Abstract
The emergence and spread of carbapenem-resistant Enterobacteriaceae (CRE) producing acquired carbapenemases have created a global public health crisis. In the United States, CRE producing the Klebsiella pneumoniae carbapenemase (KPC) are increasingly common and are endemic in some regions. Metallo-β-lactamase (MBL)-producing CRE have recently been reported in the United States among patients who received medical care in countries where such organisms are common. Here, we describe three carbapenem-resistant K. pneumoniae isolates recovered from pediatric patients at a single U.S. health care facility, none of whom had a history of international travel. The isolates were resistant to carbapenems but susceptible to aztreonam, trimethoprim-sulfamethoxazole, and fluoroquinolones. The three isolates were closely related to each other by pulsed-field gel electrophoresis and contained a common plasmid. PCR and sequence analysis confirmed that these isolates produce IMP-4, an MBL carbapenemase not previously published as present among Enterobacteriaceae in the United States.
INTRODUCTION
Enterobacteriaceae are an abundant part of the normal human gut flora and also a common cause of both health care- and community-associated infections in humans. Unfortunately, resistance to antimicrobial agents has been steadily increasing among Enterobacteriaceae, such that resistance to multiple antibiotic classes is not uncommon (19). Broad-spectrum carbapenem agents are often considered the last option for effective therapy of infections with these resistant organisms, but the emergence of carbapenem-resistant Enterobacteriaceae (CRE) over the past decade has left clinicians with few treatment options, and the spread of CRE has created a serious threat to public health.
CRE were first described in the United States more than 2 decades ago as a result of porin loss in combination with an overexpressed AmpC enzyme (4). More recently, we have seen the emergence of CRE due to production of carbapenemase enzymes that efficiently hydrolyze carbapenems and other β-lactam antibiotics. Klebsiella pneumoniae carbapenemase (KPC), an Ambler class A enzyme (35), is the most common carbapenemase produced by Enterobacteriaceae in the United States (32). The blaKPC gene is carried on a mobile genetic element and confers resistance to all β-lactam agents (5). Since it was first reported in 2001 (45), this widespread enzyme has been documented in 37 territories and states of the United States in various Enterobacteriaceae, including K. pneumoniae, Klebsiella oxytoca, Escherichia coli, Serratia marcescens, Proteus mirabilis, and Citrobacter, Enterobacter and Salmonella spp. (21, 22, 28, 32, 39). KPC-producing Enterobacteriaceae are now being recognized in numerous countries in Europe, the Middle East, and Asia, and KPC has also been documented in Acinetobacter and Pseudomonas spp. (2, 6, 33, 37, 41).
Another group of carbapenemases, the Ambler class B metallo-β-lactamases (MBLs), are of greater clinical significance worldwide (6, 34). MBLs can confer resistance to carbapenems and all β-lactam agents except aztreonam although many MBL-producing strains carry additional resistance mechanisms (e.g., extended-spectrum β-lactamases [ESBLs]) that render them resistant to aztreonam (42). Unlike KPC, MBLs have previously been unrecognized as mechanisms of carbapenem resistance among Enterobacteriaceae in the United States although they have been found in Pseudomonas aeruginosa (18, 25). Recently, two different MBL enzymes, the newly described New Delhi MBL (NDM) and Verona integron-encoded MBL (VIM), have been reported among Enterobacteriaceae in the United States. NDM has been identified in Enterobacter cloacae, K. pneumoniae, and E. coli isolates from Massachusetts, California, Illinois, and Virginia, and VIM-producing K. pneumoniae bacteria have been isolated from patients in Washington and California. In each instance, the patient carrying an MBL-producing strain had a history of recent medical care in India or Pakistan (NDM), Greece, or Italy (VIM) (7, 9, 17, 24, 29).
Here, we describe three K. pneumoniae isolates that are carbapenem-resistant due to production of an IMP-type MBL. Unlike the NDM- and VIM-producing isolates described above, the pediatric patients carrying these IMP-producing organisms had no history of travel or receipt of medical care outside the United States.
MATERIALS AND METHODS
Patient demographic data, history, and infection control investigation.
Patient information was collected by review of medical records and discussion with the primary health care providers for each patient. The identification of the novel resistance mechanisms in these three isolates took place after the inpatient clinical care of these patients had been completed, and this limited the epidemiologic investigation. No travel was identified among the patients or their mothers, but travel information among other family members was not known. Microbiology laboratory records were reviewed retrospectively to identify previously unrecognized CRE that might have represented additional cases and prospectively to identify isolates with similar antimicrobial susceptibility patterns.
Bacterial isolates.
Three K. pneumoniae urine isolates collected between November 2009 and June 2010 from infants at a single pediatric intensive care unit were submitted to the CDC for reference antimicrobial susceptibility and characterization based on unusual susceptibility profiles: intermediate or resistant to third-generation cephalosporins and carbapenems and susceptible to aztreonam, fluoroquinolones, and trimethoprim-sulfamethoxazole. An NDM-1-producing K. pneumoniae and a K. pneumoniae isolate from the same institution that was determined to not contain an MBL or KPC carbapenemase by the methods described below were included as comparators for pulsed-field gel electrophoresis only. E. cloacae strains JMI10526 and JMI3839 were included as positive and negative blaIMP controls, respectively.
Antimicrobial susceptibility.
Initial clinical testing was performed with the Vitek2 system (bioMérieux Inc., Durham, NC) and disk diffusion (11). Reference broth microdilution (BMD) was performed at the CDC using frozen in-house prepared panels (10, 12) for the following antimicrobial agents: amikacin, aztreonam, cefepime, cefotaxime, cefoxitin, ceftazidime, ciprofloxacin, colistin, doripenem, ertapenem, gentamicin, imipenem, levofloxacin, meropenem, minocycline, polymyxin B, tetracycline, tigecycline, tobramycin, and trimethoprim-sulfamethoxazole. MICs of colistin and polymyxin B were reported without interpretation, as CLSI does not have Enterobacteriaceae breakpoints for these agents. BMD panels included a series of investigational MBL screening wells containing imipenem ([IP] range, 0.25 to 1,024 μg/ml) plus the chelating agents EDTA (0.2 mM) and 1,10-phenanthroline (0.02 mM) (adapted from Migliavacca et al. [27]). A positive MBL screen was interpreted as a ≥2 doubling dilution decrease in imipenem MIC in the presence of chelating agents compared to the standard imipenem MIC. Each isolate was evaluated for carbapenemase production by the modified Hodge test (MHT) (12) with 10 μg/ml meropenem, ertapenem, and imipenem disks (BD, Franklin Lakes, NJ); K. pneumoniae ATCC BAA-1705 and BAA-1706 were included as positive and negative controls, respectively. MBL production was evaluated with Etest MBL strips (AB Biodisk, Solna, Sweden) and the direct MBL test (20) using a 10-μg imipenem disk and 0.1 mM Tris-EDTA (TE) disks (BD). K. pneumoniae ATCC BAA-2146 and P. aeruginosa ATCC 27853 were included as positive and negative controls, respectively. Etest MBL strips were performed and interpreted according to the manufacturer's instructions.
PCR amplification and sequence analysis.
PCRs for blaKPC and blaNDM were performed as described previously (24). For screening, a multiplex PCR for detection of IMP-type, VIM-type, GIM-1, SPM-1, and SIM-1 MBLs was performed as described previously (14). Isolates positive for IMP-type PCR amplicons were further characterized by PCR amplification using primers described by Yan et al. (44) for the separation of IMP-1 from IMP-2. Amplicons obtained with IMP-1-specific primers were then sequenced with the same primers using BigDye Terminator, version 3.1 (ABI, Carlsbad, CA), chemistry and an ABI 3130xl (ABI) sequence analyzer. A 448-bp blaIMP probe labeled with digoxigenin (PCR DIG Probe Synthesis Kit; Roche, Indianapolis, IN) was generated using forward primer 5′-CATGGTTTGGTGGTTCTTGT-3′ and reverse primer 5′-ATAATTTGGCGGACTTTGGC-3′ (30).
PFGE.
Relatedness of the three K. pneumoniae isolates was assessed with XbaI pulsed-field gel electrophoresis (PFGE) as previously described (21), using Bionumerics, version 5.0 (Applied Maths, Austin, TX), software with the Dice coefficient and unweighted-pair group method using average linkages (UPGMA), standardized to the Salmonella enterica serovar Braenderup H9812 standard. Isolates were also compared to the CDC database of more than 400 carbapenem-resistant K. pneumoniae strains and to contemporary carbapenemase-negative K. pneumoniae isolates from the same institution.
Plasmid analysis, transfer, and Southern blotting.
Plasmid preparations from each isolate were prepared with Qiagen Plasmid Midi kits (Qiagen, Valencia, CA), digested with HindIII (NEB, Beverly, MA), and separated by gel electrophoresis in 0.8% agarose (data not shown). Multiple plasmids were present in each strain; therefore, these plasmid preparations were used to transform E. coli DH10B cells (Invitrogen, Carlsbad, CA) by electroporation. Transformants were selected on LB agar containing 8 μg/ml ceftazidime. Transformants from each IMP-positive parent strain were screened for the presence of blaIMP by PCR with the IMP-1 family primers as described above. IMP-positive transformants were characterized by broth microdilution as described above.
Uncut parent and transformant plasmid DNAs were compared by agarose gel electrophoresis to ensure that only one plasmid was present in each of the transformants and that this plasmid corresponded in size to a plasmid band in the parent. Plasmid DNA from each parent strain and from one transformant was digested with XmnI and separated by electrophoresis in 0.75% agarose. DNA digests were transferred to Zeta-Probe nylon membranes (Bio-Rad, Hercules, CA) for Southern analysis by capillary transfer (38) and hybridized against the digoxigenin-labeled 448-bp IMP-specific probe described above using a DIG Nucleic Acid Detection Kit as recommended by the manufacturer (Roche).
RESULTS
Patient 1 is a premature male infant born at 25 weeks gestational age with a history of chronic lung disease and multiple gastrointestinal surgeries who was admitted at birth to the neonatal intensive care unit (NICU) at hospital A. He developed difficulty breathing during the 17th week of his NICU admission. Urine collected from an indwelling catheter as part of a rule-out sepsis workup grew 104 colonies/ml of a carbapenemase-producing K. pneumoniae that was negative for ESBL production and blaKPC by PCR. Due to the low numbers of organisms, no treatment specific to this organism was initiated. He remained in the NICU for more than 6 months until he was discharged home. The isolate from patient 1 is designated 1000308.
Patient 2 is a premature 5-month-old female infant born at a gestational age of 29 weeks. She was admitted to the NICU at hospital A at birth and remained there for 60 days, the first 46 of which overlapped with patient 1. Three months after she was discharged from the NICU, she returned to the hospital for a scheduled surgical repair of an atrioventricular canal defect. On postoperative day 4, a urine culture from the patient grew Enterococcus faecalis and carbapenem-resistant K. pneumoniae. The patient completed a 10-day course of ampicillin and trimethoprim-sulfamethoxazole and was discharged home. The isolate from patient 2 is designated 1002002.
Patient 3 is a male infant born at 37 weeks gestational age by emergency C-section. At birth, he required continuous positive airway pressure therapy in the delivery room and was admitted to the NICU at hospital A for continued respiratory support and monitoring. He was discharged home after a 4-day NICU admission, all four of which overlapped with patients A and B. At 5 months of age, patient C presented to the emergency department with a 2-day history of fever and drainage from his left ear. Urine collected as part of a rule-out sepsis workup grew 5,000 colonies/ml of carbapenemase-producing K. pneumoniae. The patient was prescribed amoxicillin and Tylenol/Motrin for his ear infection and discharged home. The isolate from patient 3 is designated 1002003.
Due to their unusual susceptibility patterns, K. pneumoniae isolates from each of the case patients above were submitted to the CDC for reference broth microdilution antimicrobial susceptibility testing and further characterization. Each of the isolates was intermediate or resistant to the third-generation cephalosporins and to three or more carbapenems but susceptible to aztreonam (Table 1), according to current CLSI breakpoints (13). Two isolates demonstrated a decrease of one doubling dilution in the imipenem MIC in the presence of the metal chelating agents with the MBL broth screen, and one demonstrated a decrease of two dilutions (Table 1). The isolate from patient 1 consistently yielded an imipenem MIC of 1 μg/ml and was negative for MBL production with the broth MBL screen. The modified Hodge test was positive for all three isolates with meropenem (Fig. 1A), ertapenem, and imipenem disks (data not shown).
Table 1.
Results of antimicrobial susceptibility testing in IMP-producing strains
| Antimicrobial class and agent or screening test | Susceptibility of the indicated IMP-producing K. pneumoniae parent isolate (date cultured [mo/yr])a |
Susceptibility of E. coli transformant of the indicated isolatea |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1000308 (11/09) |
1002002 (5/10) |
1002003 (6/10) |
1000308 |
1002002 |
1002003 |
Naïve recipient |
||||||||
| MIC (μg/ml) | Int | MIC (μg/ml) | Int | MIC (μg/ml) | Int | MIC (μg/ml) | Int | MIC (μg/ml) | Int | MIC (μg/ml) | Int | MIC (μg/ml) | Int | |
| Antimicrobials | ||||||||||||||
| Aminoglycocides | ||||||||||||||
| Amikacin | 2 | S | ≤1 | S | ≤1 | S | 4 | S | 2 | S | 2 | S | 4 | S |
| Gentamicin | >16 | R | 16 | R | 8 | I | >16 | R | >16 | R | >16 | R | 0.5 | S |
| Tobramycin | 8 | I | 4 | S | >16 | R | 8 | I | 8 | I | 8 | I | 1 | S |
| Aztreonam | ≤2 | S | ≤2 | S | ≤2 | S | ≤2 | S | ≤2 | S | ≤2 | S | ≤2 | S |
| Cephems | ||||||||||||||
| Cefepime | 16 | I | 4 | S | 16 | I | 4 | S | 4 | S | 2 | S | ≤1 | S |
| Cefotaxime | 32 | R | >64 | R | >64 | R | 128 | R | 32 | R | 64 | R | ≤1 | S |
| Ceftazidime | >64 | R | 64 | R | 64 | R | 128 | R | 64 | R | >128 | R | ≤1 | S |
| Cefoxitin | >32 | R | >32 | R | >32 | R | >16 | R | >16 | R | >16 | R | 8 | S |
| Carbapenems | ||||||||||||||
| Doripenem | 4 | R | 8 | R | 2 | I | 2 | I | 1 | S | 1 | S | ≤0.25 | S |
| Ertapenem | 2 | R | 4 | R | 1 | R | 1 | R | 1 | R | 1 | R | ≤0.12 | S |
| Imipenem | 1 | S | 2 | I | 2 | I | 2 | I | 1 | S | 1 | S | ≤0.5 | S |
| Meropenem | 2 | I | 4 | R | 2 | I | 2 | I | 1 | S | 1 | S | ≤0.25 | S |
| Fluoroquinolones | ||||||||||||||
| Ciprofloxacin | 0.5 | S | 0.5 | S | 0.5 | S | ≤0.25 | S | ≤0.25 | S | ≤0.25 | S | ≤0.25 | S |
| Levofloxacin | 0.5 | S | 0.5 | S | 1 | S | ≤0.25 | S | ≤0.25 | S | ≤0.25 | S | ≤0.25 | S |
| Lipopeptides | ||||||||||||||
| Polymixin | 0.5 | NA | ≤0.25 | NA | 0.5 | NA | 1 | NA | 1 | NA | 1 | NA | 1 | NA |
| Colistin | 0.5 | NA | ≤0.25 | NA | 0.5 | NA | ≤0.5 | NA | ≤0.5 | NA | ≤0.5 | NA | ≤0.5 | NA |
| Tetracyclines | ||||||||||||||
| Minocycline | ≤4 | S | ≤4 | S | 8 | I | ≤4 | S | ≤4 | S | ≤4 | S | ≤4 | S |
| Tetracycline | ≤2 | S | ≤2 | S | ≤2 | S | ≤2 | S | ≤2 | S | ≤2 | S | ≤2 | S |
| Tigecycline | ≤0.5 | S | ≤0.5 | S | ≤0.5 | S | ≤0.5 | S | ≤0.5 | S | ≤0.5 | S | ≤0.5 | S |
| Trimethoprim-sulfamethoxazole | 1/19 | S | 0.5/9.5 | S | 0.5/9.5 | S | ≤0.25/4.25 | S | ≤0.25/4.25 | S | ≤0.25/4.25 | S | ≤0.25/4.25 | S |
| Screening testsb | ||||||||||||||
| Broth MBL screen (IP/IP-EDTA-PAc) | 1/0.5 | Neg | 2/1 | Neg | 2/0.5 | Pos | 2/≤0.25 | Pos | 1/≤0.25 | Pos | 1/≤0.25 | Pos | ≤0.5/≤0.25 | Neg |
| Etest MBL (IP/IPI) | <4/<1 | IND | <4/<1 | IND | <4/<1 | IND | <4/<1 | IND | <4/<1 | IND | <4/<1 | IND | <4/<1 | IND |
Int, interpretation. For antimicrobial agents, interpretations are based on CLSI M100-S21 (13): S, susceptible; R, resistant; I, intermediate. For screening tests, a decrease of ≥2 doubling dilutions (broth MBL screen) or ≥3 doubling dilutions (Etest MBL) is considered positive (Pos). NA, not applicable; IND, indeterminate.
In the Etest, deformation of the IP and IPI ellipses (positive result) was found for all isolates and transformants; results were negative for the naïve recipient.
PA, 1,10-phenanthroline.
Fig. 1.
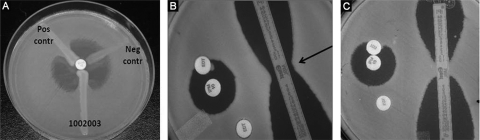
Phenotypic tests demonstrating metallo-β-lactamase production by a blaIMP-positive K. pneumoniae isolate 1002003 (A and B) and blaIMP-negative K. pneumoniae isolate 1002001 (C). (A) Modified Hodge test with meropenem demonstrating carbapenemase production by the test isolate and positive control but not the negative control. (B) Direct MBL test demonstrating subtle distortion of the zone of inhibition around the imipenem disk toward the EDTA disk and MBL Etest demonstrating deformation of the IP and IPI ellipses (arrow). (C) Example of negative results for the direct MBL test (round, undistorted zone of inhibition) and MBL Etest.
The Etest MBL was interpreted as a positive for MBL production for each isolate. The ellipse for each of the blaIMP-positive K. pneumoniae isolates would have intersected the strip below the lowest numeric value (imipenem [IP], <4 μg/ml; imipenem plus EDTA [IPI], <1 μg/ml) and demonstrated a deformed zone of inhibition (Fig. 1B) (Etest MBL package insert; AB Biodisk, Solna, Sweden). Likewise, the Direct MBL test demonstrated an extension of the zone of inhibition surrounding the imipenem disk toward the TE disk, suggesting that the isolates were producing an MBL (Fig. 1B).
PCR was performed for genes encoding the most common MBLs, including NDM, VIM, IMP, SIM, GIM and SPM, as well as for KPC, the most common carbapenemase in the United States. All three of the isolates yielded a PCR amplicon with IMP family primers (14), but no products were produced with primers to any of the other gene targets. Additional PCRs with primers specific to IMP-1 or IMP-2 subgroups of the blaIMP gene (44) demonstrated that all three isolates were carrying an allele belonging to the IMP-1 family, and sequence analysis demonstrated that it was indistinguishable from blaIMP-4 (http://www.lahey.org/Studies/).
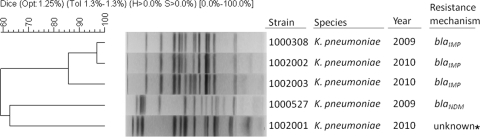
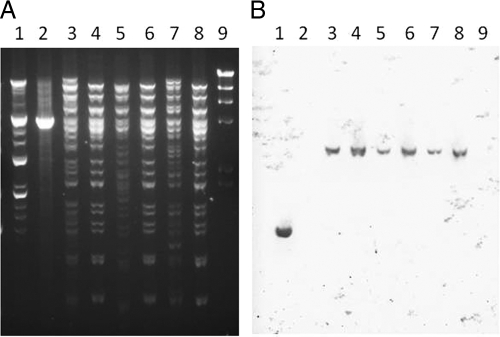
All three IMP-producing K. pneumoniae isolates were very closely related by PFGE but were unrelated to another IMP-negative carbapenemase-negative K. pneumoniae from the same institution (Fig. 2) or to other carbapenemase-producing K. pneumoniae isolates in the CDC database (data not shown). Multiple plasmids were apparent in each of the IMP-producing isolates, making it difficult to determine whether a single plasmid was common to all three isolates (Fig. 3A). Therefore, plasmid DNA from each parent was electroporated into naïve E. coli. Ceftazidime-resistant transformants from each of the IMP-positive parent strains were shown by PCR to contain blaIMP. Analysis of uncut parent and transformant plasmid DNA demonstrated the presence of a single plasmid approximately 100 kb in size in each transformant, corresponding to a plasmid band in each parent (data not shown). XmnI plasmid DNA digests (Fig. 3A) and Southern blot analysis demonstrated a single XmnI fragment of approximately 3.5 kb containing blaIMP in plasmid preparations from both parent and transformant strains (Fig. 3B).
Fig. 2.
Dendrogram showing strain relatedness of blaIMP-positive K. pneumoniae isolates compared to two contemporary blaIMP-negative carbapenem-resistant K. pneumoniae isolates from California based on pulsed-field gel electrophoresis patterns. *, isolate did not produce an MBL.
Fig. 3.
XmnI digest and Southern blot analysis of plasmid DNA demonstrating presence of multiple plasmids in blaIMP-positive K. pneumoniae isolates and a single plasmid in blaIMP-positive E. coli transformants. Lane 1, E. cloacae blaIMP positive control JMI10526; lane 2, E. cloacae blaIMP negative control JMI03839; lane 3, K. pneumoniae 1000308; lane 4, E. coli transformant of 1000308; lane 5, K. pneumoniae 1002002; lane 6, E. coli transformant of 1002002; lane 7, K. pneumoniae 1002003; lane 8, E. coli transformant of 1002003; lane 9, HindIII digest of phage lambda DNA size standard. (A) Gel electrophoresis of plasmid DNA. (B) Southern blot analysis of DNA from the experiment shown in panel A with a blaIMP-specific probe demonstrating presence of a single hybridizing band in both the parent and transformant strains.
Antimicrobial susceptibility testing was performed on the transformants, and the result was compared to that of the naïve E. coli recipient strain. Each of the transformant strains demonstrated increased MICs to cephalosporins and carbapenems but not aztreonam, consistent with IMP production. All three transformants were positive for carbapenemase production by the modified Hodge test (data not shown) and demonstrated a decrease of >2 doubling dilutions in the imipenem MIC in the presence of chelating agents compared to imipenem alone (Table 1). Each of the transformant strains also demonstrated increased MICs to tobramycin and gentamicin compared to the naïve recipient strain, suggesting that a mechanism for aminoglycoside resistance was coresident on the IMP plasmid.
Although the IMP-producing isolate from patient 1 was recognized as unusual, it would not have been classified as carbapenem resistant according to the interpretive criteria in use at the time of its isolation (11). Isolates from patients 2 and 3, however, were recovered within weeks of the release of the newer interpretive criteria, which lowered the clinical breakpoints for all of the carbapenems. According to the revised breakpoints, all three isolates demonstrated resistance to multiple carbapenem agents (12). No additional clinical isolates with similar antimicrobial susceptibility profiles were identified by review of microbiology reports from 1 month prior to the arrival of patient 1 to the present. By the time that carbapenem-resistant K. pneumoniae was recognized among these patients, all three had been discharged from the hospital and were residing at home and thus posed limited risk for further transmission in the health care setting.
DISCUSSION
The continued emergence and spread of CRE worldwide are important concerns among clinicians, infection prevention experts, and public health agencies. Internationally, MBL-type carbapenemases in particular have been of concern for some time, while KPC-producing isolates have been a far greater problem in the United States to date. This report describes three cases of probable domestically acquired IMP-producing Enterobacteriaceae and adds to reports of likely internationally acquired MBLs that have already been recognized in the United States (7, 9). Although the ultimate contribution of MBL-producing isolates to the prevalence of CRE remains to be determined, this potential increase in mechanisms highlights the need for vigorous infection prevention efforts aimed at preventing transmission.
The recent heightened attention to CRE is largely due to the emergence of the New Delhi metallo-β-lactamase, an MBL that has been strongly associated with travel and receipt of medical care on the Indian subcontinent. This has been the case among U.S. NDM-producing isolates as well; five of six patients from whom NDM-producing CRE were identified by U.S. public health agencies had a history of recent receipt of medical care in India or Pakistan (7, 17; also A. Kallen, unpublished data). Similarly, the two VIM-producing CRE identified in the United States were isolated from patients who had been recently hospitalized in Greece or Italy, countries in which these organisms have been reported (26, 35, 40). In contrast, the IMP-producing K. pneumoniae described here were from pediatric patients who had no travel history and, in the case of patient 1, had never been outside the hospital prior to CRE isolation.
IMP was originally identified in a Japanese P. aeruginosa isolate collected in 1988 that was resistant to imipenem and was named for this phenotype (active on imipenem) (43). blaIMP was located on a plasmid that readily transferred among P. aeruginosa strains, and within 5 years of that first report, blaIMP was found among Enterobacteriaceae in other Japanese hospitals in association with class 3 integrons (1, 30, 42). IMP variants have now been identified among multiple species of Gram-negative bacteria around the world (42) but have been reported infrequently in the United States and only among Pseudomonas species (18). This is the first report of this enzyme from U.S. Enterobacteriaceae; however, outbreaks caused by IMP-producing Pseudomonas have been reported from both Canada and Mexico in association with class 1 integrons (15, 16, 23, 36).
PFGE analysis demonstrated that all three IMP-producing K. pneumoniae isolates were related to each other but not to a contemporary carbapenemase-negative isolate from the same institution or to other carbapenemase-producing isolates in the CDC database. All three IMP-producing isolates harbored a common plasmid that carried blaIMP on an XmnI fragment of approximately 3.5 kb. These results and the fact that IMP-producing Enterobacteriaceae have not been previously recognized in the United States suggest a common source for these organisms. As the common link between these patients was their overlapping stays in the NICU, it is possible that transmission occurred there and that at least two of the patients remained colonized for months afterward. In the case of patient 2, IMP-producing K. pneumoniae was detected in a clinical culture collected 3 months later, and patient 3's isolate was detected as an incidental finding 5 months after his discharge. The original source and potential route of transmission of this IMP-producing K. pneumoniae isolate for these three patients are not known. It is possible that these strains are circulating widely in the community and that all three patients acquired CRE from independent sources. At least two, and possibly all three, of these cases represent asymptomatic colonization with CRE rather than true infection. It is also possible that they represent cross-transmission (e.g., by health care personnel hands) or a common environmental source. Since surveillance cultures were not obtained to identify additional cases of CRE carriage, the true extent of colonization with IMP-producing K. pneumoniae in this NICU cannot be known.
Each of the CRE described here was recognized by routine antimicrobial susceptibility testing performed in the hospital laboratory as resistant to the third-generation cephalosporins and carbapenems according to the revised (lowered) CLSI guidance for carbapenems (12), but the isolates were unusual in being susceptible to aztreonam. This antimicrobial susceptibility pattern is characteristic of MBL-producing organisms although it is not frequently encountered since most MBL-producing organisms also carry multiple other resistance mechanisms. The isolates described here are also unusual because although they carry an MBL, they remain susceptible to fluoroquinolones, trimethoprim-sulfamethoxazole, and tetracyclines. Although one isolate demonstrated intermediate susceptibility to minocycline, we believe this was likely due to variability in the test rather than acquired resistance. One of these three isolates consistently yielded an imipenem MIC within the susceptible range although it was nonsusceptible to doripenem, ertapenem, and meropenem.
The IMP-producing K. pneumoniae isolates yielded variable results with the phenotypic tests used to detect the presence of MBLs. All three were clearly positive for carbapenemase production with the MHT, but the direct MBL test, the Etest MBL, and the broth microdilution MBL screen yielded equivocal results. Two isolates demonstrated a decrease of only one doubling dilution with the broth microdilution MBL screen, which is within the acceptable variation for broth microdilution testing and would not be a reliable indicator of MBL production. Interestingly, the IMP-containing E. coli transformants were readily detected with the broth MBL screen, suggesting that some factor present in the parent strain interfered with this assay. The Etest MBL demonstrated only a subtle deformation of the ellipse in both the K. pneumoniae parent and E. coli transformant strains. Variability in the performance of phenotypic tests for detection of MBLs and difficulty with their interpretation has been described previously (29), and our results demonstrate the need to include multiple methods to ensure reliable detection of MBLs. In this case, the pattern of nonsusceptibility to all β-lactam agents except aztreonam was an important indicator of MBL presence.
At the time the first isolate was recovered (November 2009), it would have been characterized as susceptible to ertapenem, imipenem, and meropenem and nonsusceptible only to doripenem (11; http://www.accessdata.fda.gov/drugsatfda_docs/label/2009/022106s002s005lbl.pdf). Since all three patients had been discharged to their homes by the time that CRE colonization was recognized, infection prevention staff at the facility elected to perform laboratory-based surveillance for organisms with the same antimicrobial susceptibility pattern rather than active surveillance of the NICU in which these patients resided. However, consideration should be given to performing point prevalence surveys and/or active surveillance testing when novel cases of CRE infection or colonization are recognized in a health care facility (8). The emergence of this new mechanism of carbapenem resistance among Enterobacteriaceae in the United States has the potential to add to the growing burden of CRE. CRE present treatment dilemmas and are associated with high rates of mortality (31). This is particularly concerning in light of a 2009 Infectious Diseases Society of America (IDSA) report highlighting the fact that there were no antimicrobials in advanced development with activity against these organisms (3). In order to slow the emergence of these strains, aggressive infection prevention practices aimed at preventing infections with—and transmission of—these organisms should be employed. Current prevention recommendations for acute-care facilities are available from CDC (8). Foremost among these recommendations is recognizing when CRE are present in facilities so that infected or colonized patients can be put in contact precautions, and patient contacts can be screened for evidence of transmission.
In summary, this is the first published report of IMP-producing Enterobacteriaceae in the United States. The isolates appear to have been acquired domestically although the mechanism of transmission to these three patients is not known. The emergence of this and other metallo-β-lactamases (i.e., VIM and NDM) among Enterobacteriaceae in the United States has the potential to add to the already substantial burden of CRE due to the emergence and spread of KPC-producing strains. CRE are an important public health problem; in order to limit their impact, implementation of recommended interventions to prevent transmission of these organisms should be a priority for institutions where these organisms are identified.
ACKNOWLEDGMENTS
We thank JMI laboratories for providing control strains JMI10526 and JMI03839.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print on 12 October 2011.
REFERENCES
- 1. Arakawa Y., et al. 1995. A novel integron-like element carrying the metallo-beta-lactamase gene blaIMP. Antimicrob. Agents Chemother. 39:1612–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bennett J. W., Herrera M. L., Lewis J. S., II, Wickes B. W., Jorgensen J. H. 2009. KPC-2-producing Enterobacter cloacae and Pseudomonas putida coinfection in a liver transplant recipient. Antimicrob. Agents Chemother. 53:292–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boucher H. W., et al. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48:1–12 [DOI] [PubMed] [Google Scholar]
- 4. Bradford P. A., et al. 1997. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC β-lactamase, and the loss of an outer membrane protein. Antimicrob. Agents Chemother. 41:563–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bratu S., et al. 2007. Detection and spread of Escherichia coli possessing the plasmid-borne carbapenemase KPC-2 in Brooklyn, New York. Clin. Infect. Dis. 44:972–975 [DOI] [PubMed] [Google Scholar]
- 6. Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53:2227–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Centers for Disease Control and Prevention. 2010. Detection of Enterobacteriaceae isolates carrying metallo-beta-lactamase—United States, 2010. MMWR Morb. Mortal. Wkly. Rep. 59:750. [PubMed] [Google Scholar]
- 8. Centers for Disease Control and Prevention. 2009. Guidance for control of infections with carbapenem-resistant or carbapenemase-producing Enterobacteriaceae in acute care facilities. MMWR Morb. Mortal. Wkly. Rep. 58:256–260 [PubMed] [Google Scholar]
- 9. Centers for Disease Control and Prevention. 2010. Update: detection of a Verona integron-encoded metallo-beta-lactamase in Klebsiella pneumoniae—United States, 2010. MMWR Morb. Mortal. Wkly. Rep. 59:1212. [PubMed] [Google Scholar]
- 10. Clinical and Laboratory Standards Institute. 2009. Methods for dilution antimicrobial susceptibility: tests for bacteria that grow aerobically; approved standard, 8th ed M07-A8. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 11. Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial susceptibility testing; 19th informational supplement. M100-S19. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 12. Clinical and Laboratory Standards Institute. 2010. Performance standards for antimicrobial susceptibility testing; 20th informational supplement. M100-S20-U. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 13. Clinical and Laboratory Standards Institute. 2011. Performance standards for antimicrobial susceptibility testing; 21st informational supplement. M100-S21. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 14. Ellington M. J., Kistler J., Livermore D. M., Woodford N. 2007. Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J. Antimicrob. Chemother. 59:321–322 [DOI] [PubMed] [Google Scholar]
- 15. Garza-Ramos U., et al. 2008. Metallo-β-lactamase gene blaIMP-15 in a class 1 integron, In95, from Pseudomonas aeruginosa clinical isolates from a hospital in Mexico. Antimicrob. Agents Chemother. 52:2943–2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gibb A. P., et al. 2002. Nosocomial outbreak of carbapenem-resistant Pseudomonas aeruginosa with a new blaIMP allele, blaIMP-7. Antimicrob. Agents Chemother. 46:255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gupta N., Limbago B. M., Patel J. B., Kallen A. 2011. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention strategies. Clin. Infect. Dis. 53:60–67 [DOI] [PubMed] [Google Scholar]
- 18. Hanson N. D., Hossain A., Buck L., Moland E. S., Thomson K. S. 2006. First occurrence of a Pseudomonas aeruginosa isolate in the United States producing an IMP metallo-β-lactamase, IMP-18. Antimicrob. Agents Chemother. 50:2272–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kallen A. J., Srinivasan A. 2010. Current epidemiology of multidrug-resistant gram-negative bacilli in the United States. Infect. Control Hosp. Epidemiol. 31(Suppl. 1):S51–S54 [DOI] [PubMed] [Google Scholar]
- 20. Kim S. Y., Hong S. G., Moland E. S., Thomson K. S. 2007. Convenient test using a combination of chelating agents for detection of metallo-beta-lactamases in the clinical laboratory. J. Clin. Microbiol. 45:2798–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kitchel B., et al. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob. Agents Chemother. 53:3365–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kitchel B., Sundin D. R., Patel J. B. 2009. Regional dissemination of KPC-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 53:4511–4513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Laupland K. B., et al. 2005. Population-based epidemiological study of infections caused by carbapenem-resistant Pseudomonas aeruginosa in the Calgary Health Region: importance of metallo-beta-lactamase (MBL)-producing strains. J. Infect. Dis. 192:1606–1612 [DOI] [PubMed] [Google Scholar]
- 24. Limbago B. M., Rasheed J. K., Anderson K. F., Zhu W., Kallen A. J. 2010. Recognition of NDM-1 among Enterobacteriaceae in the United States, abstr. C1-675d. Abstr. 50th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC [Google Scholar]
- 25. Lolans K., Queenan A. M., Bush K., Sahud A., Quinn J. P. 2005. First nosocomial outbreak of Pseudomonas aeruginosa producing an integron-borne metallo-β-lactamase (VIM-2) in the United States. Antimicrob. Agents Chemother. 49:3538–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Luzzaro F., et al. 2004. Emergence in Klebsiella pneumoniae and Enterobacter cloacae clinical isolates of the VIM-4 metallo-β-lactamase encoded by a conjugative plasmid. Antimicrob. Agents Chemother. 48:648–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Migliavacca R., et al. 2002. Simple microdilution test for detection of metallo-β-lactamase production in Pseudomonas aeruginosa. J. Clin. Microbiol. 40:4388–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miriagou V., et al. 2003. Imipenem resistance in a Salmonella clinical strain due to plasmid-mediated class A carbapenemase KPC-2. Antimicrob. Agents Chemother. 47:1297–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mochon A. B., et al. 2011. New Delhi metallo-β-lactamase (NDM-1) producing Klebsiella pneumoniae: a case report and laboratory detection strategies. J. Clin. Microbiol. 49:1667–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Osano E., et al. 1994. Molecular characterization of an enterobacterial metallo beta-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob. Agents Chemother. 38:71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Patel G., Huprikar S., Factor S. H., Jenkins S. G., Calfee D. P. 2008. Outcomes of carbapenem-resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect. Control Hosp. Epidemiol. 29:1099–1106 [DOI] [PubMed] [Google Scholar]
- 32. Patel J. B., Rasheed J. K., Kitchel B. 2009. Carbapenemases in Enterobacteriaceae: activity, epidemiology, and laboratory detection. Clin. Microbiol. Newsl. 31:55–62 [Google Scholar]
- 33. Poirel L., Nordmann P., Lagrutta E., Cleary T., Munoz-Price L. S. 2010. Emergence of KPC-producing Pseudomonas aeruginosa in the United States. Antimicrob. Agents Chemother. 54:3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Poirel L., Pitout J. D., Nordmann P. 2007. Carbapenemases: molecular diversity and clinical consequences. Future Microbiol. 2:501–512 [DOI] [PubMed] [Google Scholar]
- 35. Queenan A. M., Bush K. 2007. Carbapenemases: the versatile beta-lactamases. Clin. Microbiol. Rev. 20:440–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Quinones-Falconi F., et al. 2010. Emergence of Pseudomonas aeruginosa strains producing metallo-beta-lactamases of the IMP-15 and VIM-2 types in Mexico. Clin. Microbiol. Infect. 16:126–131 [DOI] [PubMed] [Google Scholar]
- 37. Robledo I. E., et al. 2010. Detection of KPC in Acinetobacter spp. in Puerto Rico. Antimicrob. Agents Chemother. 54:1354–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Southern E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503–517 [DOI] [PubMed] [Google Scholar]
- 39. Tibbetts R., Frye J. G., Marschall J., Warren D., Dunne W. 2008. Detection of KPC-2 in a clinical isolate of Proteus mirabilis and first reported description of carbapenemase resistance caused by a KPC β-lactamase in P. mirabilis. J. Clin. Microbiol. 46:3080–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vatopoulos A. 2008. High rates of metallo-beta-lactamase-producing Klebsiella pneumoniae in Greece—a review of the current evidence. Euro Surveill. 13:8023. [PubMed] [Google Scholar]
- 41. Villegas M. V., et al. 2007. First identification of Pseudomonas aeruginosa isolates producing a KPC-type carbapenem-hydrolyzing β-lactamase. Antimicrob. Agents Chemother. 51:1553–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Walsh T. R., Toleman M. A., Poirel L., Nordmann P. 2005. Metallo-beta-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Watanabe M., Iyobe S., Inoue M., Mitsuhashi S. 1991. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 35:147–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yan J. J., et al. 2001. Metallo-beta-lactamases in clinical Pseudomonas isolates in Taiwan and identification of VIM-3, a novel variant of the VIM-2 enzyme. Antimicrob. Agents Chemother. 45:2224–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yigit H., et al. 2001. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]