Abstract
Diagnosis of invasive pulmonary aspergillosis (IPA) remains a major challenge to clinical microbiology laboratories. We developed rapid and sensitive quantitative PCR (qPCR) assays for genus- and species-specific identification of Aspergillus infections by use of TaqMan technology. In order to validate these assays and understand their potential diagnostic utility, we then performed a blinded study of bronchoalveolar lavage (BAL) fluid specimens from well-characterized models of IPA with the four medically important species. A set of real-time qPCR primers and probes was developed by utilizing unique ITS1 regions for genus- and species-specific detection of the four most common medically important Aspergillus species (Aspergillus fumigatus, A. flavus, A. niger, and A. terreus). Pan-Aspergillus and species-specific qPCRs with BAL fluid were more sensitive than culture for detection of IPA caused by A. fumigatus in untreated (P < 0.0007) and treated (P ≤ 0.008) animals, respectively. For infections caused by A. terreus and A. niger, culture and PCR amplification from BAL fluid yielded similar sensitivities for untreated and treated animals. Pan-Aspergillus PCR was more sensitive than culture for detection of A. flavus in treated animals (P = 0.002). BAL fluid pan-Aspergillus and species-specific PCRs were comparable in sensitivity to BAL fluid galactomannan (GM) assay. The copy numbers from the qPCR assays correlated with quantitative cultures to determine the pulmonary residual fungal burdens in lung tissue. Pan-Aspergillus and species-specific qPCR assays may improve the rapid and accurate identification of IPA in immunocompromised patients.
INTRODUCTION
Invasive pulmonary aspergillosis (IPA) is a leading cause of infectious pneumonic mortality in patients with hematological malignancies, aplastic anemia, myelodysplasia, and hematopoietic stem cell transplantation (HSCT) (14). Aspergillus fumigatus is the most common cause of invasive pulmonary aspergillosis in immunocompromised patients. Non-Aspergillus fumigatus species may also cause invasive disease (27). Aspergillus terreus is resistant to amphotericin B and causes pulmonary and disseminated infection (6, 15, 24, 28). Aspergillus flavus has been found to cause combined sinopulmonary disease and has been implicated in causing outbreaks of nosocomial aspergillosis. Aspergillus niger is usually a saprophytic organism but may cause invasive disease. Early diagnosis and initiation of antifungal therapy are important for improved outcomes.
The diagnosis of IPA remains challenging. Standard laboratory techniques for the diagnosis of IPA include direct examination, lung tissue histology, and culture of respiratory secretions. Serum galactomannan (GM) and (1→3)-β-d-glucan assays are FDA-approved assays for the diagnosis of invasive pulmonary aspergillosis. The measurement of galactomannan in bronchoalveolar lavage (BAL) fluid shows promise in enhancing the diagnosis of IPA (2, 4, 12, 18).
Under the best of circumstances, BAL fluid analysis yields a diagnosis by culture and direct exam in only approximately 50% of cases (13). Some centers report substantially lower frequencies. Saito et al. reported 0% diagnosis for patients who ultimately succumbed to IPA and whose infection was found postmortem (22).
We therefore developed rapid and sensitive real-time PCR assays for genus- and species-specific identification of Aspergillus infections by use of TaqMan technology. In order to validate these assays and understand their potential diagnostic utility, we then performed a controlled blinded study of BAL fluid and lung tissue biopsy specimens from well-characterized persistently neutropenic rabbit models of IPA with the four medically important species (15, 28). These studies establish the experimental foundation for clinical investigation of this real-time PCR system in lung transplant recipients and for the clinical application of the system in the reference laboratory (16).
MATERIALS AND METHODS
Primer and probe design.
For the design of species-specific assays, alignments of 27 to 67 ITS1 sequences for each Aspergillus species (A. fumigatus, A. flavus, A. niger, and A. terreus) were created using Geneious software (Biomatters Ltd., Auckland, New Zealand). The complete set of sequences available for each of the four species was used (National Center for Biotechnology Information [NCBI] public genetic database [GenBank]). Primers and probes that annealed to sequences of the target Aspergillus species but not to other Aspergillus spp., Penicillium spp., or other unrelated fungal genera were initially chosen. However, recent additions to GenBank revealed that the assay target region for A. niger has significant homology to five additional members of the section Nigri (Aspergillus costaricensis, Aspergillus foetidus, Aspergillus piperis, Aspergillus vandensis, and Aspergillus tubingensis [one sequence]).
In order to detect the full spectrum of potential Aspergillus pathogens, a pan-Aspergillus real-time PCR assay was developed. For design of the pan-Aspergillus real-time PCR assay, BLAST (Basic Local Alignment Search Tool) searches for homology with the 18S rRNA genes of non-Aspergillus species of fungi were conducted. For the pan-Aspergillus primers, no significant homologies with the target area were identified with any fungal species other than Penicillium spp. BLAST searches for homology with the human 18S rRNA gene were also conducted. No significant homologies for the target area were identified within the human 18S rRNA gene. Primer Express 3.0 (Applied Biosystems, Foster City, CA) was used to help select primers and probes with optimal melting temperatures for TaqMan real-time PCR on a model 7500 Fast instrument (Applied Biosystems, Foster City, CA). The primers were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA), and the probes were synthesized by Biosearch Technologies, Inc. (Novato, CA). The sequences of the primers and probes are shown in Table 1, and the locations of real-time PCR assay targets are shown in Fig. 1.
Table 1.
Primers and probes used in Aspergillus real-time PCR assays
| Assay | Primer or probe | Primer sequence (5′ → 3′)a | GenBank accession no. | Nucleotide positions |
|---|---|---|---|---|
| Pan-Aspergillus | pan-Asp-For | GTGGAGTGATTTGTCTGCTTAATTG | AF548061d | 1215–1239 |
| pan-Asp-Probe | CGGCCCTTAAATAGCCCGGTCCGb | AF548061d | 1258–1280 | |
| pan-Asp-Rev | TCTAAGGGCATCACAGACCTGTT | AF548061d | 1345–1367 | |
| A. fumigatus | A. fum-For | GCCCGCCGTTTCGAC | GU319985 | 86–100 |
| A. fum-Probe | CCCGCCGAAGACCCCAACATGb | GU319985 | 136–156 | |
| A. fum-Rev | CCGTTGTTGAAAGTTTTAACTGATTAC | GU319985 | 195–221 | |
| A. flavus | A. flav-For | CGAGTGTAGGGTTCCTAGCGA | HQ026744 | 59–79 |
| A. flav-Probe | TCCCACCCGTGTTTACTGTACCTTAGTTGCTb | HQ026744 | 88–118 | |
| A. flav-Rev | CCGGCGGCCATGAAT | HQ026744 | 133–147 | |
| A. niger | A. nig-For | GCCGGAGACCCCAACAC | GU256739 | 95–111 |
| A. nig-Probe | AATCAACTCAGACTGCACGCTTTCAGACAGb | GU256739 | 117–146 | |
| A. nig-Rev | TGTTGAAAGTTTTAACTGATTGCATT | GU256739 | 148–173 | |
| A. terreus | A. terr-For | CATTACCGAGTGCGGGTCTTTA | GU256759 | 12–33 |
| A. terr-Probe | CCCAACCTCCCACCCGTGACTATTGb | GU256759 | 37–61 | |
| A. terr-Rev | CCCGCCGAAGCAACAAG | GU256759 | 65–81 | |
| Lambda universal | λ-UIC-For | CAGCAGAACACCCCCATC | U57606 | 1162–1179 |
| internal control | λ-UIC-Probe | AACCACTACCTGAGCACCCAGTCCc | U57606 | 1207–1230 |
| λ-UIC-Rev | GTGATCGCGCTTCTCGTT | U57606 | 1249–1266 |
Primer concentrations were optimized empirically. Aspergillus target and λ-UIC primers had final reaction concentrations of 800 and 200 nM, respectively.
All Aspergillus probes possessed a 5′ 6-carboxyfluorescein (FAM) reporter dye and a 3′ dark quencher.
The λ-UIC probe possessed a 5′ VIC reporter dye and a 3′ tetramethyl rhodamine isocyanate (TAMRA) quencher.
The pan-Aspergillus oligonucleotides were designed to hybridize to the 18S rDNA region of all Aspergillus species. The sequence accession number for the pan-Aspergillus assay is based on the A. fumigatus sequence (GenBank accession no. AF548061).
Fig. 1.
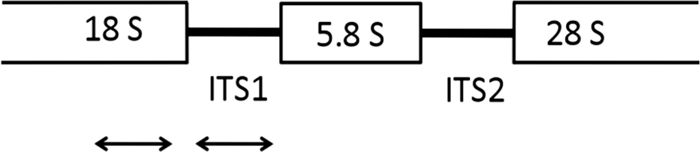
Locations of real-time PCR assay targets. An rDNA single repeat is shown. The pan-Aspergillus assay targets the 18S rRNA gene region. The species-specific assays target the ITS1 region.
Organisms.
Isolates of Aspergillus spp. and other fungi were obtained for use in in vitro development of the real-time PCR assays (Tables 2 and 3). A different panel of Aspergillus sp. isolates was studied in experimental IPA in order to validate the in vitro analytical PCR assay. Isolates of Aspergillus fumigatus (NIH 4215; ATCC MYA-1163), A. flavus (NIH 8B and 10B), A. terreus (NIH 68A and 2624; TX 94-559, 96-1290, and 98-87), and A. niger (NIH 120 and 141) were obtained from patients with invasive pulmonary aspergillosis and used in all experiments for in vitro calibration studies and experimental IPA.
Table 2.
Testing of species-specific real-time PCR assays against genomic DNAs of Aspergillus spp. and other fungi
| Fungal species | Straina | Real-time PCR assay result |
|||
|---|---|---|---|---|---|
| A. fumigatus | A. flavus | A. niger | A. terreus | ||
| Aspergillus fumigatus | NRRL 163 | + | − | − | − |
| NRRL 164 | + | − | − | − | |
| NRRL 165 | + | − | − | − | |
| NRRL 166 | + | − | − | − | |
| NRRL 5109 | + | − | − | − | |
| NRRL 5587 | + | − | − | − | |
| Aspergillus flavus | NRRL 1957 | − | + | − | − |
| NRRL 3518 | − | + | − | − | |
| NRRL 4818 | − | + | − | − | |
| Aspergillus niger | NRRL 3 | − | − | + | − |
| NRRL 326 | − | − | + | − | |
| Aspergillus terreus | NRRL 255 | − | − | − | + |
| NRRL 260 | − | − | − | + | |
| NRRL 680 | − | − | − | + | |
| NRRL 1913 | − | − | − | + | |
| NRRL 2399 | − | − | − | + | |
| Penicillium citrinum | This study | − | − | − | − |
| Penicillium chrysogenum | This study | − | − | − | − |
| Penicillium purpurogenum | This study | − | − | − | − |
| Candida albicans (SC5314) | MRL 842 | − | − | − | − |
NRRL, Northern Regional Research Laboratories (part of the USDA Agricultural Research Service Culture Collection); MRL, Mycology Reference Laboratory (part of the Center for Medical Mycology at the University Hospitals of Cleveland). Three Penicillium strains were obtained as clinical isolates from the National Cancer Institute during the course of this study.
Table 3.
Pan-Aspergillus real-time PCR assay against genomic DNAs of Aspergillus spp. and other fungi
| Fungal species | Straina | Pan-Aspergillus real-time PCR result |
|---|---|---|
| A. caesiellus | NRRL 5061 | + |
| A. candidus | NRRL 303 | + |
| A. carbonarius | NRRL 348 | + |
| A. carneus | NRRL 527 | + |
| A. clavotonanicus | NRRL 4741 | + |
| A. clavatus | NRRL 1 | + |
| A. conicus | NRRL 149 | + |
| A. deflectus | NRRL 2206 | + |
| A. flavus | NRRL 1957 | + |
| A. fumigatus | NRRL 163 | + |
| A. glaucus | NRRL 116 | + |
| A. janus | NRRL 1787 | + |
| A. japonicus | NRRL 360 | + |
| A. nidulans | NRRL 187 | + |
| A. niger | NRRL 326 | + |
| A. ochraceus | NRRL 398 | + |
| A. oryzae | NRRL 447 | + |
| A. penicillioides | NRRL 4548 | + |
| A. restrictus | NRRL 154 | + |
| A. tamari | NRRL 4911 | + |
| A. terreus | NRRL 260 | + |
| A. unguis | NRRL 216 | + |
| Candida albicans (SC5314) | MRL 842 | − |
| Candida glabrata | NRRL Y-27785 | − |
| Candida parapsilosis | NRRL Y-12969 | − |
| Cunninghamella echinulata | NRRL 1386 | − |
| Mucor circinelloides | NRRL 1405 | − |
| NRRL 223 | − | |
| Mucor indicus | NRRL 13083 | − |
| NRRL 555 | − | |
| Mucor ramosissimus | NRRL 5904 | − |
| Penicillium chrysogenum | This study | + |
| Penicillium citrinum | This study | + |
| Penicillium purpurogenum | This study | + |
| Rhizomucor pusillus | NRRL 28626 | − |
| Pneumocystis jirovecii | ATCC 50385 | − |
| Rhizomucor pusillus | NRRL 3639 | − |
| Rhizopus microsporus | NRRL 13477 | − |
| NRRL 3671 | − | |
| Rhizopus oryzae | NRRL 395 | − |
Please see the footnotes to Table 2. ATCC, American Type Culture Collection.
Growth and harvest of fungal stock cultures.
Conidia were isolated from Aspergillus spp. and Penicillium spp. grown on Sabouraud glucose agar (SGA) plates for 2 to 7 days, depending on the growth rate. Conidia were harvested by washing the cultures with phosphate-buffered saline (PBS). Conidia were then separated from hyphal elements by filtration through sterile gauze. Following centrifugation at 2,000 × g for 2 min, the conidia were washed and resuspended with PBS. Conidia were used directly as extraction positive-control material and as sources for specificity testing. Additionally, 1 × 107 A. fumigatus and A. terreus conidia were inoculated into 10 ml yeast peptone glucose medium and incubated at 37°C with shaking. Morphological changes and quantification for in vitro molecular studies were determined by direct light microscopy. Once 100% of conidia had swelled and most had initiated germ tube formation (∼5.5 h), these “germlings” were mixed in equal proportions, aliquoted, frozen at −80°C, and subsequently used in limit-of-detection (analytical sensitivity) experiments. The limit of detection is the concentration at which ≥95% of samples are positive.
Development of control for PCR inhibition and nucleic acid extraction efficiency.
In order to control for PCR inhibition and poor DNA extraction, we designed a bacteriophage lambda universal internal control (λ-UIC). The λ-UIC gene target, egfp (enhanced green fluorescent protein), derived from the jellyfish Aquifex aeolicus, is an unlikely contaminant of clinical specimens and is sufficiently foreign for this purpose. A λ-UIC lysate was formed and assayed for PFU/μl by utilizing pEGFP-C2 (Clontech, Mountain View, CA) and a Lambda Zap II predigested vector kit with Gigapack III Gold packaging extract (Agilent, Santa Clara, CA) as described in the package insert. Approximately 1 × 108 copies of λ-UIC were spiked into every specimen and extraction negative control prior to nucleic acid extraction.
Animals and immunosuppression.
Detailed methodology of the persistently neutropenic rabbit model of IPA is described elsewhere (4, 5, 19). This animal model system has been predictive of the clinical properties of other Aspergillus biomarkers, including galactomannan and (1→3)-β-d-glucan (8, 10, 17, 19, 30). Infection was confirmed by pulmonary lesion scores, histology, quantitative lung cultures, and, when applicable, computed tomography (CT) scans (29). Healthy female New Zealand White rabbits (Covance Research Products, Inc., Denver, PA) were used in all experiments. All animals were monitored under humane care and use standards in facilities accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International, according to the guidelines of the National Research Council for the care and use of laboratory animals, and under the approval of the Institutional Animal Care and Use Committee of the National Cancer Institute, Bethesda, MD. Vascular access was established by surgical placement of a Silastic tunneled central venous catheter as previously described (26). Profound and persistent neutropenia (a neutrophil concentration of <100 neutrophils/μl) and immunosuppression were established and maintained using cytarabine and methylprednisolone, as described previously (5). Antibiotics (ceftazidime, gentamicin, and vancomycin) were used for prevention of opportunistic bacterial infections during neutropenia.
Nine experimental groups were studied (see Tables 4 and 5). Untreated and treated models of IPA were investigated for each of the four species of Aspergillus, as well as a ninth group of noninfected and untreated animals.
Table 4.
Sensitivities of culture, galactomannan assay, and real-time PCR assays for detection of Aspergillus spp. in BAL fluid specimens from experimental invasive pulmonary aspergillosis
| Infection group | n | Treatment statusa | No. (%) of positive specimens |
|||
|---|---|---|---|---|---|---|
| Culture | GM assay | Real-time PCR assay |
||||
| Pan-Aspergillus | Species specific | |||||
| A. fumigatus | 24 | Untreated | 12 (50)b,d | 22 (92)d | 24 (100)b | 23 (96)b |
| A. flavus | 8 | Untreated | 8 (100) | 8 (100) | 8 (100) | 3 (38) |
| A. niger | 27 | Untreated | 27 (100) | 27 (100) | 27 (100) | 27 (100) |
| A. terreus | 14 | Untreated | 11 (79) | 6 (100)c | 14 (100) | 14 (100) |
| Total | 73 | Untreated | 58 (79)e | 63 (86) | 73 (100)e | 67 (92) |
| A. fumigatus | 18 | Treated | 0 (0)f | 5 (28) | 7 (39)f | 8 (44)f |
| A. flavus | 19 | Treated | 13 (68)g | 19 (100) | 19 (100)g | 15 (79) |
| A. niger | 12 | Treated | 12 (100) | 12 (100) | 12 (100) | 12 (100) |
| A. terreus | 8 | Treated | 8 (100) | QNSi | 8 (100) | 8 (100) |
| Total | 62 | Treated | 39 (62)h | 36 (73) | 52 (83)h | 49 (78) |
| No infection | 12 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
Infected rabbits were administered amphotericin B deoxycholate at 1 mg/kg of body weight/day for several days prior to assay (for A. fumigatus, 4 to 10 days; for A. flavus, 4 to 9 days; for A. niger, 5 to 13 days; and for A. terreus, 4 to 12 days).
P < 0.0007.
BAL fluid samples from six untreated A. terreus-infected rabbits were tested by GM assay. All six were positive.
P = 0.003.
P < 0.0001.
P < 0.008.
P = 0.02.
P = 0.01.
QNS, the quantity of BAL fluid left was not sufficient for GM assay.
Table 5.
Sensitivities of culture and real-time PCR assays for detection of Aspergillus spp. in lung tissue specimens from experimental invasive pulmonary aspergillosis
| Species | n | Treatment statusa | No. (%) of positive specimens |
||
|---|---|---|---|---|---|
| Culture | Real-time PCR assay |
||||
| Pan-Aspergillus | Species specific | ||||
| A. fumigatus | 14 | Untreated | 13 (93) | 14 (100) | 14 (100) |
| A. terreus | 6 | Untreated | 6 (100) | 6 (100) | 6 (100) |
| Total | 20 | Untreated | 19 (95) | 20 (100) | 20 (100) |
| A. fumigatus | 11 | Treated (AMB at 2.5 mg/kg/day) | 3 (27) | 6 (55) | 7 (64) |
| 8 | Treated (AMB at 5.0 mg/kg/day) | 4 (50) | 6 (75) | 5 (63) | |
| A. terreus | 6 | Treated (RVC) | 5 (83) | 6 (100) | 6 (100) |
| Total | 25 | Treated | 12 (48) | 18 (72) | 18 (72) |
A. fumigatus-infected rabbits were administered liposomal amphotericin B (AMB) at either 2.5 or 5 mg/kg/day for several days prior to assay (6 to 12 days). A. terreus-infected rabbits were administered ravuconazole (RVC) at 10 mg/kg/day for several days prior to assay (12 days).
Inoculation.
For each experiment, an inoculum of 1 × 108 to 1.25 × 108 conidia of an isolate of A. fumigatus, A. flavus, A. terreus, or A. niger in a volume of 250 μl to 350 μl was prepared and determined by colony counts. Endotracheal inoculation was performed on day 2 of experiments, while rabbits were under general anesthesia, as described previously (5). Antifungal therapy was initiated 24 h postinoculation in treated animals.
Quantitative cultures of lung tissue and BAL fluid.
Lung tissue from each rabbit was sampled postmortem and cultured by standard excision of tissue from each lobe. Each fragment was weighed individually, placed in a sterile polyethylene bag (Whirl-Pak bags; Nasco, Fort Atkinson, WI), and homogenized with sterile normal saline (NS) for 30 s per tissue sample (Stomacher 80; Tekmar Corp., Cincinnati, OH). Lung homogenate dilutions (10−1 and 10−2) were prepared in sterile NS. Aliquots (100 μl) of homogenates and homogenate dilutions were plated onto SGA plates and incubated at 37°C for the first 24 h and then placed at room temperature for another 24 h. The number of CFU of Aspergillus spp. was counted and recorded for each lobe, and the CFU/g was calculated. A finding of one colony of Aspergillus spp. was considered positive. Lung tissue specimens were obtained from 20 untreated Aspergillus-infected rabbits and 25 treated Aspergillus-infected rabbits.
BAL was performed postmortem on each lung preparation, as previously described (19), by the instillation of 10 ml of sterile NS into the clamped trachea with a sterile 12-ml syringe and subsequent withdrawal. The instillations were repeated twice. The lavage fluid was then centrifuged for 10 min at 400 × g. Part of the supernatant was discarded, leaving the pellet and 2 ml of supernatant, which was then vortexed. Aliquots of 100 μl of BAL fluid and 100 μl of a dilution (10−1) of this fluid were cultured on 5% SGA plates. Specimens of BAL fluid were obtained from archived samples from previous studies of IPA using A. fumigatus (20, 21) and A. terreus (28), as well as from newly conducted experiments on IPA for all four species. BAL fluid specimens were obtained from 12 noninfected rabbits, 73 untreated Aspergillus-infected rabbits, and 62 treated Aspergillus-infected rabbits. Postmortem-collected BAL fluid samples were divided into two portions and used for GM and PCR studies. Aliquots of BAL fluid and 500- to 1,000-μl tissue homogenate samples were stored at −70°C and subsequently shipped on dry ice to Viracor-IBT Laboratories. The identities of specimens were coded alphanumerically and blinded for the organism and treatment group.
Galactomannan assay.
The BAL fluid samples were analyzed for galactomannan concentrations by using the Platelia Aspergillus enzyme-linked immunosorbent assay (Bio-Rad Laboratories, France) performed according to the manufacturer's instructions, as previously described for serum galactomannan determination (4, 19). Peroxidase-linked monoclonal rat antibody EB-A2 was used to sensitize the microplate wells and bind the Aspergillus galactomannan antigen. The optical density (OD) of each BAL fluid specimen and control was measured by using a microplate spectrophotometer (Multiscan MMC/340) at 450-nm and 620-nm wavelengths. Enzyme immunoassay data were expressed as the BAL fluid galactomannan index (GMI). The GMI for each BAL fluid specimen is equal to the OD of the BAL fluid sample divided by the OD of a threshold serum control (1 ng/ml of galactomannan) provided in the test kit.
Nucleic acid extraction from fungal isolates and BAL fluids.
Nucleic acid extraction from BAL fluid was performed using a Roche MagNA Pure automated extraction system (large-volume DNA isolation kit). In a 2-ml microcentrifuge tube, 600 μl of 710- to 1,180-μm glass beads (Sigma-Aldrich, St. Louis, MO), 200 μl Roche bacterium/fungus lysis buffer, and 40 μl Roche proteinase K (Indianapolis, IN) were combined. For nucleic acid extraction from fungal isolates, approximately 1 × 105 germinated conidia were added to the 2-ml microcentrifuge tube as a positive control. For nucleic acid extraction from BAL fluid, the specimen was thawed and 400 μl was added to the reaction tube. The mixture was bead beaten in a FastPrep24 instrument (MP Biomedicals, Solon, OH) twice at 6.5 m/s for 60 s and then incubated at 56°C for 15 min. After a brief spin to remove drops from the lid, the lysate was transferred from the beads to a MagNA Pure sample cartridge. Using 600 μl PBS, residual lysate was collected by transferring bead washes into the sample cartridge. An aliquot of 5 μl of λ-UIC was added to each well of the sample cartridge. The samples were extracted at an elution volume of 100 μl. An extraction negative control and germinated conidia positive control were included in each extraction run.
Nucleic acid extraction of tissue homogenate.
An AllPrep DNA/RNA minikit (Qiagen, Germantown, MD) was utilized for extractions, with some modifications. Five hundred to 1,000 μl tissue homogenate was pelleted at 15,000 × g for 5 min, and the pellet was resuspended in either 350 or 700 μl RLT Plus buffer containing β-mercaptoethanol. Three hundred fifty microliters of suspension and 5 μl of bacteriophage λ-UIC were added to a MagNA Lyser green bead tube. This sample was bead beaten with a Precellys 24 instrument (Bertin Technologies, Montigny-le-Bretonneux, France) at 6,500 rpm for 60 s. The tube was gently centrifuged to collect the suspension, which was then extracted using a Qiagen QIACube instrument and AllPrep DNA/RNA mini kit reagents. The final elution volume was 100 μl. An extraction negative control and germinated conidia positive control were included in each extraction run.
Real-time PCR.
The real-time PCR general formulation mix was 5 μl of a 6× primer-probe mix and 15 μl of 2× ABI Fast mix, which contains a proprietary mix of DNA polymerase, deoxynucleoside triphosphates (dNTPs), buffers, and salts (Applied Biosystems, Foster City, CA), 0.09% bovine serum albumin (Sigma-Aldrich, St. Louis, MO), and 10 μl of template (extracted nucleic acids, quantitation standards, etc.). An ABI 7500 Fast instrument was used, with initial denaturation at 95°C for 20 s followed by 45 repetitions of a two-step amplification cycle of denaturation at 95°C for 3 s and annealing/extension at 60°C for 30 s. Thermal cycling was followed by cooling down to room temperature. Quantitation standards (serial dilutions of the Aspergillus assay target gene cloned into the pCR2.1 plasmid vector [Invitrogen, Carlsbad, CA], ranging from 5 × 101 to 5 × 107 copies per reaction) were run in conjunction with each set of samples. Eluates were analyzed in duplicate real-time PCR wells. Quantitative values used to establish linearity, dynamic range, and lower limits of detection of the assay were derived from wells with crossover thresholds (CT values) of <40. The positivity cutoff used in experimental IPA was defined as both wells having CT values of <40.
Statistical analysis.
Differences in proportions were analyzed using Fisher's exact test. Differences in means were analyzed utilizing the Mann-Whitney t test (two-tailed). A P value of <0.05 was considered to be statistically significant. Summary data are reported as means ± standard errors of the means (SEM).
RESULTS
In vitro performance testing of specificity, linearity, dynamic range, and limit of detection.
The Aspergillus PCR assays were examined for specificity by utilizing a panel of species (Tables 2 and 3). The Aspergillus species-specific and genus-specific primers were designed to maximize and minimize 3′ homology to the target species and nontarget species, respectively. The species-specific assays were examined using genomic extracts from at least two isolates of A. fumigatus, A. flavus, A. niger, A. terreus, and Penicillium spp. Candida albicans genomic DNA was also tested. The species-specific assays detected only the target species, as summarized in Table 2. The pan-Aspergillus real-time PCR assay detected all Aspergillus spp. tested (Table 3). However, it also detected all Penicillium species tested (Penicillium citrinium, Penicillium chrysogenum, and Penicillium purpurogenum), but it did not detect the five Candida species tested (C. albicans, Candida glabrata, Candida krusei, Candida parapsilosis, and Candida tropicalis) or Pneumocystis jirovecii. To assess the potential for cross-reactivity with other molds, the pan-Aspergillus, A. fumigatus, and A. terreus assay oligonucleotides were analyzed using BLAST searches against other major hyaline and pigmented mold pathogens and were determined to be not cross-reactive with the following genera: Fusarium, Scedosporium, Pseudallescheria, Paecilomyces, Bipolaris, Curvularia, Alternaria, Cladosporium, Cladophialophora, and Phyllophora. Rare matches were encountered in Paecilomyces BLAST searches using the pan-Aspergillus assay oligonucleotides as query sequences. Paecilomyces marquandii, Paecilomyces variotii, and several Byssochlamys sp. (teleomorph of Paecilomyces) sequences matched the pan-Aspergillus assay sequences, but the matches were infrequent. Most of the available sequences of the suspect Paecilomyces spp. did not match the pan-Aspergillus assay sequences.
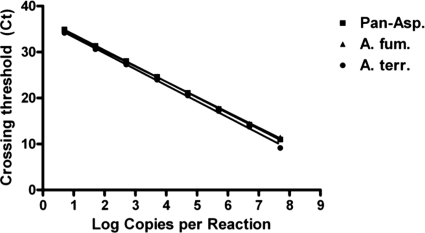
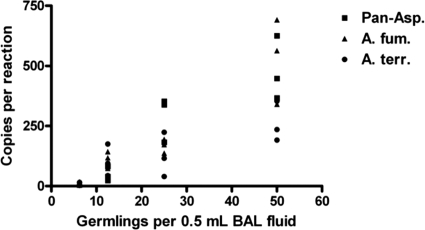
The assays reliably detected as few as 5 copies of purified plasmid standard target and were linear from 5 copies to 5 × 107 copies (Fig. 2). The PCR efficiency was calculated by the following equation: e = [10^(−1/slope)] − 1, where e and “slope” indicate PCR efficiency and the slope of the assay's line in Fig. 2, respectively. The PCR efficiencies were 95.8%, 99.6%, and 93.5% for the pan-Aspergillus, A. fumigatus, and A. terreus assays, respectively. To assess the linearity of the full process (nucleic acid extraction and real-time PCR), 50, 25, 13, and 7 germlings were spiked into 0.5 ml BAL fluid and processed by nucleic acid extraction and real-time PCR. Ten replicates were tested for each of the 3 assays to determine the limit of detection. For clarity, results for a limited number of replicates (three) for each assay are shown in Fig. 3. The full process applied to human BAL fluid spiked with Aspergillus germlings had a limit of detection of 7 germlings per extraction (10 detected out of 10 tested).
Fig. 2.
Real-time PCR linearity and dynamic range. Five to 5 × 107 copies of each assay's target region were introduced into real-time PCR wells and analyzed to determine crossing thresholds. The log copy number detected per reaction was plotted versus the crossing threshold for pan-Aspergillus (squares), A. fumigatus (triangles), and A. terreus (circles).
Fig. 3.
Limit of detection. A mixture of A. fumigatus and A. terreus germlings was introduced at low concentrations (7 to 50 germlings) into BAL fluid, followed by nucleic acid extraction. The eluates were tested by each of the Aspergillus PCR assays. The number of copies detected per reaction was plotted against the number of organisms per 0.5 ml BAL fluid for pan-Aspergillus (squares), A. fumigatus (triangles), and A. terreus (circles).
Detection of Aspergillus species by culture, galactomannan assay, and real-time PCR with BAL fluid.
Qualitative results of culture, galactomannan assay, and real-time PCR with BAL fluid for rabbits with experimental IPA are summarized in Table 4. The BAL fluid specimens with negative results and the extraction negative controls for the run were required to have threshold crossing cycles of <35 for negative results to be valid and for acceptance of the extraction run, respectively. All such specimens and controls met this criterion. Pan-Aspergillus and species-specific real-time PCRs with BAL fluid were more sensitive than culture for detection of IPA caused by A. fumigatus in untreated (P < 0.0007) and treated (P < 0.003) animals. For infections caused by non-Aspergillus fumigatus species, culture and pan-Aspergillus PCR of BAL fluid yielded similar sensitivities for untreated and treated animals (Table 4). The A. fumigatus, A. niger, and A. terreus species-specific assays yielded similar results to those of the pan-Aspergillus assays performed on the same samples (Table 4). The A. flavus assay, on the other hand, was positive for only 38% of the untreated A. flavus-infected rabbits (Table 4).
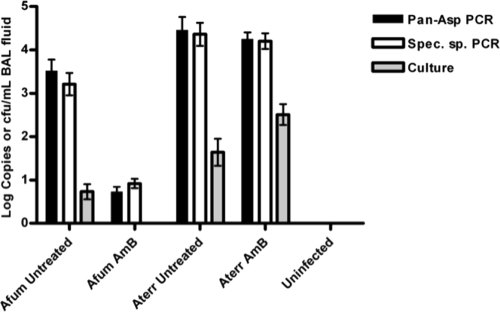
The quantitative results of culture and real-time PCR data are summarized in Fig. 4. There was a significant difference in copies/ml BAL fluid between the untreated and treated groups for A. fumigatus but not for amphotericin-resistant A. terreus (Fig. 4). Comparing galactomannan testing results with culture and real-time PCR results revealed that galactomannan results generally agreed with the real-time PCR results and that galactomannan assay was therefore more sensitive than culture.
Fig. 4.
Detection of rDNA copies in BAL fluid, showing efficacy of amphotericin B (AmB) treatment in an experimental model of invasive pulmonary aspergillosis. Copies/ml and CFU/ml of BAL fluid are shown for real-time PCR assays (pan-Aspergillus [black bars] and A. fumigatus or A. terreus species-specific [white bars] assays) and culture (gray bars), respectively. Utilizing the A. fumigatus and pan-Aspergillus assays, there was a statistically significant difference in numbers of copies/ml of BAL fluid between the untreated and treated groups for A. fumigatus (P < 0.0001) but not for A. terreus (P = 0.813) (two-tailed Mann-Whitney t test).
Detection of Aspergillus species by culture and real-time PCR with lung tissue.
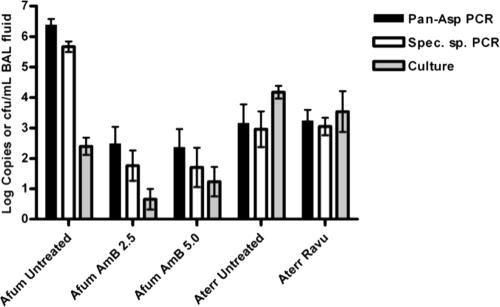
Given the results of reduced specificity of the real-time PCR assay for A. niger and limited sensitivity of the assay for A. flavus, we proceeded with fungal tissue burden assessments in experimental models for only A. fumigatus and A. terreus. Qualitative results of culture and real-time PCR with lung tissue from rabbits with experimental IPA are summarized in Table 5. Pan-Aspergillus and species-specific real-time PCRs with tissue yielded higher rates of positivity for detection of IPA caused by A. fumigatus than those observed for culture for treated animals, but the differences were not statistically significant. The quantitative results of culture and real-time PCR data are summarized in Fig. 5. The tissue specimens with negative results and the extraction negative controls for the run were required to have threshold crossing cycles of <35 for negative results to be valid and for acceptance of the extraction run, respectively. All such specimens and controls met this criterion. There was a significant difference in copies/ml tissue homogenate between the untreated and treated groups for A. fumigatus but not for A. terreus (Fig. 5). With the exception of untreated A. fumigatus-infected rabbits, gene target copy detection in the real-time PCR assays correlated with quantitative cultures of pulmonary residual fungal burdens in lung tissue.
Fig. 5.
Detection of rDNA copies in tissue homogenates, showing efficacy of amphotericin B (AmB) treatment (A. fumigatus) and ravuconazole (Ravu) treatment (A. terreus) in an experimental model of invasive pulmonary aspergillosis. Log copies per tissue sample and log CFU/g tissue are shown for real-time PCR assays (pan-Aspergillus [black bars] and A. fumigatus or A. terreus species-specific [white bars] assays) and culture (gray bars), respectively. Utilizing the two-tailed Mann-Whitney t test, there was a statistically significant difference in numbers of copies per tissue extraction between the untreated and treated groups for A. fumigatus (P < 0.0001 for amphotericin B at 2.5 mg/kg of body weight/day and P < 0.0003 for amphotericin B at 5.0 mg/kg/day) but not for A. terreus (P = 0.8988).
DISCUSSION
We developed a set of real-time PCR assays for the detection of Aspergillus spp. by utilizing pan-Aspergillus primers as well as species-specific primers for A. fumigatus, A. flavus, A. niger, and A. terreus and then validated these systems in animal models of invasive pulmonary aspergillosis for each of these species. Infections caused by each of the four species were detected successfully by the pan-Aspergillus and species-specific real-time PCR assays, using BAL fluid from animals with experimental IPA. Through a series of iterative quality control studies, primer-probe designs, and correlation with the four models of experimental pulmonary aspergillosis, the final repertoire of assays consists of the pan-Aspergillus assay as well as the species-specific assays for A. fumigatus and A. terreus. These studies led to the availability of assays to health care providers in the United States for their application to direct patient care through Viracor-IBT Laboratories.
These laboratory studies also provided the experimental foundation for clinical investigation of the real-time PCR system with archived BAL fluid specimens from a large cohort of lung transplant recipients. The findings of the laboratory animal studies presented here were predictive of the outcome (16). That study analyzed the performance of the pan-Aspergillus and A. fumigatus- and A. terreus-specific real-time PCR assays and included 16 cases of proven or probable IPA and 97 negative controls. The Aspergillus PCR system detected 100% of cases of proven or probable IPA, with specificities of 89%, 96%, and 99% for the pan-Aspergillus, A. fumigatus, and A. terreus assays, respectively.
For the purposes of clinical and epidemiological studies, the European Organization for the Research and Treatment of Cancer/Mycoses Study Group (EORTC/MSG) established criteria for proven and probable IPA (3). The presence of defined thresholds of galactomannan in serum and BAL fluid and of (1→3)-β-d-glucan in serum is considered an acceptable microbiological criterion for diagnosis of probable IPA. However, diagnostic PCR was not included as a suitable microbiological criterion due to the absence of validation and standardization. The assay reported here has been validated rigorously in laboratory animal model systems and, more recently, in clinical specimens. If standardization in additional centers is achieved, this Aspergillus PCR system should be considered for inclusion in the next iteration of EORTC/MSG definitions for diagnosis of invasive fungal infections (3).
The rationale for selection of the four Aspergillus species in these studies was predicated upon their clinical importance. A. fumigatus constitutes the most common cause of invasive pulmonary and disseminated aspergillosis. A. flavus is the second most common cause of invasive aspergillosis. However, in some institutions, A. flavus has been the most common cause of invasive disease, particularly in the setting of nosocomial outbreaks (7, 25). A. niger is a less common cause of acute invasive disease but is a frequent cause of chronic aspergillosis, especially aspergilloma (23). Finally, A. terreus, which has been described as the second most common cause of IPA in some institutions, causes invasive pulmonary and disseminated aspergillosis and is usually resistant to amphotericin B (28).
Although several quantitative, real-time PCR (qPCR) assays have been reported for diagnosis of IPA, many lack specificity (11). Moreover, due to differences in antifungal susceptibility, species-specific identification is important to improve outcomes.
The pan-Aspergillus assay is specific for more than 100 Aspergillus spp. and does not amplify genomic DNA from Candida spp. or members of the Mucorales. Moreover, BLAST searches did not demonstrate significant homology with other medically important pathogenic fungi. However, the primers and probes did produce a reaction with genomic DNAs of Penicillium spp. While penicillia are rarely the cause of documented invasive disease, they may be a source of laboratory contamination as well as a cause of tracheobronchial colonization, particularly in patients with damaged airways. However, based upon the findings of the real-time PCR study of lung transplant recipients, the clinical significance of this reactivity with genomic DNAs of Penicillium spp. was relatively small (16). That study found that among 11 lung transplant recipients colonized by Penicillium spp., only one patient's BAL fluid specimen reacted with the pan-Aspergillus real-time PCR assay. These data suggest that the hyphal burden of the organism is relatively low or that Penicillium is present principally in the form of conidia. The real-time PCR system was comparable in sensitivity to BAL fluid galactomannan assay for treated and untreated rabbits. The two assays may be complementary in a clinical setting by permitting rapid species identification. The real-time PCR system allows a rapid turnaround time that is comparable to that of BAL fluid galactomannan assay, thus permitting simultaneous availability of antigen molecular data to clinicians.
The clinical performance of the assay was determined in a study of 150 lung transplant recipients, including 16 with proven or probable IPA, 26 with Aspergillus colonization, 11 with non-Aspergillus mold colonization, and 97 negative controls (16). The sensitivities of the pan-Aspergillus and A. fumigatus assays were 100% and 85%, respectively. The sensitivity of the A. terreus assay could not be determined due to too few A. terreus cases (the single A. terreus case was found to be positive by the assay). The specificities of the pan-Aspergillus, A. fumigatus, and A. terreus assays were 88%, 96%, and 99%, respectively.
For acceptable specificity, the sequence design required that the A. flavus reverse primer be short (Table 1). While the A. flavus assay performed in acceptable fashion with purified quantitation standards, subsequent testing revealed that it performed poorly with nucleic acids extracted from specimens (Table 4). To test whether the short (15 bp) reverse primer was being absorbed nonspecifically, an alternate, 24-bp reverse primer was designed. Using a genomic template, the new reverse primer rescued the detection to match that of the pan-Aspergillus real-time PCR assay. Unfortunately, this new reverse primer reduced the specificity of the assay for A. flavus and thus could not be included in a redesigned assay. Problems in designing species-specific molecular Aspergillus assays stem from discrepancies (taxonomic problems as well as misidentifications and possible sequencing errors) in the NCBI GenBank public database. Problems in primer and probe design also emerge in Aspergillus taxonomy from identifications made using different methods (traditional versus molecular) (1).
One hundred to 10,000 times higher values were measured for numbers of copies detected (via real-time PCR) than numbers of CFU detected (via culture) for analyses of BAL fluid (Fig. 4). Similar results were obtained with analyses of tissue homogenates for the untreated A. fumigatus-infected group only (Fig. 5). These differences are owed in part to the presence of many ribosomal DNA (rDNA) gene targets for PCR for every organism detectable by culture. One report of 6 A. fumigatus clinical isolates indicated a range of 38 to 91 rDNA copies per genome (9). In addition, many genomes likely correspond to each CFU due to the presence of nonviable and viable organisms in specimens. Molecular identification of Aspergillus spp. is best accomplished by using primers and probes targeting several different gene loci. However, such a multiplexed assay would be complex and expensive for a clinical laboratory.
The real-time PCR system demonstrated greater sensitivity than culture for detection of A. fumigatus in BAL fluid (Table 4). Pan-Aspergillus PCR was also more sensitive than culture for detection of A. flavus in treated animals. The same-day turnaround time (8 to 12 h from specimen receipt) affords a further advantage toward timely initiation of antifungal therapy, as culture usually requires several days to generate a result. While the real-time PCR and galactomannan assay sensitivities generally agreed for BAL fluid, the species-specific real-time PCR assays provide additional information to guide antifungal drug selection. This is relevant to the question of whether A. terreus, which is intrinsically resistant to amphotericin B, is the infectious agent. For those medical centers or reference laboratories conducting molecular diagnostic procedures but not galactomannan assays within their facility, the pan-Aspergillus and species-specific PCR assays may improve diagnostic yields over those with conventional culture methods.
Determination of the real-time PCR performance using blood specimens is an important task, as collection of blood is less invasive than that of BAL fluid or tissue. We are developing methods toward efficient extraction of Aspergillus DNA from blood for this evaluation. Pan-Aspergillus and species-specific real-time PCR assays may improve the rapid and accurate identification of IPA in immunocompromised patients. To evaluate the utility of the real-time PCR system presented here, alterations and optimizations are being developed, to be followed by in vivo and clinical evaluations.
This report highlights the value of validation of diagnostic assays in well-characterized laboratory animal models, including the persistently neutropenic rabbit models of IPA, in predicting the performance of novel diagnostic methods for patients. Given the paucity of well-characterized clinical samples and the uncontrolled variability of host factors in patients with IPA, the use of a robust panel of samples from well-characterized animal models can be a powerful tool for development of new assays for detection of aspergillosis and other uncommon mycoses.
Footnotes
Published ahead of print on 5 October 2011.
REFERENCES
- 1. Balajee S. A., et al. 2009. Molecular identification of Aspergillus species collected for the Transplant-Associated Infection Surveillance Network. J. Clin. Microbiol. 47:3138–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Clancy C. J., et al. 2007. Bronchoalveolar lavage galactomannan in diagnosis of invasive pulmonary aspergillosis among solid-organ transplant recipients. J. Clin. Microbiol. 45:1759–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. De Pauw B., et al. 2008. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin. Infect. Dis. 46:1813–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Francesconi A., et al. 2006. Characterization and comparison of galactomannan enzyme immunoassay and quantitative real-time PCR assay for detection of Aspergillus fumigatus in bronchoalveolar lavage fluid from experimental invasive pulmonary aspergillosis. J. Clin. Microbiol. 44:2475–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Francis P., et al. 1994. Efficacy of unilamellar liposomal amphotericin B in treatment of pulmonary aspergillosis in persistently granulocytopenic rabbits: the potential role of bronchoalveolar d-mannitol and serum galactomannan as markers of infection. J. Infect. Dis. 169:356–368 [DOI] [PubMed] [Google Scholar]
- 6. Hachem R. Y., et al. 2004. Aspergillus terreus: an emerging amphotericin B-resistant opportunistic mold in patients with hematologic malignancies. Cancer 101:1594–1600 [DOI] [PubMed] [Google Scholar]
- 7. Hahn T., et al. 2002. Efficacy of high-efficiency particulate air filtration in preventing aspergillosis in immunocompromised patients with hematologic malignancies. Infect. Control Hosp. Epidemiol. 23:525–531 [DOI] [PubMed] [Google Scholar]
- 8. Hayden R., et al. 2008. Galactomannan antigenemia in pediatric oncology patients with invasive aspergillosis. Pediatr. Infect. Dis. J. 27:815–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Herrera M. L., Vallor A. C., Gelfond J. A., Patterson T. F., Wickes B. L. 2009. Strain-dependent variation in 18S ribosomal DNA copy numbers in Aspergillus fumigatus. J. Clin. Microbiol. 47:1325–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hope W. W., et al. 2010. The initial 96 hours of invasive pulmonary aspergillosis: histopathology, comparative kinetics of galactomannan and (1→3)-β-d-glucan, and consequences of delayed antifungal therapy. Antimicrob. Agents Chemother. 54:4879–4886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hope W. W., Walsh T. J., Denning D. W. 2005. Laboratory diagnosis of invasive aspergillosis. Lancet Infect. Dis. 5:609–622 [DOI] [PubMed] [Google Scholar]
- 12. Husain S., et al. 2008. Performance characteristics of the platelia Aspergillus enzyme immunoassay for detection of Aspergillus galactomannan antigen in bronchoalveolar lavage fluid. Clin. Vaccine Immunol. 15:1760–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kahn F. W., Jones J. M., England D. M. 1986. The role of bronchoalveolar lavage in the diagnosis of invasive pulmonary aspergillosis. Am. J. Clin. Pathol. 86:518–523 [DOI] [PubMed] [Google Scholar]
- 14. Kontoyiannis D. P., et al. 2010. Prospective surveillance for invasive fungal infections in hematopoietic stem cell transplant recipients, 2001–2006: overview of the Transplant-Associated Infection Surveillance Network (TRANSNET) Database. Clin. Infect. Dis. 50:1091–1100 [DOI] [PubMed] [Google Scholar]
- 15. Lass-Florl C. 2010. In vitro susceptibility testing in Aspergillus species: an update. Future Microbiol. 5:789–799 [DOI] [PubMed] [Google Scholar]
- 16. Luong M. L., et al. 2011. Comparison of an Aspergillus real-time polymerase chain reaction assay with galactomannan testing of bronchoalveolar lavage fluid for the diagnosis of invasive pulmonary aspergillosis in lung transplant recipients. Clin. Infect. Dis. 52:1218–1226 [DOI] [PubMed] [Google Scholar]
- 17. Marr K. A., et al. 2004. Detection of galactomannan antigenemia by enzyme immunoassay for the diagnosis of invasive aspergillosis: variables that affect performance. J. Infect. Dis. 190:641–649 [DOI] [PubMed] [Google Scholar]
- 18. Musher B., et al. 2004. Aspergillus galactomannan enzyme immunoassay and quantitative PCR for diagnosis of invasive aspergillosis with bronchoalveolar lavage fluid. J. Clin. Microbiol. 42:5517–5522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Petraitiene R., et al. 2001. Antifungal activity and pharmacokinetics of posaconazole (SCH 56592) in treatment and prevention of experimental invasive pulmonary aspergillosis: correlation with galactomannan antigenemia. Antimicrob. Agents Chemother. 45:857–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Petraitiene R., et al. 2004. Efficacy, safety, and plasma pharmacokinetics of escalating dosages of intravenously administered ravuconazole lysine phosphoester for treatment of experimental pulmonary aspergillosis in persistently neutropenic rabbits. Antimicrob. Agents Chemother. 48:1188–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Petraitis V., et al. 2005. Efficacy and safety of generic amphotericin B in experimental pulmonary aspergillosis. Antimicrob. Agents Chemother. 49:1642–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Saito H., Anaissie E. J., Morice R. C., Dekmezian R., Bodey G. P. 1988. Bronchoalveolar lavage in the diagnosis of pulmonary infiltrates in patients with acute leukemia. Chest 94:745–749 [DOI] [PubMed] [Google Scholar]
- 23. Smith N. L., Denning D. W. 2011. Underlying conditions in chronic pulmonary aspergillosis, including simple aspergilloma. Eur. Respir. J. 37:865–872 [DOI] [PubMed] [Google Scholar]
- 24. Steinbach W. J., et al. 2004. Infections due to Aspergillus terreus: a multicenter retrospective analysis of 83 cases. Clin. Infect. Dis. 39:192–198 [DOI] [PubMed] [Google Scholar]
- 25. Vonberg R. P., Gastmeier P. 2006. Nosocomial aspergillosis in outbreak settings. J. Hosp. Infect. 63:246–254 [DOI] [PubMed] [Google Scholar]
- 26. Walsh T. J., Bacher J., Pizzo P. A. 1988. Chronic Silastic central venous catheterization for induction, maintenance and support of persistent granulocytopenia in rabbits. Lab. Anim. Sci. 38:467–471 [PubMed] [Google Scholar]
- 27. Walsh T. J., Groll A. H. 2001. Overview: non-fumigatus species of Aspergillus: perspectives on emerging pathogens in immunocompromised hosts. Curr. Opin. Investig. Drugs 2:1366–1367 [PubMed] [Google Scholar]
- 28. Walsh T. J., et al. 2003. Experimental pulmonary aspergillosis due to Aspergillus terreus: pathogenesis and treatment of an emerging fungal pathogen resistant to amphotericin B. J. Infect. Dis. 188:305–319 [DOI] [PubMed] [Google Scholar]
- 29. Walsh T. J., et al. 2009. Diagnostic imaging of experimental invasive pulmonary aspergillosis. Med. Mycol. 47(Suppl. 1):S138–S145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Walsh T. J., et al. 2004. Detection of galactomannan antigenemia in patients receiving piperacillin-tazobactam and correlations between in vitro, in vivo, and clinical properties of the drug-antigen interaction. J. Clin. Microbiol. 42:4744–4748 [DOI] [PMC free article] [PubMed] [Google Scholar]