Abstract
Monitoring HIV drug resistance is an important component of the World Health Organization's global HIV program. HIV drug resistance testing is optimal with commercially available clinically validated test kits using plasma; however, that type of testing may not be feasible or affordable in resource-constrained settings. HIV genotyping from dried blood spots (DBS) with noncommercial (in-house) assays may facilitate the capture of HIV drug resistance outcomes in resource-constrained settings but has had varying rates of success. With in-house assays for HIV reverse transcriptase, we evaluated the yield of genotyping DBS samples collected from HIV-infected children who were enrolled in two clinical trials conducted in sub-Saharan Africa (median HIV viral load, 5.88 log10 HIV RNA copies/ml; range, 4.04 to 6.99). Overall, HIV genotypes were obtained for 94 (89.5%) of 105 samples tested (95% and 84% from clinical trials #1 and #2, respectively); however, successful analysis of 15 (16.1%) of the 94 samples required repeat testing using a different set of primers on previously synthesized cDNA. The yield of genotyping was lower on the DBS that were stored suboptimally from clinical trial #2 (56% versus 88% for optimally stored). Concordance with plasma genotypes derived using a clinically validated, commercial kit-based assay (ViroSeq HIV-1 genotyping system) was also assessed in a subset of children with paired testing. For 34 samples with paired DBS and plasma genotypes, there was 100% concordance for major drug resistance mutations. DBS genotyping using in-house assays provides an alternative for antiretroviral drug resistance testing in children in resource-constrained regions but may require region-specific optimization before widespread use.
INTRODUCTION
With the scale-up of programs for using antiretroviral drugs to prevent and treat HIV infection in children globally, the prevalence of HIV drug resistance is likely to increase. Surveillance of HIV drug resistance is an important component of the World Health Organization (WHO) global drug resistance and prevention strategy (2, 15). The availability of HIV drug resistance testing is limited in resource-constrained settings due to the high cost of such assays as well as their need for plasma samples, which requires processing of whole blood within hours of collection to minimize the degradation of viral nucleic acid. The use of clinically validated, kit-based genotyping assays that are commercially available may further limit HIV drug resistance testing in such settings due to cost and the possible lack of a steady supply of test reagents. Use of dried blood spots (DBS) rather than plasma samples and of in-house assays rather than commercial test kits may facilitate monitoring of HIV drug resistance in infected persons residing in resource-constrained settings (2–5, 7–9).
The use of in-house assays that amplify HIV reverse transcriptase (HIV-RT) and protease separately and include a nested PCR step may improve the sensitivity of genotyping DBS over that obtained with commercially available assays which amplify HIV-RT and protease in a single amplicon (>1,500 bp) with a single round of PCR (2, 4). The success levels of HIV genotyping of DBS with in-house and commercial assays (Trugene or ViroSeq) are reported as between 90 and 100% and between 57% to 83%, respectively (2). We previously reported a success rate of 94% of genotyping DBS from subtype B HIV-infected youths with an in-house assay. A high concordance between results obtained with this in-house assay was found between DBS and plasma samples. In that study, the 12 samples tested had a median viral load of 17,792 copies/ml (range of <50 to 105,000 copies/ml) (17). In this study, the success of genotyping pretreatment samples collected as DBS from children infected with non-subtype-B HIV who were likely to have HIV drug resistance due to exposure to nevirapine (NVP) for prevention of mother-to-child HIV transmission was examined. We report on the necessity for additional testing with different primers to improve the overall yield of genotyping DBS samples from different geographic regions.
MATERIALS AND METHODS
Study population.
DBS samples were collected from HIV-infected infants and children enrolled in two clinical trials: (i) the Six Week Extended Nevirapine Trial (SWEN trial; ClinicalTrials.gov number NCT00074399) (1, 12), and (ii) the P1060 trial (ClinicalTrials.gov number NCT00307151) (11). The SWEN trial, conducted in Uganda, India, and Ethiopia, compared the effectiveness of single-dose nevirapine (sdNVP) to that of up to 6 weeks of extended daily infant nevirapine (NVP) prophylaxis for the prevention of postnatal HIV transmission (1). The P1060 trial, conducted in South Africa, Malawi, Tanzania, India, Zambia, Uganda, and Zimbabwe, compared the efficacy of a NVP-based regimen to that of a protease inhibitor-based regimen for the treatment of HIV-infected children who did or did not receive sdNVP prophylaxis at birth (11). The following sets of DBS samples were analyzed: (i) for the SWEN trial, 58 DBS samples collected from Ethiopian infants at 6 months of age, and (ii) for the P1060 trial, 49 DBS samples collected from sdNVP-exposed children aged 6 to 22 months. DBS samples tested in this study were collected prior to the initiation of antiretroviral drugs for the treatment of HIV infection.
Collection and storage of DBS samples.
In both clinical trials, EDTA-anticoagulated whole blood (50 μl) was spotted and air dried onto each of five circles of filter paper (Whatman No. 903; Florham Park, NJ). DBS cards were stored according to instructions defined by the clinical trial. For the SWEN trial, DBS were stored at study sites for 1 to 6 years at −20°C to −40°C; DBS cards from the P1060 trial were stored for 0.3 to 1.8 years at −20°C to −40°C. DBS cards from both sites were shipped to the Johns Hopkins University (JHU) at ambient temperature. DBS were stored in their original packing until processing at 4°C (SWEN samples) and −20°C (P1060 samples). Some of the DBS cards from the P1060 trial were assessed at the time of processing as suboptimal since they were not in air-impermeable bags with silica desiccant as recommended in the guidelines by the WHO (16).
Viral load testing.
In the SWEN trial, HIV viral load tests were performed in a CLIA-certified laboratory at JHU using a single spot from a DBS card (Roche Amplicor HIV-1 monitor test, version 1.5; Roche Diagnostics, Branchburg, NJ). Spots from the same DBS card were used for HIV genotyping. In the P1060 trial, HIV viral load tests were performed using plasma samples collected at the same time as the samples that were used to prepare DBS. In the P1060 trial, viral loads were determined in different laboratories using one of several commercially available test kits, as previously described (11).
Extraction of nucleic acid from DBS.
Spots were punched from DBS cards using a 0.5-in. (1.2-cm) custom-made hole puncher (Online River, LLC, Greenwich, CT) and were deposited into a tube containing 9 ml of lysis buffer (bioMérieux, Durham, NC). In the SWEN trial, the two spots (prepared with a total of 100 μl of blood) were used for genotyping as recommended by the WHO (16). In the P1060 trial, however, five spots were used based on one of the study objectives to assess the emergence of HIV drug resistance at 4 weeks during antiretroviral therapy, when plasma HIV RNA is expected to be significantly lower than pretreatment levels. Five spots of whole blood are nearly equivalent to 100 μl of plasma. Nucleic acid (RNA and DNA) was extracted from the first 10 samples in the SWEN trial by using a manual NucliSENS silica-based extraction method; the rest of the samples were processed using a semiautomated NucliSENS silica-based extraction method (bioMérieux, Durham, NC). Two DBS samples from the SWEN trial were excluded from analysis because of technical problems that arose during the manual nucleic acid extraction step. Since the rest of the spots were consumed for infant diagnosis and viral load testing, no additional spots were available for testing, and the remaining 56 samples from this trial were analyzed. For extraction (both manual and automated), the tubes were rocked at room temperature for 1 h. Nucleic acid was then isolated using silica beads according to the manufacturer's instructions and was eluted using 60 μl of elution buffer in a tube containing 40 U of RNAseOUT (Invitrogen, Carlsbad, CA).
HIV genotyping.
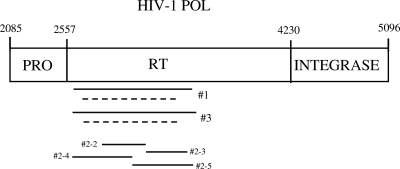
The success rates of genotyping from DBS were compared using three different genotyping methods (referred to here as assay #1, assay #2, and assay #3) (Table 1; Fig. 1). Given the lower-than-expected frequency of amplification of non-subtype-B samples from the SWEN trial using our previously published assay (assay #1) (Table 1) (17), we used a second approach (assay #2) that included even smaller overlapping HIV amplicons (227 to 382 bp) (10, 13); that approach had been used successfully to genotype non-subtype-B plasma samples using ultradeep pyrosequencing from children in a previous study (10, 13). A third assay was subsequently developed that used a combination of primers from assay #2 for the first round of PCR and the nested PCR primers and conditions for assay #1. HIV RNA was first reverse transcribed to produce cDNA (Table 1). An aliquot of the cDNA was then amplified using one of the three methods described in Table 1. In the SWEN trial, for samples that were not successfully amplified using assay #1, the cDNA was reamplified using assay #2 (10, 13). Given the improved performance of genotyping DBS when assay #2 was used to retest samples that were not amplified using assay #1, a new PCR amplification assay (assay #3) was developed and used for the second clinical trial (P1060) as the first test method. The cDNA was reamplified using assay #2 if negative, as was done for samples from the SWEN trial.
Table 1.
Primers and conditions used for HIV genotyping from DBSa
| Assay type | Step | Primer name | Primer sequence (5′–3′) | Product size (bp) | HXB2 position | Incubation conditions |
|---|---|---|---|---|---|---|
| Assay #1b | RT | 3outRTb | GGCTGTACTGTCCATTTA | NA | 3277–3260 | 50°C × 50 min, 85°C × 5 min |
| 1st round PCR | 5outRTb (F) | GTCCTRTTGAAACTGTAC | 720 | 2557–3277 | 94°C × 3 min; 32 cycles of 94°C × 30 s, 55°C × 30 s, 68°C × 45 s; 68°C × 5 min | |
| 3outRTb (R) | GGCTGTACTGTCCATTTA | |||||
| 2nd round PCR | 5innRTb (F) | ATGGCCCAAAAGTYAAAC | 663 | 2599–3262 | 94°C × 3 min; 32 cycles of 94°C × 30 s, 52°C × 30 s, 68°C × 45 s; 68°C × 5 min | |
| 3innRTb (R) | TTATCAGGATGGAGTTCA | |||||
| Assay #2c | RT | cDNA template the same as for initial assaye | ||||
| Amplicon 2 | 2F-HIVC (F) | GCCTCCCTCGCGCCATCAGGGAAGTTCAATTAGGRATACCACACCC | 247 | 2813–3060 | 94°C × 3 min; 40 cycles of 94°C × 15 s, 55°C × 20 s, 72°C × 30 s; 72°C × 8 min | |
| 2R-HVC (R) | GCCTTGCCAGCCCGCTCAGAGGGCTCTAAGATTYTTGTCAT | |||||
| Amplicon 3 | 3F-HIVC (F) | GCCTCCCTCGCGCCATCAGAGAGCCCTTTAGAGCAMAAAAYCCAGA | 227 | 3057–3284 | ||
| 3R-HIVC (R) | GCCTTGCCAGCCCGCTCAGCTGTATAGGCTGTACTGTCCATTTGTC | |||||
| Amplicon 4 | 4F-HIVC (F) | GCCTCCCTCGCGCCATCAGGTACCAGTAAAATTAAAGCCAGGAATGG | 382 | 2571–2953 | ||
| 4R-HIVC (R) | GCCTTGCCAGCCCGCTCAGATACTAGGTATGGTGAATGCAGTATAYTT | |||||
| Amplicon 5 | 5F-HIVC (F) | GCCTCCCTCGCGCCATCAGCACCAGGGATTAGATATCARTAYAATGT | 363 | 2965–3328 | ||
| 5R-HIVC (R) | GCCTTGCCAGCCCGCTCAGAACTTCTGTATATCATTGACAGTCCA | |||||
| Assay #3d | RT | 5Rcdna | AACTTCTGTATATCATTGACAGTCCA | NA | 3328–3303 | 42°C × 50 min, 70°C × 15 min |
| 1st round PCR | 4F-HIVC (F) | GCCTCCCTCGCGCCATCAGGTACCAGT AAAATTAAAGCCAGGAATGG | 757 | 2571–3328 | 94°C × 3 min; 40 cycles of 94°C × 30 s, 55°C × 30 s, 72°C × 90 s; 72°C × 8 min | |
| 5R-HIVC (R) | GCCTTGCCAGCCCGCTCAGAACTTCTG TATATCATTGACAGTCCA | |||||
| 2nd round PCR | Same as assay #1 |
Primers used in amplification were also used as the sequencing primers. NA, not applicable; F, forward; R, reverse.
Previously published method (17).
Multiple short amplicons were amplified using 25-μl reaction mixtures containing the four different primer pairs at a final concentration of 0.4 μM and 2.5 U of FastStart high-fidelity enzyme blend (Roche Applied Science, Mannheim, Germany) (13). Assay #2 was used to repeat genotyping for samples that failed to amplify using assay #1 or assay #3.
The first-round of PCR for assay #3 in a 50-μl reaction mixture containing the amplicon 4 forward primer, 4F-HIVC, and amplicon 5 reverse primer, 5R-HIVC, at a final concentration of 0.4 μM and 2.5 U of FastStart high-fidelity enzyme blend (Roche Applied Science, Mannheim, Germany).
Assay #2 used the same cDNA template that was used in the first assay.
Fig. 1.
Amplicons produced using in-house HIV genotyping assays (see text). The positions of amplicons in HIV genotyping assays described in Table 1 are shown. Solid lines show the first-round PCR amplicons, and dotted lines show the amplicons produced in nested PCR. Three assays were used for analysis (#1, #2, and #3). Assay #2 produces four short, overlapping amplicons (#2-2, #2-3, #2-4, and #2-5). PRO, HIV protease; RT, HIV reverse transcriptase. Numbers above the diagram indicate coordinates in HXB2.
Amplified DNA was analyzed by agarose gel electrophoresis, purified using the QIAquick PCR purification kit (Qiagen, Valencia, CA), and sequenced using an ABI 3730 capillary electrophoresis instrument (Applied Biosystems, South San Francisco, CA) with HIV reverse transcriptase-specific sequencing primers (Table 1), as previously described (17).
Plasma samples from the P1060 trial were also analyzed in a previous study using the ViroSeq HIV-1 genotyping system (Celera, Alameda CA) (11). The ViroSeq system detects mutations in HIV protease and HIV reverse transcriptase; the methods used for DBS analysis in this report provide information only about mutations in HIV reverse transcriptase. Testing using plasma and DBS were performed by different personnel in different laboratories. Resistance test results were not compared until all of the testing was completed.
Analysis of HIV drug resistance mutations.
HIV sequences derived from DBS genotyping were aligned to subtype C HIV reference sequences from the Los Alamos database (http://www.HIV.lanl.gov/content/index/) using Sequencher 4.9 software (GeneCodes, Ann Arbor, MI). Polymorphisms were visualized in BioEdit 7.0.9 (Ibis Bioscience, Carlsbad, CA). HIV subtypes were assessed using neighbor-joining phylogenetic trees synthesized in Mega 4.0 (http://www.megasoftware.net/). These methods were also used to compare the sequences from DBS samples and to compare sequences from DBS and plasma samples. Nonnucleoside reverse transcriptase inhibitor (NNRTI) and nucleoside reverse transcriptase inhibitor (nRTI) drug resistance mutations were interpreted using the Stanford Drug Resistance Database (http://HIVdb.stanford.edu/).
Statistical analysis.
Statistical analysis was performed using Stata version 11. Fisher's exact tests were used to compare differences in performance of genotyping using the various assays.
RESULTS
HIV genotyping of DBS samples.
Table 2 shows the source of the DBS samples used for analysis, the duration of sample storage at the study sites, the viral loads of the samples, and the results of HIV genotyping. Amplification was successful for 41 (73%) of 56 of the samples from the SWEN trial using assay #1 only. A second amplification using assay #2 on previously synthesized cDNA was successful for 12 of the 15 samples that did not amplify with assay #1. Using this combined approach (assay #1 followed by assay #2), 53 (95%) of the 56 samples from the SWEN trial were successfully genotyped with complete coverage of known sites of drug resistance in up to amino acid 260 in HIV reverse transcriptase. The three samples that were retested were considered to be amplification failures, even though PCR products were detected by gel electrophoresis, because only partial coverage of HIV reverse transcriptase was obtained.
Table 2.
Yield of HIV genotyping of DBS samples collected in clinical trials in sub-Saharan Africa
| Clinical trial | Sample subset | Median duration (yr) of sample storageb | No. of samplesc | Median log10 HIV viral load (range) | Yield (%) of HIV genotyping usinga: |
Overall recovery (%) with retestingd | |
|---|---|---|---|---|---|---|---|
| Assay #1 | Assay #3 | ||||||
| SWEN | All | 3.3 | 56 | 5.9 (4.0–7.0) | 41/56 (73%) | NA | 53/56 (95%) |
| P1060 | All | 1 | 49 | 5.8 (4.6–6.9) | NA | 38/49 (78%) | 41/49 (84%) |
| Optimal storage | 0.5 | 33 | 6.0 (4.6–6.9) | NA | 29/33 (88%) | 30/33 (91%) | |
| Suboptimal storage | 1.2 | 16 | 5.9 (5.5–6.4) | NA | 9/16 (56%) | 11/16 (69%) | |
NA, not applicable.
Duration of storage at study sites prior to sample shipment.
Samples and sample storage are described in Materials and Methods.
Assay #2 was performed if assay #1 or #3 failed to yield genotype (see Materials and Methods).
Based on these results, assay #3 was used to first test samples from the second clinical trial. Using assay #3, 38 (78%) of 49 samples from the P1060 trial were successfully genotyped. Because some of the samples from the P1060 trial were not properly stored (see Materials and Methods), we compared the genotyping success rates for samples that were compared with those that were not properly stored (Table 2). The proportion of samples successfully genotyped using assay #3 was higher for samples that were stored properly than for samples that were not stored properly (29/33 = 88% versus 9/16 = 56%; P = 0.03) (Table 2). The 11 samples from the P1060 trial that failed to amplify with assay #3 were retested using assay #2 as was done for samples from the first clinical trial; 3 (27%) of those 11 samples were successfully amplified. Using this combined approach (assay #3 followed by assay #2), we were able to genotype 30 (91%) of the 33 samples that were stored properly and 11 (69%) of the 16 samples that were not stored properly (overall success rate, 41/49 = 84%) (Table 2).
Detection of drug resistance in DBS samples.
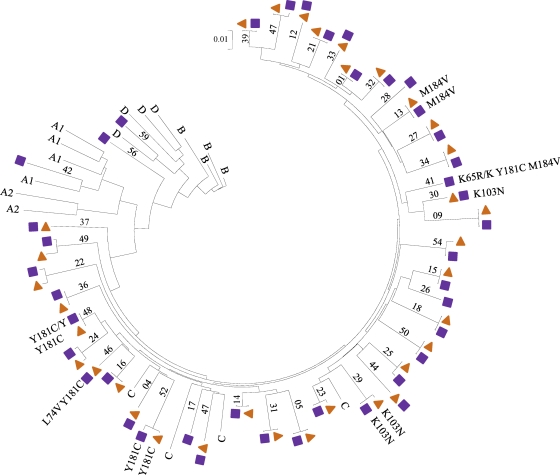
As expected, NNRTI resistance mutations were detected in samples from infants in the SWEN trial who received nevirapine for the prevention of mother-to-child transmission of HIV. Overall, 43% (23/53) of the infants had NNRTI mutations detected (Y181C [n = 9], Y188C [n = 5], K103N [n = 4], G190A [n = 4], and V106M [n = 1]). Antiretroviral drug resistance mutations were also detected in 7 (17%) of 41 samples from the P1060 trial (NNRTI mutations: Y181C [n = 4] and K103N [n = 2]; NRTI mutations: M184V [n = 2], K65R [n = 1], and L74V [n = 1]; one child had K65R, Y181C, and M184V, and a second child had L74V and Y181C [Fig. 2 ]).
Fig. 2.
Phylogenetic relationships of sequences obtained from matched dried-blood-spot and plasma samples (P1060 trial). A phylogenetic tree constructed using sequences obtained using DBS samples (n = 41; 41 symbols) or plasma samples (n = 34; 34 symbols) is shown; 34 of the samples are matched (prepared from the same whole-blood collection). Reference sequences for subtype C are indicated with “C.” Mutations detected in the samples are shown.
Comparison between DBS and plasma genotypes.
A single blood collection was used to prepare the DBS and plasma samples for a subset of infants in the P1060 trial. HIV genotyping results were obtained for 41 (84%) of the 49 DBS samples analyzed (Table 1); genotyping results from the ViroSeq HIV-1 genotyping system were obtained previously from plasma samples matched to 34 of those 41 samples (Fig. 2) (11). Phylogenetic analysis of HIV sequences obtained from analysis of DBS and plasma samples grouped together as expected, confirming the absence of sample mix-ups (Fig. 2).
Drug resistance mutations were detected in samples from 6 of 34 children who had results obtained for both DBS and plasma. In all six cases, the same major drug resistance mutations were detected in both samples (Fig. 2). In one sample, an amino acid substitution at position 215 of HIV reverse transcriptase (T215I) was detected in plasma by using the ViroSeq HIV genotyping system but was not detected in the DBS sample by using in-house assay #3.
DISCUSSION
In this study, we evaluated the yield of HIV genotyping of DBS samples obtained from HIV-infected children enrolled in clinical trials in sub-Saharan Africa (1, 11). HIV genotypes for reverse transcriptase were successfully obtained for 94 (90%) of 105 samples analyzed, despite collection and long-term storage in countries with differing temperature, humidity, and storage conditions. However, it is important to emphasize that three different primer sets were required to achieve this success rate, with 16.1% (15 of 94 samples) failing to amplify on the first attempt and requiring a second step of testing (one-step PCR on previously synthesized cDNA with a different method that produced shorter, overlapping amplicons). While incorporation of repeat testing with multiple short amplicons enhanced the recovery of genotypes in the samples tested from the first clinical trial (tested using assay #1), switching to assay #3, which included a combination of the outermost primers of the short, overlapping primers that we previously reported for pyrosequencing of subtype C HIV reverse transcriptase (10) and involved testing of samples from five countries (South Africa, Uganda, Malawi, Zimbabwe, and Tanzania), increased the yield of genotyping to nearly 90% for samples that were stored optimally. The addition of the repeat testing increased the success rate by 3% for optimally stored samples and 13% for those that were suboptimally stored, highlighting the importance of optimal storage conditions for the genotyping of DBS. Importantly, with assay #3, a subset of the samples from the P1060 trial had matched plasma samples that were analyzed previously using an FDA-cleared HIV genotyping kit. For the pretreatment samples that were analyzed with both methods in a blind manner, results from DBS genotyping were 100% concordant with plasma genotyping for major drug-resistant mutations obtained using the FDA-cleared genotyping kit. Together, these findings suggest that the primers used in assay #3 may be suitable for application in sub-Saharan Africa.
There are several limitations to this study. First, all of the samples tested were obtained from infants and children who were not receiving antiretroviral therapy at the time of sample collection. Therefore, HIV viral loads in the samples were high, facilitating HIV nucleic acid recovery and amplification from DBS samples. The proportion of samples successfully genotyped with DBS may be lower when samples with lower viral loads (e.g., those obtained at the time of treatment failure) are tested. Second, most of the samples analyzed did not have drug resistance mutations; further evaluation of the sensitivity for detecting mutations using DBS samples is needed, using samples with a variety of drug resistance mutations present at different levels in the viral population. Third, the methods used in this report provide information for HIV reverse transcriptase only. Methods that include analysis of HIV-1 protease are needed, particularly since the WHO now recommends protease inhibitor-based regimens for infants and children with prior exposure to nevirapine prophylaxis (14); NVP-based regimens are widely used for prophylaxis in resource-constrained countries. Analysis of both HIV reverse transcriptase and protease from DBS samples is likely to require amplification of multiple amplicons because HIV RNA can become degraded during storage and/or extraction; in contrast, commercially available assays that use plasma for analysis typically obtain information for both HIV protease and HIV reverse transcriptase from a single amplicon. Fourth, the methods used in this report rely on nested PCR for DNA amplification; nested PCR can increase the risk of sample cross-contamination and may bias the proportion of viral variants represented in the amplified DNA product. Finally, we used three different methods for RT-PCR (different primer sets and amplification conditions); different numbers of DBS spots were also used for testing. Therefore, the amounts of blood used for analysis varied among the samples tested.
Nevertheless, the overall success of obtaining genotypes in this study and the high concordance with plasma genotypes coupled with the fact that many laboratories in resource-constrained settings are already familiar with use of DBS samples for infant diagnostic testing (6) suggest that in-house DBS genotyping assays provide an alternative for antiretroviral drug resistance testing in children in resource-constrained regions but may require region-specific optimization before widespread use.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (NIH) (R01-HD057784-04 to D.P., R01-AI38576 to A.R., and U01-AI068632 [BRS-IMPCT-Q-00100-T014 and BRS-IMPCT-Q-06-00100-T003] to S.H.E.) and the Elizabeth Glaser Pediatric AIDS Foundation (Elizabeth Glaser Scientist Award to D.P.). Overall support for the International Maternal Pediatric Adolescent AIDS Clinical Trials (IMPAACT) group was provided by the National Institute of Allergy and Infectious Diseases (NIAID) (U01 AI068632), the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), and the National Institute of Mental Health (NIMH) (AI068632). This work was supported by the Statistical and Data Analysis Center at Harvard School of Public Health under the NIAID cooperative agreements 5 U01 AI41110 (with the Pediatric AIDS Clinical Trials Group [PACTG]) and 1 U01 AI068616 (with the IMPAACT group). Support of the sites was provided by the NIAID and the NICHD International and Domestic Pediatric and Maternal HIV Clinical Trials Network, funded by the NICHD (contract number N01-DK-9-001/HHSN267200800001C).
We thank the participants of the Ethiopian SWEN trial and the IMPAACT P1060 trial and the study teams from both trials for assistance with sample and data management.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Published ahead of print on 28 September 2011.
REFERENCES
- 1. Bedri A., et al. 2008. Extended-dose nevirapine to 6 weeks of age for infants to prevent HIV transmission via breastfeeding in Ethiopia, India, and Uganda: an analysis of three randomised controlled trials. Lancet 372:300–313 [DOI] [PubMed] [Google Scholar]
- 2. Bertagnolio S., Parkin N. T., Jordan M., Brooks J., Garcia-Lerma J. G. 2010. Dried blood spots for HIV-1 drug resistance and viral load testing: a review of current knowledge and WHO efforts for global HIV drug resistance surveillance. AIDS Rev. 12:195–208 [PubMed] [Google Scholar]
- 3. Bertagnolio S., et al. 2007. HIV-1 drug resistance surveillance using dried whole blood spots. Antivir. Ther. 12:107–113 [PubMed] [Google Scholar]
- 4. Buckton A. J. 2008. New methods for the surveillance of HIV drug resistance in the resource poor world. Curr. Opin. Infect. Dis. 21:653–658 [DOI] [PubMed] [Google Scholar]
- 5. Cassol S. A., et al. 1996. Dried blood spots collected on filter paper: an international resource for the diagnosis and genetic characterization of human immunodeficiency virus type-1. Mem. Inst. Oswaldo Cruz 91:351–358 [DOI] [PubMed] [Google Scholar]
- 6. Fiscus S. A., et al. 2006. HIV-1 viral load assays for resource-limited settings. PLoS Med. 3:e417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hamers R. L., Smit P. W., Stevens W., Schuurman R., Rinke de Wit T. F. 2009. Dried fluid spots for HIV type-1 viral load and resistance genotyping: a systematic review. Antivir. Ther. 14:619–629 [PubMed] [Google Scholar]
- 8. Johannessen A., Troseid M., Calmy A. 2009. Dried blood spots can expand access to virological monitoring of HIV treatment in resource-limited settings. J. Antimicrob. Chemother. 64:1126–1129 [DOI] [PubMed] [Google Scholar]
- 9. McNulty A., et al. 2007. Evaluation of dried blood spots for human immunodeficiency virus type 1 drug resistance testing. J. Clin. Microbiol. 45:517–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moorthy A., et al. 2011. Induction therapy with protease-inhibitors modifies the effect of nevirapine resistance on virologic response to nevirapine-based HAART in children. Clin. Infect. Dis. 52:514–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Palumbo P., et al. 2010. Antiretroviral treatment for children with peripartum nevirapine exposure. N. Engl. J. Med. 363:1510–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Persaud D., et al. 2011. Slower clearance of nevirapine resistant virus in infants failing extended nevirapine prophylaxis for prevention of mother-to child HIV-transmission. AIDS Res. Hum. Retroviruses 27:823–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Simen B. B., et al. 2009. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J. Infect. Dis. 199:693–701 [DOI] [PubMed] [Google Scholar]
- 14. World Health Organization 2010. Antiretroviral therapy for HIV infection in infants and children: towards universal access. Recommendations for a public health approach. World Health Organization, Geneva, Switzerland: [PubMed] [Google Scholar]
- 15. World Health Organization 2010. HIV drug resistance early warning indicators. World Health Organization, Geneva, Switzerland [Google Scholar]
- 16. World Health Organization 2010. WHO manual for HIV drug resistance testing using dried blood spot specimens. World Health Organization, Geneva, Switzerland [Google Scholar]
- 17. Ziemniak C., George-Agwu A., Moss W. J., Ray S. C., Persaud D. 2006. A sensitive genotyping assay for detection of drug resistance mutations in reverse transcriptase of HIV-1 subtypes B and C in samples stored as dried blood spots or frozen RNA extracts. J. Virol. Methods 136:238–247 [DOI] [PubMed] [Google Scholar]