 | 83/89 | 98.2 | 107/109 | 96.0 | 94.1 | 16/17 | 99.4 | 180/181 | 99.0 | |
The comparator HC2 test used in the first study does not distinguish between C. trachomatis and N. gonorrhoeae infection. Of 124 samples positive for C. trachomatis or N. gonorrhoeae by HC2, 110 (88.7%) were positive by the new assay. Of 143 samples negative by HC2, 141 (94.0%) were negative by the new assay.
The second study investigated first-catch urine, PreservCyt, and SurePath samples previously tested by the APTIMA Combo 2 assay and cervical and vaginal swabs previously tested by the BD ProbeTec assay. One hundred forty-nine samples of these specimen types were tested in a blinded fashion. In this study, there was 100% total agreement for N. gonorrhoeae detection between the new assay and APTIMA Combo 2 with first-catch urine (30/30), PreservCyt (29/29), or SurePath (30/30) samples. One hundred percent total agreement of the new assay for N. gonorrhoeae detection with BD ProbeTec was seen with vaginal (30/30) and cervical (30/30) swabs. There was 100% total agreement between the new assay and APTIMA Combo 2 for detection of C. trachomatis in urine (30/30) and SurePath (30/30) samples. One PreservCyt sample was APTIMA positive, new-assay negative for detection of C. trachomatis. Positive agreement with APTIMA Combo 2 for detection of C. trachomatis in PreservCyt medium was 93.8% (15/16), with a negative agreement of 100% (14/14), for a total agreement of 96.6%. One cervical swab sample and one vaginal swab sample were BD ProbeTec positive, new-assay negative for detection of C. trachomatis. Positive agreement with BD ProbeTec for C. trachomatis detection in cervical swabs was 94.7% (18/19), while positive agreement for vaginal swabs was 92.9% (13/14). For both swab types, negative agreement was 100% (11/11 and 16/16, respectively), and total agreement was 96.7%.
The third study evaluated agreement between the new assay and APTIMA Combo 2 for first-catch urine and PreservCyt samples. This study aimed to verify the results of the previous study with a larger sample size. The operator was not blinded to sample status in this study. Urine samples were collected and tested in APTIMA Combo 2 prior to being designated for testing with the new assay. Neat urine samples were stored for an average of 2.10 days (standard deviation = 0.4 days) prior to being added to the new-assay sample transport medium. Total agreement between the new assay and APTIMA Combo 2 for C. trachomatis detection in first-catch urine specimens was 95.8%, with a positive agreement of 92.9% (92/99) and a negative agreement of 98.3% (113/115). Total agreement between the new assay and APTIMA Combo 2 for N. gonorrhoeae detection in first-catch urine specimens was 95.3%, with a positive agreement of 75.0% (18/24) and a negative agreement of 97.9% (186/190). Total agreement between the new assay and APTIMA Combo 2 for N. gonorrhoeae detection in PreservCyt samples was 99.0%, with a positive agreement of 94.1% (16/17) and a negative agreement of 99.1% (180/181). Total agreement between the new assay and APTIMA Combo 2 for C. trachomatis detection in PreservCyt samples was 96.0%, with a positive agreement of 93.3% (83/89) and a negative agreement of 98.2% (107/109).
DISCUSSION
tHDA technology (1) was previously applied to the detection of several infectious organisms (10, 12, 15). Specific modifications made to the technology for efficient multiplex amplification and detection were described by Doseeva et al. (6a), which enable combined amplification and detection of three targets and an amplification control in a single reaction. The tHDA assay presented here uses these modifications to detect C. trachomatis and N. gonorrhoeae in multiplex. The assay detects the multicopy N. gonorrhoeae opa gene (11) and has targets on both the genome and cryptic plasmid of C. trachomatis (13). The cryptic plasmid target lies outside the region deleted in the Swedish variant of C. trachomatis (14). The use of multiple targets renders the assay less prone to false-negative results, which may be caused by mutations or deletions in the target genes. The new assay demonstrated the ability to detect 0.05 IFU/ml for C. trachomatis and 0.003 CFU/ml for N. gonorrhoeae. This level of detection is comparable or superior to those claimed by commercially available assays. For example, limits of detections for C. trachomatis according to their package inserts are 1 IFU/assay for APTIMA Combo 2, 5 to 200 EBs/assay for BD ProbeTec, 1 IFU/assay for Roche COBAS Amplicor, and 320 DNA copies/assay for Abbott RealTime CT/NG. The package inserts list limits of detections for N. gonorrhoeae as 50 cells/assay for APTIMA Combo 2, 5 to 25 cells/assay for BD ProbeTec, 5 CFU/assay for Roche COBAS Amplicor, and 320 DNA copies/assay for Abbott RealTime CT/NG. Studies of the analytical sensitivities of these assays have shown that some of them vary in practice from the claims in their package inserts; for example, Chong et al. showed that the APTIMA Combo 2 test had a limit of detection of 0.01 EB per reaction (6).
To determine if the analytical sensitivity and specificity of the new assay would translate into similar performance on clinical samples, the assay was used to test residual material from samples previously assayed by other FDA-cleared, commercially available tests. This was carried out in a series of small studies. Total agreement between the new assay and comparator assays varied somewhat depending on the sample type and target detected. The lowest total agreement was 94.0% with HC2 CT/GC, a signal amplification-based technology. This was primarily due to positive HC2 CT/GC results that were not detected by either tHDA or qPCR. This could be due to some cross-reactivity of the probes used in the HC2 CT/GC test, as documented in the product insert. The samples were not tested with the more specific HC2 CT-ID or HC2 GC-ID test. In the second study, positive and negative agreements were very high between the new assay and both APTIMA Combo 2 and BD ProbeTec, regardless of sample type. Though only limited numbers of samples were tested in the second study, positive samples were overrepresented in order to evaluate the ability of the assay to detect infection. In the third study, though overall agreements were good, positive agreement for N. gonorrhoeae in first-catch urine specimens was substantially lower than seen in the second study. This may be explained in part by the study design. Urine samples were first tested by APTIMA Combo 2, and only after results were generated were the urine specimens decanted into the stability medium, with an average delay of 2.1 days. Nucleic acids are not highly stable in urine (2, 8), and the delay between sample collection and addition of stabilizer may have contributed to target degradation, which led to the lower positive agreement seen for N. gonorrhoeae. However, good positive and negative agreements were seen between the new assay and APTIMA Combo 2 for C. trachomatis detection in first-catch urine specimens, indicating that N. gonorrhoeae may be more sensitive than C. trachomatis to storage in neat urine. Overall agreements between the new assay and APTIMA Combo 2 for C. trachomatis in first-catch urine specimens were high (>95%). Good agreement was also seen for C. trachomatis or N. gonorrhoeae (96.0% and 99.0%, respectively) in PreservCyt samples. Overall, the tHDA assay exhibits a performance comparable to those of commercially available assays and has potentially improved work flow and throughput advantages.
The common and often asymptomatic nature of many infections has led to calls for screening programs to detect and treat infected individuals. An assay with good test characteristics and a simple work flow is ideally suited to perform the high-volume testing that is required for screening programs. Isothermal amplification can enable high-throughput automation, eliminating the need for expensive and bulky thermocycling equipment. Collecting data in a homogenous endpoint mode enables sample detection with a simple fluorometer, also expediting high-throughput automation. Sequence-specific sample preparation based on hybrid capture technology and magnetic beads removes inhibitors and unwanted sequences, while still being compatible with a wide variety of media. The sequence-specific sample preparation is robust and simple and requires no complicated equipment, and manual sample extraction for 48 samples is complete in approximately half an hour. The sample preparation technique is also amenable to automation on any magnetic bead handling system, and amplification proceeds in the presence of the magnetic beads. This technology could potentially be well suited for future development in point-of-care applications, where the simplicity of the assay protocol and lack of thermocycling are benefits. For point-of-care applications, the time to first results becomes more important. Real-time amplification monitoring in these studies showed a mean amplification time before detection of signal of 28.0 min, and optimization could reduce that further. In conclusion, the tHDA assay presented here has performance characteristics similar to those of commercially available assays, including some of the most sensitive and specific tests on the market (4, 5). We demonstrated the feasibility of a tHDA-based isothermal assay for detection of C. trachomatis and N. gonorrhoeae in clinical specimens. Additional studies need to be conducted to assess further the clinical performance of the assay.
ACKNOWLEDGMENTS
We acknowledge Karolina Upton, Tom Forbes, and Kim McPartland for their assistance in developing the technology used in the studies presented here.
All work was funded by Qiagen.
Footnotes
Published ahead of print on 28 September 2011.
REFERENCES
- 1. An L., et al. 2005. Characterization of a thermostable UvrD helicase and its participation in helicase-dependent amplification. J. Biol. Chem. 280:28952–28958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cannas A., et al. 2009. Implications of storing urinary DNA from different populations for molecular analyses. PLoS One 4:e6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Centers for Disease Control and Prevention 2010. Sexually transmitted disease surveillance, 2009. Centers for Disease Control and Prevention, Department of Health and Human Services, Atlanta, GA [Google Scholar]
- 4. Chernesky M., et al. 2006. High analytical sensitivity and low rates of inhibition may contribute to detection of Chlamydia trachomatis in significantly more women by the APTIMA Combo 2 assay. J. Clin. Microbiol. 44:400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chernesky M., et al. 2007. Abilities of APTIMA, AMPLICOR, and ProbeTec assays to detect Chlamydia trachomatis and Neisseria gonorrhoeae in PreservCyt ThinPrep liquid-based Pap samples. J. Clin. Microbiol. 45:2355–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chong S., et al. 2003. Specimen processing and concentration of Chlamydia trachomatis added can influence false-negative rates in the LCx assay but not in the APTIMA Combo 2 assay when testing for inhibitors. J. Clin. Microbiol. 41:778–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a. Doseeva V., et al. 13 October 2011. Multiplex isothermal helicase-dependent amplification assay for detection of Chlamydia trachomatis and Neisseria gonorrhoeae. Diagn. Microbiol. Infect. Dis. doi:10.1016/j.diagmicrobio.2011.08.021 [DOI] [PubMed] [Google Scholar]
- 7. Gaydos C. A., Theodore M., Dalesio N., Wood B. J., Quinn T. C. 2004. Comparison of three nucleic acid amplification tests for detection of Chlamydia trachomatis in urine specimens. J. Clin. Microbiol. 42:3041–3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ingersoll J., et al. 2008. Stability of Trichomonas vaginalis DNA in urine specimens. J. Clin. Microbiol. 46:1628–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Johnson R. E., et al. 2000. Evaluation of nucleic acid amplification tests as reference tests for Chlamydia trachomatis infections in asymptomatic men. J. Clin. Microbiol. 38:4382–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim H. J., et al. 2011. A rapid and simple isothermal nucleic acid amplification test for detection of herpes simplex virus types 1 and 2. J. Clin. Virol. 50:26–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meyer T. F., Gibbs C. P., Haas R. 1990. Variation and control of protein expression in Neisseria. Annu. Rev. Microbiol. 44:451–477 [DOI] [PubMed] [Google Scholar]
- 12. Motré A., Kong R., Li Y. 2011. Improving isothermal DNA amplification speed for the rapid detection of Mycobacterium tuberculosis. J. Microbiol. Methods 84:343–345 [DOI] [PubMed] [Google Scholar]
- 13. Palmer L., Falkow S. 1986. A common plasmid of Chlamydia trachomatis. Plasmid 16:52–62 [DOI] [PubMed] [Google Scholar]
- 14. Ripa T., Nilsson P. A. 2007. A Chlamydia trachomatis strain with a 377-bp deletion in the cryptic plasmid causing false negative nucleic acid amplification tests. Sex. Transm. Dis. 34:255–256 [DOI] [PubMed] [Google Scholar]
- 15. Tang W., et al. 2010. Nucleic acid assay system for tier II laboratories and moderately complex clinics to detect HIV in low-resource settings. J. Infect. Dis. 201:S46–S51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vincent M., Xu Y., Kong H. 2004. Helicase-dependent isothermal DNA amplification. EMBO Rep. 5:795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Workowski K. A., Berman S. M. 2010. Sexually transmitted diseases treatment guidelines, 2010. MMWR Recommend. Rep. 59:1–110 [PubMed] [Google Scholar]