Abstract
In order to evaluate the discriminatory power of different methods for genotyping of Mycobacterium tuberculosis complex (MTBC) isolates, we compared the performance of (i) IS6110 DNA fingerprint typing, (ii) spoligotyping, and (iii) 24-loci mycobacterial interspersed repetitive units-variable number of tandem repeats (MIRU-VNTR) typing in a long-term study on the epidemiology of tuberculosis (TB) in Schleswig-Holstein, the northernmost federal state of Germany. In total, we analyzed 277 MTBC isolates collected from patients between the years 2006 and 2010. The collection comprised a broad spectrum of 13 different genotypes, among which strains of the Haarlem genotype (31%) were most prominent, followed by strains belonging to the Delhi and Beijing lineages (7% and 6%, respectively). On the basis of IS6110 restriction fragment length polymorphism (RFLP) and spoligotyping analyses, 211 isolates had unique patterns (76%) and 66 isolates (24%) were in 20 clusters. MIRU-VNTR combined with spoligotyping analyses revealed 202 isolates with unique patterns (73%) and 75 isolates in 18 clusters (27%). Overall, there was 93.1% concordance between the typing results obtained; 198 strains were identified as unique, and 60 isolates were clustered by both typing combinations (including all 31 isolates with confirmed epidemiological links). Of the remaining 19 isolates with discrepant results, 15 were falsely clustered by MIRU-VNTR (six Beijing genotype strains) and four were clustered by IS6110 RFLP (low IS6110 copy number) only. In conclusion, in the study population investigated, a minority of isolates, especially of the Beijing genotype, clustered by standard 24-loci MIRU-VNTR and without an obvious epidemiological link may require second-line typing by IS6110 RFLP or hypervariable MIRU-VNTR loci.
INTRODUCTION
Tuberculosis (TB) remains one of most devastating diseases worldwide, and it is responsible for a tremendous number of deaths due to the lack of access to effective and rationally delivered drug therapy. Globally, there were an estimated 9.4 million incident cases of TB in 2009 (19). This is an increase from 8.3 million cases in 2000. In Germany, the incidence of TB remains low, with a total of 4,444 new cases reported to the Robert Koch Institute in 2009 (5.4 per 100,000 inhabitants [14]). Noteworthy, foreign citizens have a disproportionately higher risk for tuberculosis infection in Germany: in 2009, the incidence was 21.0 per 100,000 inhabitants in comparison to 3.8 per 100,000 inhabitants with German citizenship.
The federal state of Schleswig-Holstein is the northernmost federal state of Germany, with 2.8 million inhabitants. Its tuberculosis incidence rate remains below the overall German incidence (3.2 per 100,000 inhabitants). However, Kiel, the capital of Schleswig-Holstein, which has about 238,000 residents, displayed a more-than-doubled incidence of 6.7 in 2009 (14). Another interesting aspect about TB in Schleswig-Holstein is its close proximity to Hamburg, the second-largest city in Germany (over 1.8 million inhabitants) and eighth-largest in the European Union. It contains the third-largest port in Europe, which renders the city a huge transshipment center for both goods and humans from all over the world. This is reflected by a TB incidence of 10.0 per 100,000 inhabitants in 2009, the highest rate of all federal states in Germany.
Population-based epidemiological studies of TB have aimed to trace putative TB transmission chains by identifying patients with identical isolates and linking these molecular typing results to individual contact data. Three predominant state-of-the-art molecular typing methods exist: (i) restriction fragment length polymorphism (RFLP) typing based on the IS6110 insertion sequence, (ii) spoligotyping, and (iii) the 24-loci mycobacterial interspersed repetitive unit-variable number tandem repeats (MIRU-VNTR) typing (2, 16). Taken on their own, all methods have their particular disadvantages: IS6110 fingerprinting is laborious, time-consuming, and requires highly standardizes procedures; spoligotyping has the lowest discrimination power to differentiate strains; and MIRU-VNTR remains a rather new procedure. Pilot population-based studies have shown that MIRU-VNTR's discriminatory power and predictive value for tracing TB transmission was close to that of IS6110 RFLP in some populations (1, 13); however, this needs to be further evaluated in different settings. In any case, the combination of spoligotyping and MIRU-VNTR is likely to be the new gold standard method for large population-based studies due to their rapid and simple applicability (1, 13).
Therefore, we aimed to validate the performance of MIRU-VNTR typing in a long-term epidemiological study, including all patients with culture-confirmed TB reported to the regional district public health departments from 2006 to the beginning of 2010. We compared clustering rates of RFLP and MIRU typing and linked available epidemiological and contact tracing data to validate MIRU- and RFLP-associated clusters.
MATERIALS AND METHODS
Data collection.
Patient data were collected prospectively by trained public health staff using a standardized questionnaire.
Strain collection and DNA techniques.
Identification of isolates as belonging to the Mycobacterium tuberculosis complex (MTBC) was done by using the Genotype MTBC assay according to the manufacturer's instructions (Hain Lifescience, Nehren, Germany). Extraction of genomic DNA from mycobacterial strains and DNA fingerprinting with IS6110 as a probe was performed by using standardized protocols as described elsewhere (17). Furthermore, all isolates were analyzed by the spoligotyping technique described by Kamerbeek et al. (10). For MIRU-VNTR genotyping, 24 loci were amplified by PCR analysis as described previously (13, 16). Briefly, analyses were performed by using multiplex PCRs, the ROX-labeled MapMarker 1,000-bp-size standard (BioVentures, Inc., Murfreesboro, VT), and the ABI 3130xl sequencer with 16 capillaries (Applied Biosystems, Foster City, CA). Sizing of the PCR fragments and assignment of the various VNTR alleles was done by using customized GeneScan and Genotyper software packages (Applied Biosystems). In the recent round of the external quality control for 24-loci MIRU-VNTR typing performed by the European Centre for Disease Prevention and Control (ECDC) and/or the Dutch National Institute for Public Health and the Environment (RIVM) in 2010, we obtained typing results matching 100% with the reference data (30 out of 30 strains tested) and a 100% intralaboratory reproducibility.
Computer analysis.
The IS6110 fingerprints, spoligotyping patterns, and MIRU-VNTR typing results of the M. tuberculosis complex isolates were analyzed by the BioNumerics program (Windows XP, version 6.5; Applied Maths, Kortrijk, Belgium) as described previously (12, 17). The IS6110 patterns were digitized and analyzed for similarity by using the Dice coefficient and a band-matching tolerance of 1.2%. MIRU-VNTR data were imported from Excel spreadsheets and analyzed using the categorical coefficient. For both methods, similarity trees were calculated using the unweighted pair group method with arithmetic averages (UPGMA) as instructed by the manufacturer. Clusters were defined as groups of patients infected with M. tuberculosis strains showing either identical restriction fragment length polymorphism (RFLP) patterns (the same number of IS6110 bands at identical positions) and spoligotyping patterns or identical MIRU-VNTR and spoligotyping patterns.
RESULTS
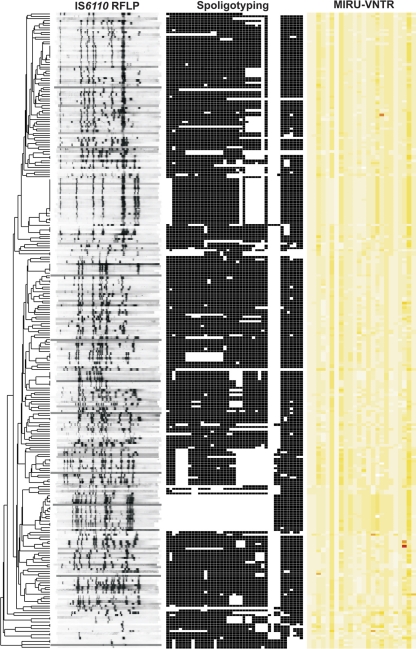
The study comprised 277 MTBC isolates obtained from TB patients documented in Schleswig-Holstein between 2006 and 2010. For the first step, we investigated the population structure and classified all strains in major phylogenetic lineages of the MTBC through combined analysis of IS6110 DNA fingerprint, 24-loci MIRU-VNTR, and spoligotyping patterns (Fig. 1) using a reference strain collection and the tools available at the MIRU-VNTRplus webpage (http://www.miru-vntrplus.org) (18).
Fig. 1.
IS6110 DNA fingerprinting, spoligotyping, and 24-loci MIRU-VNTR typing patterns of 277 strains analyzed in this study. The IS6110 band positions are normalized so that banding patterns of all strains are mutually comparable. The repeat unit numbers of 24 MIRU loci are displayed in yellow shades with cutoffs ranging from 0 (white) to 35 (red) units.
Based on these analyses, we determined a broad spectrum of 13 different phylogenetic lineages in the study population. Eighty-seven strains (31%) belong to the M. tuberculosis Haarlem lineage, 19 strains (7%) to the Delhi lineage, 17 strains (6%) to the Beijing lineage, 14 (5%) to the LAM (Latin American-Mediterranean) lineage, seven (2.5%) to the EAI (East African-Indian) lineage, six (2%) to the Ural lineage, three (1%) to the TUR (LAM7-Turkey) lineage, three (1%) to the Cameroon lineage, three (1%) to the S-type lineage, and one strain (0.4%) to the Uganda lineage. Moreover, one strain (0.4%) was classified as Mycobacterium africanum West African 1, one strain (0.4%) as Mycobacterium caprae, and four (1.5%) as Mycobacterium bovis. In addition, 111 strains (40%) could not be linked to a previously described lineage and were classified as “not defined.”
To assess the discriminatory power and epidemiological significance of IS6110 fingerprinting and spoligotyping versus MIRU-VNTR and spoligotyping, we performed cluster analyses with both combinations and subsequently correlated the clustering results to epidemiological data available from contact tracing information. As expected, IS6110 RFLP and MIRU-VNTR typing alone resulted in much higher numbers of distinct types than spoligotyping alone (Table 1). In addition, spoligotyping did not subdivide any cluster defined based either on IS6110 RFLP or MIRU-VNTR typing. Therefore, the analysis revealed 231 or 220 distinct patterns for IS6110 RFLP or MIRU-VNTR typing, respectively, regardless of the additional inclusion of the spoligotyping results. Using IS6110 RFLP (and spoligotyping) analyses, 211 isolates (76%) displayed unique patterns, and 66 isolates (24%) were included in 20 clusters. MIRU (and spoligotyping) analyses revealed 202 isolates (73%) with unique patterns and 75 samples (27%) clustered in 18 clusters.
Table 1.
Clustering results by genotyping method
| Method | No. of unique isolates | No. of clustered isolates | No. of clusters | No. of distinct types | Clustering rates (%) |
|---|---|---|---|---|---|
| IS6110 RFLP | 211 | 66 | 20 | 231 | 16.6 |
| Spoligotyping | 89 | 188 | 27 | 116 | 58.1 |
| MIRU-VNTR (24 loci) | 202 | 75 | 18 | 220 | 20.5 |
| IS6110 RFLP + spoligotyping | 211 | 66 | 20 | 231 | 16.6 |
| MIRU-VNTR (24 loci) + spoligotyping | 202 | 75 | 18 | 220 | 20.5 |
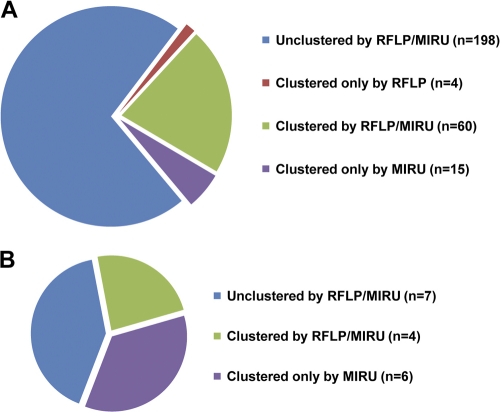
Overall, there was a good concordance when the clustering results of the strains with IS6110 fingerprinting combined with spoligotyping and MIRU typing combined with spoligotyping were compared. In total, 198 isolates remained unique by both combinations of RFLP and spoligotyping or MIRU typing and spoligotyping, whereas 79 isolates were clustered by one or both genotyping combinations (Fig. 2 A). The vast majority of clusters consisted of 2 to 5 persons (Table 2). Interestingly, out of 21 clusters, 10 (48%) were formed by strains not allocated to an as-yet-described MTBC lineage. The only clade bearing more than two clusters was Haarlem, with four clusters.
Fig. 2.
Clustering numbers of IS6110 RFLP and MIRU-VNTR typing. (A) Out of 277 isolates, 198 isolates remained unclustered, whereas 79 isolates were clustered. The latter could be subdivided into the following three groups: clustered by RFLP typing, clustered by MIRU typing, and clustered by both. (B) Clustering rates of RFLP and MIRU genotyping of the Beijing clade within the Schleswig-Holstein study.
Table 2.
Identified clusters
| Cluster | No. of patients with strains identified by: |
Epidemiologyb | Coveragec | Genotype | |
|---|---|---|---|---|---|
| RFLP + spoligotyping | MIRU + spoligotyping | ||||
| 1 | 2 | 2a | (2) | Haarlem | |
| 2 | 6 | 6 | 2 | 100 | Haarlem |
| 3 | 3 | 4 | 2 (1) | 100 | Not defined |
| 4 | 15 | 20 | 6 (7) | 100 | Haarlem |
| 5 | 2 | 0 | M. bovis | ||
| 6 | 2 | 2 | Haarlem | ||
| 7 | 4 | 4 | 4 | 100 | Not defined |
| 8 | 3 | 3 | 2 | 100 | Not defined |
| 9 | 2 | 0 | Delhi | ||
| 10 | 2 | 2 | Not defined | ||
| 11 | 2 | 2 | Not defined | ||
| 12 | 2 | 2 | Not defined | ||
| 13 | 5 | 5 | 5 | 100 | Not defined |
| 14 | 2 | 2 | Turkmenistan | ||
| 15 | 2 | 3 | 2 | 100 | Delhi |
| 16 | 2 | 2 | 2 | 100 | Not defined |
| 17 | 2 | 2 | Not defined | ||
| 18 | 2 | 7 | 2 | 100 | Beijing |
| 19 | 4 | 4 | 4 | 100 | LAM |
| 20 | 2 | 3 | Beijing | ||
| 21 | 0 | 2 | Not defined | ||
| Unclustered | 211 | 202 | Various | ||
Both patients from cluster 1 were grouped into cluster 4 by MIRU-VNTR genotyping.
No. of proven epidemiological connections. Additional unproven, but likely, connections are in parentheses.
Percent confirmation by IS6110 RFLP and MIRU-VNTR typing of epidemiological connections revealed by contact tracing.
Overall, 12 clusters (57%) comprised exactly the same patients (numbers and identities) when analyzed by IS6110 and spoligotyping or MIRU typing and spoligotyping. If the total numbers of patients whose isolates are clustered by both combinations are considered, this accounts for a total of 62 patients in 17 clusters covering 94% of all cases clustered by IS6110 RFLP and spoligotyping persons and 89% of all cases clustered by MIRU-VNTR typing and spoligotyping. Of utmost importance is that all isolates from patients with a proven epidemiological link by contact tracing were included among these cases and identically clustered by IS6110 RFLP and MIRU-VNTR typing. Altogether, for 31 persons with clustered isolates, an epidemiological link was confirmed by contact tracing; for 10 additional persons, those connections seemed to be obvious due to their environment and stated contacts (Table 2).
However, the number of isolates clustered only by MIRU typing and spoligotyping (n = 15) exceeded the number of isolates clustered only by IS6110 and spoligotyping (n = 4). To compare the crude resolution powers in a more quantitative manner, we calculated the Wallace coefficients (RFLP and spoligotyping, 0.988 [0.979 to 0.996]; MIRU and spoligotyping, 0.587 [0.399 to 0.775]). This means that if two strains were clustered by IS6110 and spoligotyping in our study, they had about a 98% chance of being clustered by MIRU typing and spoligotyping, while conversely, this was only about a 59% chance. Overall, MIRU-VNTR analysis revealed fewer clusters, but those clusters included more patients, thereby increasing the clustering rate.
Only two IS6110 and spoligotyping clusters were split by MIRU typing and spoligotyping (cluster 5 and cluster 9) (Table 2), both in cases of strains with low IS6110 band numbers (M. tuberculosis Delhi lineage and M. bovis). In both cases, the epidemiological information suggested an absence of epidemiological links, since the respective patients had no known relation and used to live in different counties.
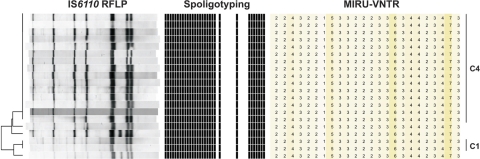
A total of six MIRU-VNTR clusters were split by IS6110 and spoligotyping, including the largest cluster of the Schleswig-Holstein study (IS6110 and spoligotyping cluster 4) (Fig. 3). While IS6110 and spoligotyping cluster 4 contained 15 persons, MIRU typing and spoligotyping unified those within cluster 1 (n = 2) and three additional patients, resulting in a cluster containing 20 patients. Interestingly, cluster 1 and the three previously unclustered persons possessed almost identical IS6110 fingerprints compared to those in cluster 4: all five patterns differed by only one band. This largest cluster was located in Kiel, the biggest city and the capital of Schleswig-Holstein, underlining that it has the highest incidence rate in Schleswig-Holstein. We found that drugs and alcohol abuse were the most important predispositions, beside close relationships, for spreading the infection within this and all other clusters.
Fig. 3.
IS6110 DNA fingerprinting, spoligotyping, and 24-loci MIRU-VNTR typing patterns of cluster 1 (C1), cluster 4 (C4), and strains differing by only one band.
Cluster 21, identified by MIRU typing and spoligotyping analysis, was subdivided by more divergent IS6110 fingerprint patterns, displaying a difference of two bands (resulting in patterns of 9 and 11 bands total). Similarly, both typing combinations identified two Beijing clusters (clusters 18 and 20); however, MIRU-VNTR included one (cluster 20) and five (cluster 18) additional patient strains differing by one to three IS6110 bands in both clusters (Fig. 2B). Notably, no epidemiological links were reported for the Beijing strains added by MIRU-VNTR typing.
DISCUSSION
In-depth evaluation of new typing methods in different settings is crucial to correctly interpret typing results obtained in studies with different designs. In comparative population-based evaluations, we and others found a similar if not slightly better performance of 15- or 24-loci MIRU-VNTR typing compared to IS6110 RFLP typing for discrimination of clinical isolates and detection of transmission chains in TB patient populations (13). Few subsequent studies compared MIRU-VNTR typing with the “gold standard” IS6110 RFLP typing, because the latter is rather laborious and more difficult to perform (6, 7, 15). The majority of the studies confirmed the initial finding that 24-loci MIRU typing can be used for MTBC genotyping with a similar performance as IS6110 typing, but some studies suggested a lower discriminatory power of 24-loci MIRU-VNTR typing in settings where Beijing strains are dominant (7, 15). However, for the latter comparative evaluations, no in-depth contact tracing information was available to further evaluate the capability of a particular genotyping method to detect recent transmission chains correctly but distinguish unrelated strains with sufficient discrimination. Here, we evaluated the resolution power of MIRU-VNTR typing and spoligotyping in comparison to IS6110 RFLP and spoligotyping over a 4-year period in an state in northern Germany with a low incidence of TB, and we used in-depth contact tracing information to assess the epidemiological significance of the molecular results obtained with both methods.
Under stringent conditions with a population of 95% high-copy-number IS6110 RFLP fingerprint patterns, we found an overall high degree of concordance (93%) between IS6110 RFLP and 24-loci MIRU-VNTR typing for defining clustered and unique isolates. The addition of spoligotyping did not influence the discriminatory power of either of the two other methods but was rather useful for supporting phylogenetic classifications predicted by MIRU-VNTR typing, as previously suggested (1, 12). Importantly, all clusters with a confirmed recent transmission link were detected by both MIRU-VNTR and IS6110 RFLP typing.
However, our data reveal an overall higher resolution power of IS6110 RFLP typing. MIRU-VNTR analyses resulted in slightly fewer clusters, however, comprising a larger number of patients which were subdivided by IS6110 RFLP typing. Only two low-band IS6110 clusters were split by MIRU-VNTR typing, while six MIRU-VNTR clusters were split by IS6110 RFLP typing. This resulted in only four isolates clustered by IS6110 RFLP typing only, but 15 isolates clustered by MIRU-VNTR typing only. Accordingly, the difference in discriminatory power is also reflected by the Wallace coefficients, indicating that RFLP-clustered data had a very high chance (98%) of being clustered by MIRU-VNTR typing, while MIRU typing-clustered strains had only a 59% chance of being clustered by RFLP typing.
Interestingly, the identified discrepancies between analyses of IS6110 fingerprint and MIRU-VNTR typing appeared to be related to strains of particular phylogenetic lineages. Not surprisingly, the IS6110 low-copy-number M. bovis cluster was clearly split by MIRU-VNTR typing (4, 11, 13).
The most prominent difference in discriminatory power was found for strains of the Beijing lineage. Although the two Beijing clusters (cluster 18 and cluster 20) (Table 2) were detected by both methods, MIRU-VNTR typing grouped more isolates together, especially in cluster 18. These results are in accordance with data from previous studies using MIRU-VNTR typing for genotyping of strain collections comprising a higher proportion of Beijing strains. It has already been shown that 12-loci MIRU-VNTR typing was not sufficient to discriminate Beijing family strains (6, 9). Similarly, Hanekom and colleagues reported that even 15- and 24-loci MIRU-VNTR typing has a reduced discriminatory power compared to IS6110 typing for strains of the Beijing lineage in Cape Town, South Africa, and that the inclusion of more variable loci might be necessary to reach the same level as that with IS6110 RFLP typing (7). As an exception, Shamputa et al. found a similar discriminatory power of 24-loci MIRU-VNTR and IS6110 RFLP typing on Korean Beijing strains and suggested that 24-loci-based MIRU-VNTR typing is a likely suitable alternative to RFLP to differentiate clinical isolates in their setting dominated by Beijing strains (15).
Finally and most importantly, we analyzed the correlation between contact tracing information and clustering data. Overall, 31 definite transmission links were found in 10 clusters (Table 2). All of them were clustered by both typing methods, further proving the capability of 24-loci MIRU-VNTR typing and IS6110 RFLP typing to reveal recent transmission chains in population-based epidemiological studies (13). The epidemiological links verified by MIRU-VNTR typing were distributed in clusters formed by strains of several phylogenetic lineages, such as Beijing, LAM, Delhi, and Haarlem. As such, the sensitivity for detection of recent transmission chains appears to be independent from the phylogenetic lineage, at least in a low-incidence setting. However, the sole use of 24-loci MIRU-VNTR typing in our study would have resulted in a higher number of patients in clusters that would have been reported to public health offices. This leads to a lower percentage of confirmed transmission links (47% in the case of IS6110 RFLP and 41% in the case of MIRU-VNTR typing) in the clustered cases and, consequently, to a lower specificity of 24-loci MIRU-VNTR typing for detection of recent transmission in this setting. This effect might be higher in countries with a high burden of strain Beijing, e.g., the former Soviet Union, clearly limiting the applicability of 24-loci MIRU typing as the sole method for highly discriminatory genotyping of clinical isolates in such study settings.
Besides providing in-depth information on the performance of the two genotyping techniques, our study also provided new insights in TB epidemiology in Germany. To our knowledge, this was the first long-term study in a state of a country with a low incidence of TB, which successfully confirmed epidemiological information by three different typing methods.
Interestingly, the close proximity to Hamburg also affected the outcome of this study, since the Haarlem strain, which has been responsible for the largest cluster, is also continuously found in Hamburg (our unpublished observation), thus confirming the close interaction between metropolitan and more rural areas. Further efforts are necessary to link putative transmission chains, regardless of district boundaries.
In addition, our study represents the first in-depth analysis of the population structure of TB in the study area. Our data confirm a rather diverse population structure with a total of 13 different genotypes in northern Germany. However, by far the largest single group of strains belonged to the M. tuberculosis Euro-American Haarlem lineage. The overall distribution of genotypes in Schleswig-Holstein was in line with other distribution analyses based on spoligotyping in Europe (3). Interestingly, the majority of strains could not be clearly matched to a previously defined genotype and was rendered “undefined.” Considering MIRU-VNTR classification and spoligotyping patterns, they are likely to belong mainly to the Euro-American super lineage (5). This indicates, in accordance with previous investigations (8), that the diversity in the Euro-American lineage is underestimated and needs further investigation.
In conclusion, in the study population investigated, 24-loci MIRU-VNTR typing (and spoligotyping) displayed a reduced discriminatory power in comparison to IS6110 RFLP typing (and spoligotyping). As this is mainly due to the reduced discrimination of MIRU-VNTR typing in Beijing strains, the inclusion of more variable loci might be necessary for strains of particular lineages.
ACKNOWLEDGMENTS
We thank Philip Supply for fruitful discussion and advice and Matthias Merker for critical reading. Furthermore, we thank I. Radzio, T. Struwe-Sonnenschein, and F. Sührck for excellent technical assistance.
Parts of this work were supported by the Schleswig-Holsteinische Gesellschaft zur Verhütung und Bekämpfung der Tuberkulose und der Lungenkrankheiten e.V.
Footnotes
Published ahead of print on 12 October 2011.
REFERENCES
- 1. Allix-Béguec C., Fauville-Dufaux M., Supply P. 2008. Three-year population-based evaluation of standardized mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 46:1398–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barnes P. F., Cave M. D. 2003. Molecular epidemiology of tuberculosis. N. Engl. J. Med. 349:1149–1156 [DOI] [PubMed] [Google Scholar]
- 3. Brudey K., et al. 2006. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cowan L. S., Mosher L., Diem L., Massey J. P., Crawford J. T. 2002. Variable-number tandem repeat typing of Mycobacterium tuberculosis isolates with low copy numbers of IS6110 by using mycobacterial interspersed repetitive units. J. Clin. Microbiol. 40:1592–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gagneux S., Small P. M. 2007. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect. Dis. 7:328–337 [DOI] [PubMed] [Google Scholar]
- 6. Guo Y.-L., et al. 2011. Genotyping and drug resistance patterns of Mycobacterium tuberculosis strains in five provinces of China. Int. J. Tuberc. Lung Dis. 15:789–794 [DOI] [PubMed] [Google Scholar]
- 7. Hanekom M., et al. 2008. Discordance between mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing and IS6110 restriction fragment length polymorphism genotyping for analysis of Mycobacterium tuberculosis Beijing strains in a setting of high incidence of tuberculosis. J. Clin. Microbiol. 46:3338–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Homolka S., et al. 2008. High genetic diversity among Mycobacterium tuberculosis complex strains from Sierra Leone. BMC Microbiol. 8:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kam K. M., et al. 2005. Utility of mycobacterial interspersed repetitive unit typing for differentiating multidrug-resistant Mycobacterium tuberculosis isolates of the Beijing family. J. Clin. Microbiol. 43:306–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kamerbeek J., et al. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mazars E., et al. 2001. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. U. S. A. 98:1901–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Niemann S., et al. 2010. Mycobacterium tuberculosis Beijing lineage favors the spread of multidrug-resistant tuberculosis in the Republic of Georgia. J. Clin. Microbiol. 48:3544–3550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oelemann M. C., et al. 2007. Assessment of an optimized mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing system combined with spoligotyping for population-based molecular epidemiology studies of tuberculosis. J. Clin. Microbiol. 45:691–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Robert Koch Institute 2009. Tuberculosis surveillance in Germany. Data of the tuberculosis report 2009. Robert Koch Institute, Berlin, Germany: http://www.rki.de/cln_226/nn_274324/DE/Content/InfAZ/T/TuberkuloseDownload/TB2009__Data,templateld=raw,property=publicationFile.pdf/TB2009_Data.pdf [Google Scholar]
- 15. Shamputa I. C., Lee J., Allix-Béguec C., Cho E.-J., Lee J. I., et al. 2010. Genetic diversity of Mycobacterium tuberculosis isolates from a tertiary care tuberculosis hospital in South Korea. J. Clin. Microbiol. 48:387–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Supply P., et al. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 44:4498–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van Embden J. D., et al. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weniger T., Krawczyk J., Supply P., Niemann S., Harmsen D. 2010. MIRU-VNTRplus: a web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucleic Acids Res. 38:W326–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. World Health Organization 2010. Global tuberculosis control: WHO report 2010. World Health Organization, Geneva, Switzerland: http://whqlibdoc.who.int/publications/2010/9789241564069_eng.pdf [Google Scholar]