Abstract
Sequence-based typing (SBT) is the internationally recognized standard method for genotyping Legionella pneumophila. To date all strains of serogroup 1 (SG1) and some of SGs 2 to 14 yield a seven-allele profile and can be assigned a sequence type (ST). However, for some strains belonging to SGs 2 to 14, the targeted region of the neuA gene could not be amplified using the published standard primers. We determined the DNA sequence of a neuA gene homolog located in the lipopolysaccharide synthesis locus of strain Dallas-1E. By using newly designed degenerate consensus primers based on the neuA homolog in strains Dallas-1E, Philadelphia-1, Paris, Lens, and Corby, we were able to obtain DNA sequences for all 48 non-SG1 strains which were untypeable by the standard method. Our data show that the neuA gene is present in all L. pneumophila strains but differs significantly in some non-SG1 strains at both the DNA and amino acid levels. The new primers can be used to amplify and sequence the neuA gene in all strains and can substitute for the standard primers. This offers the possibility of assigning an ST to all strains of L. pneumophila.
INTRODUCTION
Legionellae are ubiquitous Gram-negative bacteria which occupy natural and manmade aquatic environments. They are the causative agents of Legionnaires' disease, which occurs as sporadic or epidemic cases of pneumonia acquired by inhalation or aspiration of legionellae from different environmental sources, such as cooling towers and potable water and warm water supplies. Currently, the genus Legionella comprises more than 50 species and more than 70 serogroups, with Legionella pneumophila serogroup 1 (SG1) causing the majority of human infections (11, 12; J. P. Euzeby, List of Prokaryotic Names with Standing in Nomenclature [http://www.bacterio.cict.fr/l/legionella.html]). Based on DNA homology studies, the species L. pneumophila is subdivided into three subspecies, i.e., L. pneumophila subsp. pneumophila, L. pneumophila subsp. fraseri, and L. pneumophila subsp. pascullei (2).
Monoclonal antibody (MAb) subgrouping (12) and sequence-based typing (SBT) (8, 15) are the established epidemiological typing methods for comparison of clinical and environmental isolates of L. pneumophila. Typing data showing that linked clinical and environmental strains are indistinguishable by these typing methods can be used to support or refute evidence for establishing the source of infection.
The seven target sequences include the neuA gene located within the lipopolysaccharide (LPS) synthesis locus of L. pneumophila. This gene has been previously shown to encode an N-acylneuraminate cytidylyltransferase, an enzyme involved in the biosynthesis of the LPS in L. pneumophila (9). By using the published neuA primers (15), standard SBT could be applied in all strains belonging to SG1 and to some other SGs. However, in some non-SG1 L. pneumophila strains, the neuA gene could not be amplified (1, 11, 16, 20). It was considered likely therefore that this was due to either the heterogeneity of neuA DNA sequence preventing primer binding or the absence of the gene in some of these non-SG1 strains, as suggested by microarray analysis (3). The possibility of the absence of a gene used in typing schemes was shown recently for Haemophilus influenzae. Due to a deletion of an operon encoding a fuculokinase, which is part of the seven-locus sequence typing scheme (17), a multilocus sequence typing (MLST) scheme could not be applied.
Therefore, the purpose of this study was to determine the degree of heterogeneity, if any, of the neuA genes, with the intention of designing new primer sets which could be used to amplify and sequence neuA in all L. pneumophila strains.
MATERIALS AND METHODS
Legionella strains.
Altogether 117 L. pneumophila strains were used in this study (Table 1). These strains were originally isolated from patients or water systems and were obtained from the authors' strain collections, the American Type Culture Collection (ATCC), Manassas, VA, and the National Collection of Type Cultures (NCTC), Health Protection Agency, London, United Kingdom (Table 1).
Table 1.
Allele numbers for 117 L. pneumophila strains isolated from clinical and environmental samples from different sources and regions by using the standard neuA primers and the new consensus primers
| Strain | Source countrya | Isolate typeb | SG |
neuA allele no. |
|
|---|---|---|---|---|---|
| Using standard primersc | Using new consensus primersd | ||||
| Dallas-1E ATCC 33216 | USA | E | 5 | F | 201 |
| MICU-B | USA | E | 5 | F | 201 |
| ATCC 33735 | |||||
| U8W | USA | E | 5 | F | 201 |
| ATCC 33737 | |||||
| W227-1 | Germany | E | 5 | F | 202 |
| H064160534-4 | England | E | 8 | F | 203 |
| Concord 3 ATCC 35096 | USA | C | 8 | F | 203 |
| 1169-MN-H ATCC 43703 | USA | C | 14 | F | 203 |
| W10-219 | Germany | E | 8 | F | 203 |
| W10-287 | Germany | E | 8 | F | 203 |
| H082960038 | England | E | 10 | F | 204 |
| L07-220-1 | Germany | C | 4 | F | 204 |
| H064160536-3 | England | E | 10 | F | 205 |
| Los Angeles-1 ATCC 33156 | USA | C | 4 | F | 206 |
| W10-144 | Germany | E | 4 | F | 206 |
| H042740084 | England | E | 6 | F | 207 |
| H083580006 | England | E | 10 | F | 207 |
| L00-295 | Germany | C | 2 | F | 207 |
| L02-135 | Germany | C | 4 | F | 207 |
| L08-403 | Germany | C | 10 | F | 207 |
| L08-404 | Germany | C | 10 | F | 207 |
| LC0395 | Belgium | E | 10 | F | 207 |
| LC0569 | England | C | 8 | F | 207 |
| LC0606 | England | E | 8 | F | 207 |
| LC1132 | England | C | 10 | F | 207 |
| LC6813 | England | E | 10 | F | 207 |
| LC6817 | England | E | 10 | F | 207 |
| ST-247 | Germany | E | 3 | F | 207 |
| ST-247-8 | Germany | E | 3 | F | 207 |
| W01-1979-4 | Germany | E | 3 | F | 207 |
| W03-788 | Germany | E | 10 | F | 207 |
| W03-843-1 | Germany | E | 15 | F | 207 |
| W04-306-1 | Germany | E | 2 | F | 207 |
| W07-1175 | Germany | E | 10 | F | 207 |
| W08-259-1 | Germany | E | 3 | F | 207 |
| W08-259-2 | Germany | E | 2 | F | 207 |
| W08-632 | Germany | E | 10 | F | 207 |
| W-1188 | Germany | E | 3 | F | 207 |
| W330-21 | Germany | E | 3 | F | 207 |
| W-567 | Germany | E | 2 | F | 207 |
| W671-1 | Germany | E | 2 | F | 207 |
| WS-47-3 | Germany | E | 3 | F | 207 |
| W10-341 | Germany | E | 2 | F | 207 |
| W08-130 | Germany | E | 2–14 cross-reacting | F | 208 |
| W08-433-1 | Germany | E | 4 | F | 208 |
| W10-286 | Germany | E | 4 | F | 208 |
| W05-531 | Germany | E | 13 | F | 209 |
| W10-960 | Germany | E | 13 | F | 210 |
| Chicago 8 ATCC 33823 | USA | E | 7 | F | 211 |
| L08-477 | Germany | C | 1 | 9 | 9 |
| Oxford-1 (NCTC 11287) | England | C | 6 | 9 | 9 |
| H081760005 | NK | C | 1 | 9 | 9 |
| W07-508-2 | Germany | E | 12 | 9 | 9 |
| Charite 7 | Germany | E | 2 | 9 | 9 |
| Togus-1 ATCC 33154 | USA | C | 2 | 8 | 8 |
| Charite 15 | Germany | E | 2 | 8 | 8 |
| W10-285 | Germany | E | 2 | 8 | 8 |
| RR08000504 | England | E | 4 | 7 | 7 |
| Riesa-1 | Germany | E | 3 | 7 | 7 |
| H082680013 | England | C | 1 | 6 | 6 |
| L08-482 | Germany | C | 1 | 6 | 6 |
| L08-532 | Germany | C | 1 | 6 | 6 |
| RV 34-08 | France | C | 1 | 6 | 6 |
| EUL 140e | Spain | C | 1 | 5 | 5 |
| 4163 | Italy | C | 1 | 4 | 4 |
| NIIB 2481 | Japan | C | 6 | 4 | 4 |
| NMEX 35 | Portugal | E | 35 | 35 | |
| 09.5804 | England | E | 1 | 33 | 33 |
| LG 0919 3016 | France | C | 1 | 31 | 31 |
| HL 0046 3008 | France | C | 1 | 30 | 30 |
| RR09000328 | Czech Republic | C | 1 | 3 | 3 |
| RV 35-08 | UK | E | 1 | 3 | 3 |
| W08-439 | Germany | E | 10 | 3 | 3 |
| W08-882 | Germany | E | 6 | 3 | 3 |
| W08-883 | Germany | E | 6 | 3 | 3 |
| H090520323 | NK | C | 28 | 28 | |
| LG 0737 1032 | France | C | 1 | 26 | 26 |
| LG 0837 5013 | France | C | 1 | 26 | 26 |
| 80519019001 | NLD | C | 1 | 25 | 25 |
| Lansing-3 | USA | C | 15 | 24 | 24 |
| RR07000791 | England | E | 1 | 23 | 23 |
| H094060813 | NK | C | 1 | 22 | 22 |
| H052900085 | Greece | C | 1 | 20 | 20 |
| H080600491 | England | E | 1 | 2 | 2 |
| L07-362 | Germany | C | 1 | 2 | 2 |
| W08-434-1 | Germany | E | 1 | 2 | 2 |
| H074080523 | England | C | 1 | 19 | 19 |
| NIIB 0141 | Japan | C | 1 | 18 | 18 |
| NIIB 2301 | Japan | C | 1 | 18 | 18 |
| EUL 163 | Austria | C | 2-14 cross-reacting | 17 | 17 |
| EUL 45 | Italy | C | 1 | 16 | 16 |
| H080780061 | England | E | 1 | 15 | 15 |
| L03-346 | Germany | E | 1 | 15 | 15 |
| W10-210 | Germany | E | 3 | 15 | 15 |
| RV 32-08 | England | C | 1 | 15 | 15 |
| RV 36-08 | England | E | 1 | 15 | 15 |
| EUL 13 | Scotland | C | 1 | 14 | 14 |
| H081480590 | England | E | 13 | 13 | |
| L08-422 | Germany | C | 3 | 13 | 13 |
| H082520166 | England | C | 1 | 12 | 12 |
| W10-995 | Germany | E | 1 | 12 | 12 |
| H083080428 | England | E | 1 | 11 | 11 |
| L08-371 | Germany | C | 1 | 11 | 11 |
| L08-486 | Germany | C | 1 | 11 | 11 |
| W08-394-4 | Germany | E | 1 | 11 | 11 |
| W08-444 | Germany | E | 1 | 11 | 11 |
| W08-446 | Germany | E | 1 | 11 | 11 |
| W08-449-1 | Germany | E | 1 | 11 | 11 |
| W08-449-2 | Germany | E | 1 | 11 | 11 |
| W10-291 | Germany | E | 3 | 11 | 11 |
| RR09000315 | Czech Republic | C | 1 | 10 | 10 |
| Philadelphia-1 ATCC 33152 | USA | C | 1 | 1 | 1 |
| L08-498 | Germany | C | 1 | 1 | 1 |
| L08-555-1 | Germany | C | 1 | 1 | 1 |
| RV 33-08 | England | E | 1 | 1 | 1 |
| W08-445 | Germany | E | 1 | 1 | 1 |
| W08-447 | Germany | E | 1 | 1 | 1 |
| H082940035 | Italy | C | 1 | 1 | 1 |
NK, not known (country of origin of isolate unknown due to a history of travel).
E, environmental; C, clinical.
Standard primers for neuA from the European Working Group for Legionella Infections (15). F, failed (no PCR amplification with the standard primers).
This study.
EUL, European Union Legionella culture collection (8).
Typing of Legionella strains.
All strains were typed by using monoclonal antibody subgrouping (12) and sequence-based typing (8, 15). Previous attempts to determine the neuA allele using the standard neuA primers were unsuccessful for 48 of these strains (indicated in Table 1 by F, for fail).
Determination of the DNA sequence of the LPS locus in strain Dallas-1E.
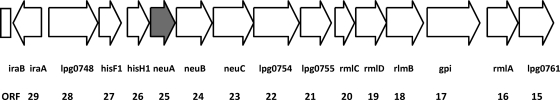
The entire sequence of the LPS locus, including the neuA gene, was determined in L. pneumophila strain Dallas-1E (ATCC 33216). This strain belongs to L. pneumophila subsp. fraseri, reacts serologically as SG5, and was originally isolated from an environmental specimen in the United States. We hypothesized that the neuA gene of this strain might be representative for heterogeneity within this region. In order to analyze the region of the Dallas-1E LPS locus between the homolog of iraB (lpg0746) and rmlA (lpg0760) (Fig. 1), including neuA, DNA fragments ranging from 1.5 to 4.0 kbp were amplified with different primer combinations and directly sequenced. Some primers have been previously described (18), while new ones were designed for the Dallas-1E strain (see Table S1 in the supplemental material).
Fig. 1.
Schematic representation of the gene organization in the LPS synthesis locus in strain Philadelphia-1 including the neuA gene (bp 817030 to 924915 in the genome). Open reading frames (ORFs) are represented as open boxes, with arrowheads indicating the orientation. The design of the open reading frames was based on published sequences (6). The filled box indicates the neuA gene.
Design of new neuA primers.
To prove the presence of the neuA gene in all strains, consensus primers neuAcons-up (positions 185 to 206; 5′-ATG GDG CYT CWG TDC CHT GG-3′) and neuA-cons-do2 (positions 618 to 594; 5′-CTR TYT ARW GCC CAA TCS ATT GG-3′) were designed using published neuA sequences of the SG1 strains Philadelphia-1 (6) Lens, Paris (4), and Corby (10) and the newly found sequence from strain Dallas-1E.
Nucleotide sequence accession numbers.
The complete sequence of the LPS region of strain Dallas-1E was deposited in the GenBank under accession number FN256429. Partial sequences of the neuA gene were deposited under GenBank accession numbers FR750545 to FR750553.
RESULTS
Bioinformatics analysis of the Legionella LPS locus in strain Dallas-1E.
All single-stranded sequences having phred scores of >20 were aligned, and contig assembly was performed using the computer programs Seqman, version 8.1.2 (DNA STAR, Madison, WI), and BioNumerics, version 5.0 (Applied Maths, Sint-Martens-Latem, Belgium). The complete sequence of the LPS region of strain Dallas-1E was deposited in the GenBank under accession number FN256429. The analysis revealed that a neuA homolog of 699 bp (232 amino acids) is present in Dallas-1E. Comparison of this nucleotide sequence with the sequences available from complete genome sequences of L. pneumophila using blastn (21; http://blast.ncbi.nlm.nih.gov/Blast.cgi) yielded maximum identification scores of 68% with strains Paris (NC_006368.1), Philadelphia-1 (NC_002942.5), Alcoy (NC_014125.1), Corby (NC_009494.2), and Lens (NC_006369.1). At the amino acid level, analysis using blastp revealed 159/232 (69%) identities and 194/232 (84%) positives (this is the number and fraction of residues for which the alignment scores have positive values) with Philadelphia-1 and 89/229 (39%) identities and 124/229 (54%) positives with NeuA of Neisseria meningitidis, the only bacterial N-acylneuraminate cytidylyltransferase whose three-dimensional (3D) structure has been characterized to date (14). NeuA proteins of L. pneumophila Philadelphia-1, Dallas-1E, and N. meningitidis were clearly identified as belonging to the cytidylyltransferase (CTP transf 3) family of proteins when their amino acid sequences were submitted to Pfam (http://pfam.sanger.ac.uk), a database of 12,273 protein families (7).
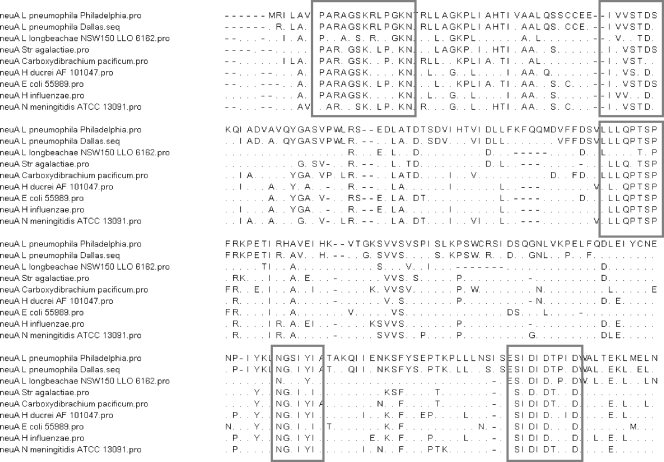
When the deduced amino acid sequence was aligned by the Lipman-Pearson method using DNA Star software, several regions with identity or high homology could be found (Fig. 2). Interestingly, the homology between the sequenced neuA gene of L. pneumophila (Philadelphia-1 and Dallas-1E) and of another Legionella species, Legionella longbeachae (strain NSW150), was only 27% and thus significantly lower than that of non-Legionella bacteria. Functionally, the neuA gene from L. pneumophila could complement a neuA-negative Escherichia coli mutant (13). Thus, it seems reasonable to suggest that the neuA genes of strains Philadelphia-1 and Dallas-1E are functionally equivalent.
Fig. 2.
Amino acid alignment by the Lipmann-Pearson (DNA Star program) method of the neuA gene regions of two L. pneumophila strains, L. longbeachae NSW150, and five other bacterial species showing regions of high similarity (>6 amino acids): Carboxydibrachium pacificum DSM 12653 (accession number ABXP01000000; identity, 40%), Desulfomicrobium baculatum DSM 4028 (accession number NC_013173; identity, 43%), Geobacter lovleyi (accession number NC_01081; identity, 38%), E. coli 55989 (accession number NC_011748; identity, 34%), and N. meningitidis ATCC 13091 (accession number AEEF01000113; identity, 38%). In the sequences, periods indicate residues that differ from the sequence of Philadelphia-1.
Homologies of <70% at both the DNA and amino acid levels suggest that this genomic region may have been acquired by horizontal gene transfer of mobile genetic elements among different Legionella strains (5).
Evaluation of new neuA primers.
Based on these data, we postulated the existence of neuA genes in all strains of L. pneumophila even though in some non-SG1 strains these neuA homolog sequences may have a lower level of similarity. Therefore, the consensus primers were used to amplify an internal fragment of neuA in 117 L. pneumophila strains (Table 1).
From all strains, the neuA gene was successfully amplified with the new consensus primers described above by employing an annealing temperature of 55°C and a primer concentration of 60 pmol per reaction mixture. All strains included in this study yielded readable DNA sequences with the newly designed consensus primers. These sequences were assembled and analyzed manually using BioNumerics, version 5.0 (Applied Maths, St. Martems-Latem, Belgium), for the new neuA alleles or using the automated online Legionella SBT Sequence Quality Tool (http://www.hpa-bioinformatics.org.uk/cgi-bin/legionella/sbt/seq_assemble_legionella1.cgi) (19) (Table 1) in the case of known neuA alleles.
We further analyzed these internal fragments of the neuA genes corresponding to nucleotide positions 229 to 583 of the Philadelphia-1 neuA gene. We note that a triplet present at positions 269 to 271 (GAA corresponding to glutamic acid at position 91) in Dallas-1E and MICU-B (L. pneumophila subsp. pascullei ATCC 33735) is not present in the neuA gene homolog obtained from the other non-SG1 strains. Due to this deletion, the analyzed fragment was 354 bp in length for these two strains and 351 bp in the other non-SG1 strains. Altogether, 11 new allele types of neuA were determined. They are available under GenBank accession numbers FR750545 to FR750553. For the purposes of this study, they were numbered from 201 to 211 (in order to distinguish them from the standard neuA alleles, of which there are currently 39 numbered from 1 to 39). The neuA allele type 207 was obtained from the majority of strains which could not be assigned a neuA allele using the standard primers (Table 1).
Relatedness of the neuA gene in strains belonging to different subspecies and serogroups.
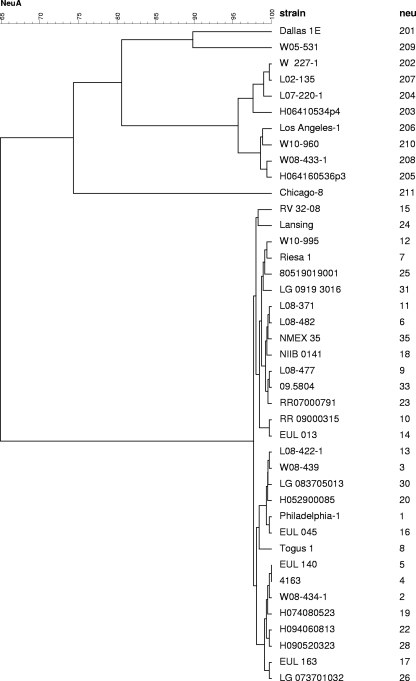
In both of our laboratories, the sequence of the 354-bp fragment of the neuA gene for the strain Dallas-1E homolog was determined (and 100% identical), demonstrating the robustness of the method. This sequence was designated new neuA allele 201. In Fig. 3, a dendrogram based on the unweighted-pair group method using average linkages (UPGMA) shows the genetic relatedness of the neuA sequences. As shown for the complete neuA gene, the 354-bp fragments differed markedly between strain Dallas-1E and strain Philadelphia-1. In general, the new neuA alleles 201 to 211 showed greater heterogeneity than found among the standard neuA alleles, designated 1 to 39 (Fig. 3). It can be demonstrated that among the three subspecies of L. pneumophila, strain Dallas-1E, which is representative of L. pneumophila subsp. fraseri, showed 100% homology to strains MICU-B and U8W (L. pneumophila subsp. pascullei) (2). In contrast, only 80% homology is present between the neuA genes of strains Dallas-1E and Los Angeles-1, both belonging to L. pneumophila subsp. fraseri (2). Also of interest, the neuA gene of strain Lansing 3 serogroup 15 (L. pneumophila subsp. fraseri) has 98% homology to the corresponding gene in strain Philadelphia-1 (L. pneumophila subsp. pneumophila) (2). The greatest difference among the strains not having the standard neuA gene was found with strain Chicago 8 (SG7; ATCC 33823) (Fig. 3). Our results demonstrated no correlation between the neuA sequences in the three subspecies of L. pneumophila and the serogroup, and they suggest that this region has not coevolved with the whole genome.
Fig. 3.
UPGMA dendrogram showing the alignment of 354/351-bp fragment sequences of the neuA alleles for L. pneumophila strains by using the software package BioNumerics, version 5.0. For each allele one example is shown.
DISCUSSION
Our results demonstrate clearly that the neuA gene involved in LPS synthesis is present in all strains but exists with remarkable heterogeneity. These data are consistent with the observation in the published microarray study, which did not find any evidence of the presence of the neuA gene in strain Dallas-1E using a threshold defined to a DNA similarity of ≥80% (3).
Previously published data (15) showed the utility of the neuA gene in SBT. It substantially increased the discriminatory ability for typing L. pneumophila SG1. Based on our new consensus primers, successful amplification and sequencing of the neuA gene were achieved with all strains tested. Even though these new neuA alleles were heterogenous compared to the original alleles, the use of newly designed primers did allow assignment of new neuA allele numbers. This offers the potential of improving both the level of discrimination and the ability to assign a sequence type (ST) for all strains of L. pneumophila. The ST, while based on an allelic string, has become a useful and practical shorthand descriptor in epidemiological investigations.
In conclusion, if a novel primer set such as that described here was used to amplify neuA genes from those non-SG1 strains where the amplification of the standard neuA failed, it could be used to obtain an allele designation for a neuA homolog and, thus, an ST where currently this is not possible. This proposal will be discussed among the Legionella typing community to see if such changes to the laboratory protocols and online software would be acceptable.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Kerstin Lück and Susan Menzel for technical assistance and Tim Harrison for constructive comments on the manuscript.
This study was supported by the Robert Koch Institute/German Federal Ministry of Health grant 1369-351.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 28 September 2011.
REFERENCES
- 1. Amemura-Maekawa J., et al. 2010. Characterization of Legionella pneumophila isolates from patients in Japan according to serogroups, monoclonal antibody subgroups and sequence types. J. Med. Microbiol. 59:653–659 [DOI] [PubMed] [Google Scholar]
- 2. Brenner D. J., et al. 1988. Legionella pneumophila serogroup Lansing 3 isolated from a patient with fatal pneumonia, and descriptions of L. pneumophila subsp. pneumophila subsp. nov., L. pneumophila subsp. fraseri subsp. nov., and L. pneumophila subsp. pascullei subsp. nov. J. Clin. Microbiol. 26:1695–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cazalet C., et al. 2008. Multigenome analysis identifies a worldwide distributed epidemic Legionella pneumophila clone that emerged within a highly diverse species. Genome Res. 18:431–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cazalet C., et al. 2004. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36:1165–1173 [DOI] [PubMed] [Google Scholar]
- 5. Charpentier X., Kay E., Schneider D., Shuman H. A. 2011. Antibiotics and UV radiation induce competence for natural transformation in Legionella pneumophila. J. Bacteriol. 193:1114–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chien M., et al. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305:1966–1968 [DOI] [PubMed] [Google Scholar]
- 7. Finn R. D., et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gaia V., et al. 2005. Consensus sequence-based scheme for epidemiological typing of clinical and environmental isolates of Legionella pneumophila. J. Clin. Microbiol. 43:2047–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Glaze P. A., Watson D. C., Young N. M., Tanner M. E. 2008. Biosynthesis of CMP-N,N-diacetyllegionaminic acid from UDP-N,N-diacetylbacillosamine in Legionella pneumophila. Biochemistry 47:3272–3282 [DOI] [PubMed] [Google Scholar]
- 10. Glöckner G., et al. 2008. Identification and characterization of a new conjugation/type IVA secretion system (trb/tra) of Legionella pneumophila Corby localized on two mobile genomic islands. Int. J. Med. Microbiol. 298:411–428 [DOI] [PubMed] [Google Scholar]
- 11. Harrison T. G., Afshar B., Doshi N., Fry N. K., Lee J. V. 2009. Distribution of Legionella pneumophila serogroups, monoclonal antibody subgroups and DNA sequence types in recent clinical and environmental isolates from England and Wales (2000–2008). Eur. J. Clin. Microbiol. Infect. Dis. 28:781–791 [DOI] [PubMed] [Google Scholar]
- 12. Helbig J. H., et al. 2002. Pan-European study on culture-proven Legionnaires disease: distribution of Legionella pneumophila serogroups and monoclonal subgroups. Eur. J. Clin. Microbiol. Infect. Dis. 21:710–716 [DOI] [PubMed] [Google Scholar]
- 13. Lüneberg E., et al. 2000. Cloning and functional characterization of a 30 kb gene locus required for lipopolysaccharide biosynthesis in Legionella pneumophila. Int. J. Med. Microbiol. 290:37–49 [DOI] [PubMed] [Google Scholar]
- 14. Mosimann S. C., et al. 2001. Structure of a sialic acid-activating synthetase, CMP-acylneuraminate synthetase in the presence and absence of CDP. J. Biol. Chem. 276:8190–8196 [DOI] [PubMed] [Google Scholar]
- 15. Ratzow S., Gaia V., Helbig J. H., Fry N. K., Lück P. C. 2007. Addition of neuA, the gene encoding N-acylneuraminate cytidylyl transferase, increases the discriminatory ability of the consensus sequence-based scheme for typing Legionella pneumophila serogroup 1 strains. J. Clin. Microbiol. 45:1965–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reimer A. R., Au S., Schindle S., Bernard K. A. 2010. Legionella pneumophila monoclonal antibody subgroups and DNA sequence types isolated in Canada between 1981 and 2009: laboratory component of national surveillance. Eur. J. Clin. Microbiol. Infect. Dis. 29:191–205 [DOI] [PubMed] [Google Scholar]
- 17. Ridderberg W., Fenger M. G., Nrskov-Lauritsen N. 2010. Haemophilus influenzae may be untypable by the multilocus sequence typing scheme due to a complete deletion of the fucose operon. J. Med. Microbiol. 59:740–742 [DOI] [PubMed] [Google Scholar]
- 18. Thürmer A., Helbig J. H., Jacobs E., Lück P. C. 2009. PCR-based serotyping of Legionella pneumophila. J. Med. Microbiol. 58:588–595 [DOI] [PubMed] [Google Scholar]
- 19. Underwood A. P., Bellamy W., Afshar B., Fry N. K., Harrison T. G. 2006. Development of an online tool for the European Working Group for Legionella Infections sequence based typing, including automatic quality assessment and data submission, p. 163–166 In Cianciotto N. P., et al. (ed.), Legionella: state of the art 30 years after its recognition . ASM Press, Washington, DC [Google Scholar]
- 20. Vergnes M., et al. 2011. Insertion sequences as highly resolutive genomic markers for sequence type 1 Legionella pneumophila Paris. J. Clin. Microbiol. 49:315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Z., Schwartz S., Wagner L., Miller W. 2000. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7:203–214 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.