Abstract
We tested the activities of rifampin (RIF) and rifaximin (RFX) against 180 Clostridium difficile clinical isolates selected from Canadian and Italian culture collections. MICs were determined by CLSI agar dilution for both drugs and by Etest for RIF. Sixteen of 85 Italian isolates (18.8%) showed high-level resistance to both rifamycins (MICs, >16 μg/ml), compared to 2 of 95 (2.1%) Canadian isolates. Two new rpoB mutations were identified in rifamycin-resistant isolates. RIF susceptibility by Etest correlated completely with susceptibility to both rifamycins determined by agar dilution.
TEXT
Recently described strains of Clostridium difficile that are associated with greater morbidity and mortality (12, 15) pose a significant clinical challenge, since treatment failure and therapy relapse are more frequent (12, 20), although this has not been universally seen (26). The current standard treatment for Clostridium difficile infection (CDI) is oral metronidazole or vancomycin (1), but more effective treatments are needed, because decreased susceptibility to metronidazole has been reported (2, 17, 19). Rifaximin (RFX) is being investigated as an alternative therapy for CDI (8, 13, 14), but C. difficile isolates have demonstrated resistance to RFX (6, 9, 10a, 18). Antibiotic susceptibility testing of C. difficile by anaerobic agar dilution (the CLSI recommended method) is labor-intensive and time-consuming. Etest strips exist for antibiotic susceptibility testing of rifampin (RIF), a rifamycin that is related to RFX. It remains controversial whether the RIF Etest can reliably predict susceptibility to RFX (10a, 18).
We determined the MICs for 2 rifamycins (RIF and RFX) with a large number of C. difficile clinical isolates and assessed if RIF Etest susceptibility predicted RIF and RFX agar dilution results. We also compared rifamycin resistance among C. difficile isolates from Montreal (Canada) and the Istituto Superiore di Sanità (ISS) in Rome, Italy, since rifamycin use differs dramatically in these 2 countries. A molecular analysis was performed on resistant isolates.
A convenience sample of 180 clinical isolates of C. difficile from patients with CDI was selected for analysis from culture collections at the Jewish General Hospital (JGH) in Montreal, Canada (n = 95) and the ISS (n = 85) (Table 1). A total of 1,303 toxin-positive stools were submitted from JGH patients over the same 7-year period (1989 and 2004 to 2009), but no information was available on the number of toxin-positive stools in the population served by the ISS during their 22-year collection period (1987 to 2008). Two control strains were included, as per CLSI guidelines (5): nontoxigenic C. difficile ATCC 700057 and Bacteroides fragilis ATCC 25285.
Table 1.
C. difficile isolates included in the present analysis, by site and year
| Source | No. of C. difficile isolates by yr |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1987–1989 | 1993–2003 | 2004 | 2005 | 2006 | 2007 | 2008 | 2009 | Total | |
| JGH | 30 | 0 | 10 | 11 | 11 | 11 | 12 | 10 | 95 |
| ISS | 10 | 8 | 1 | 33 | 13 | 9 | 11 | 0 | 85 |
Antimicrobial susceptibility testing was performed by agar dilution for RIF and RFX (Sigma-Aldrich Canada Ltd., Oakville, ON, Canada) according to the methods described for CLSI M11-A7 (MIC range, 0.002 to 16 μg/ml) (5). Antibiotics were dissolved in methanol, and serial 1:2 dilutions were performed in brucella broth (RIF) or 0.1 M phosphate buffer (pH 7.4) with 0.45% sodium dodecyl sulfate (RFX). Antibiotic dilutions were added to brucella blood agar supplemented with 5% laked sheep blood, 5 μg/ml of hemin, and 1 μg/ml vitamin K1 (SBA). Ten microliters of a 1 McFarland suspension of C. difficile was inoculated onto nonprereduced SBA supplemented with antibiotics and incubated anaerobically at 37°C for 48 h. MIC values were read as the lowest concentration of antibiotic showing a marked reduction in growth compared to an antibiotic-free control plate. Isolates were considered susceptible (MIC, ≤0.016 μg/ml), intermediate (MIC, >0.016 and <16 μg/ml), or resistant (MIC, ≥16 μg/ml). No CLSI guidelines exist for determining susceptibility of C. difficile to rifamycins, so these ranges were determined based on the natural breakpoints observed in the current and previous studies (6, 9, 10a, 18). Susceptibility to RIF was also evaluated using Etest strips (AB Biodisk, Sweden) on SBA according to the manufacturer's instructions, using a 1 McFarland suspension of C. difficile on SBA. An Etest strip for RFX does not currently exist.
The method described by S. R. Curry et al. (6) was used to identify the rpoB mutations in strains resistant to RIF. Briefly, primers CDrpoB2F and CDrpoB2R were used to amplify a 200-bp region of the rpoB gene known to contain point mutations associated with resistance to RIF. Both DNA strands of the purified PCR products were then sequenced and analyzed for mutations.
Pulsed-field gel electrophoresis (PFGE) typing was performed for all isolates as previously described (7). Gels were visualized using Image Master software (Bio-Rad Laboratories, Canada), and relatedness was based on previously described criteria (25).
Resistance to the rifamycin antibiotics was more prevalent in the C. difficile isolates from the ISS (MIC90, >16 μg/ml) than in the isolates from the JGH (MIC90, <0.002 μg/ml) (Table 2). Sixteen of 85 (18.8%) isolates from Italy and 2/95 (2.1%) isolates from Canada were resistant to the rifamycin antibiotics (Fisher's exact test, P = 0.0001). Two further isolates from the ISS collection displayed intermediate susceptibilities, but with MICs in the susceptible range (RIF Etest MICs, 0.12 and 0.5 μg/ml; RIF agar dilution MICs, 0.016 and 0.5 μg/ml; RFX agar dilution MICs, 0.25 and 4 μg/ml).
Table 2.
Comparison of susceptibility to RIF and RFX of C. difficile strains from the JGH (Montreal, Canada) and the ISS (Rome, Italy)
| Antibiotic | Source | No. (%) of resistant isolates | MIC data (μg/ml) |
||
|---|---|---|---|---|---|
| MIC50 | MIC90 | Range | |||
| RIF | JGH | 2 (2.4) | <0.002 | <0.002 | <0.002–>16 |
| ISS | 16 (18.8) | >16 | |||
| Total | 18 (11.3) | 0.05 | |||
| RFX | JGH | 2 (2.4) | 0.004 | 0.008 | <0.002–>16 |
| ISS | 16 (18.8) | >16 | |||
| Total | 18 (11.3) | 4 | |||
We identified 4 single-locus polymorphisms (SLP) to the rpoB gene in the rifamycin-resistant isolates (Table 3). Two SLPs identified in our study had not been previously described in resistant isolates of C. difficile. The new SLP S488P was found in an isolate with intermediate susceptibility. The new substitution S498T in association with R505K was found in a resistant isolate and has not been previously described in C. difficile. We identified the SLP H502N in a highly resistant isolate from Canada. No rpoB mutations were identified in one of the rifamycin-resistant isolates from Canada.
Table 3.
rpoB mutations detected in the C. difficile isolates resistant to rifamycins
| Substitution | No. of isolates | Source (n) | Rifamycin susceptibilityb |
|---|---|---|---|
| H502N/R505K | 7 | ISS (7) | R |
| R505K | 6 | ISS (6) | R |
| S488Pa | 1 | ISS (1) | I |
| S498T,a R505K | 3 | ISS (3) | R |
| H502N | 2 | ISS (1), JGH (1) | I/R |
Mutation not previously described for C. difficile.
R, resistant; I, intermediate.
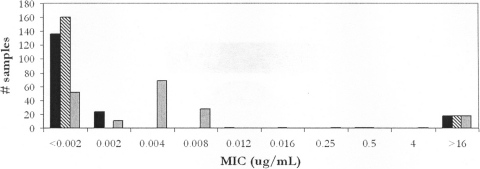
Agar dilution is the reference method for anaerobic antibiotic susceptibility testing (5), but it is labor-intensive and requires highly trained staff, while the Etest method is relatively simple. RIF susceptibility for all 180 isolates was 100% concordant between the two methods, with complete concordance between the results for RIF (both agar dilution and Etest) and RFX (agar dilution) (Fig. 1).
Fig. 1.
Correlation of RIF susceptibility (by Etest) with RIF and RFX susceptibility (by agar dilution). The graph shows MIC values for 180 isolates of C. difficile for RIF as determined by Etest (black bars) and agar dilution (striped bars) and for RFX as determined by agar dilution (gray bars).
Thirty-four of 95 Montreal isolates and 0 of 85 Italian isolates were NAP1 or closely related. Four isolates were not typeable by PFGE. None of the NAP1 isolates was resistant to the rifamycins. Rifamycin-resistant isolates were distributed within 13 PFGE types.
In Italy, rifamycin antibiotics have been in use for over 2 decades (21). In particular, RFX is licensed as a treatment for hepatic encephalopathy, diarrhea, and travelers' diarrhea and as prophylaxis for gastrointestinal surgery (11, 23, 24). In comparison, RFX is not licensed for use in Canada, and average RIF use at the JGH (2004 to 2009) was only 515 g (858 defined daily doses) per year. Since most CDI at the JGH are health care associated, this indicates a very low level of JGH patient exposure to RIF. Our study found a statistically significant 8-fold-higher frequency of rifamycin resistance in the Italian isolates compared to those from the JGH, suggesting that resistance to rifamycins may occur as a result of selective pressure after RFX exposure. Unlike recent studies (6, 18), we found no greater rifamycin resistance in the NAP1 isolates, perhaps because the 34 NAP1 isolates from the JGH were unlikely to have been exposed to rifamycins. RFX-resistant isolates in other studies were from the United States, where RFX is used to treat travelers' diarrhea, enteric Escherichia coli infections (11), and hepatic encephalopathy (22).
It is unclear how RFX in vitro resistance correlates with clinical outcomes, since it achieves fecal drug levels of approximately 8,000 μg/ml (10, 18), and other intestinal factors may affect this concentration (4, 10, 18).
We demonstrated that susceptibility to RIF by either Etest or agar dilution correlated completely with susceptibility to RFX. Thus, rifamycin class susceptibility in C. difficile can be assessed using the relatively easy RIF Etest method, consistent with the findings of O'Connor et al. (18). The divergent study (10a), which showed that RIF (agar dilution and Etest) and RFX (agar dilution) susceptibilities did not correlate, used Mueller-Hinton agar instead of brucella blood agar, as recommended by CLSI and the Etest manufacturer. The medium used for antimicrobial susceptibility testing can strongly influence the MIC of the antibiotic (5). Our results suggest that the RIF Etest is a valid method for the determination of RIF and RFX susceptibilities of C. difficile, showing complete correlation with agar dilution.
Resistance to the rifamycin class of antibiotics has been putatively linked to 8 SLPs in the rpoB gene of the RNA polymerase locus (6, 18) and another 12 SLPs in the rifamycin binding pocket (3). We identified 2 mutations, S488P and S498T, in the rpoB gene that had not been previously described in C. difficile, although a different mutation at S488 (S488T) has been described. Position S498 in the C. difficile rpoB gene corresponds to A477 in Staphylococcus aureus, which has been linked to rifamycin resistance. Both new SLPs occur in amino acids in Taq polymerase that do not interact directly with rifamycins in the binding pocket (3, 16), suggesting that the most likely mechanism of action is structural modification of the binding pocket and lowered affinity of rpoB for rifamycins. The other two previously described mutations found in our study, H502N and R505K, are believed to affect the direct interaction of rpoB with the rifamycins. Interestingly, the H502N mutation has been associated with intermediate resistance to rifamycins, but never to high-level resistance (6, 18), as we found, which may indicate a separate mechanism of resistance in C. difficile. These SLPs in the rpoB gene are conserved in many genera of rifamycin-resistant bacteria, as demonstrated by sequence alignments (6, 16, 18).
In conclusion, we have documented significantly different rates of rifamycin resistance in C. difficile from two countries with highly disparate rifamycin use, suggesting that selective pressure may play a role. We also demonstrated that RIF susceptibility by Etest correlates completely with RIF and RFX susceptibilities by agar dilution.
Acknowledgments
Mark Miller received research funding from and is on the advisory board for Optimer Pharma, is on the advisory board for Salix, and received research funding from Cubist and Actelion.
Footnotes
Published ahead of print on 12 October 2011.
REFERENCES
- 1. Abramowicz M. (ed.), 2011. Treatment of Clostridium difficile infection. Med. Lett. Drugs Ther. 53:14–15 [PubMed] [Google Scholar]
- 2. Baines S. D., et al. 2008. Emergence of reduced susceptibility to metronidazole in Clostridium difficile. J. Antimicrob. Chemother. 62:1046–1052 [DOI] [PubMed] [Google Scholar]
- 3. Campbell E. A., et al. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901–912 [DOI] [PubMed] [Google Scholar]
- 4. Cellai L., Colosimo M., Marchi E., Venturini A. P., Zanolo G. 1984. Rifaximin (L/105), a new topical intestinal antibiotic: pharmacokinetic study after single oral administration of 3H-rifaximin to rats. Chemioterapia (Basel) 3:373–377 [PubMed] [Google Scholar]
- 5. Clinical and Laboratory Standards Institute 2008. Methods for antimicrobial susceptibility testing of anaerobic bacteria; approved standard, 6th ed. M11-A7. CLSI, Wayne, PA [Google Scholar]
- 6. Curry S. R., et al. 2009. High frequency of rifampin resistance identified in an epidemic Clostridium difficile clone from a large teaching hospital. Clin. Infect. Dis. 48:425–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fawley W. N., Wilcox M. H. 2002. Pulsed-field gel electrophoresis can yield DNA fingerprints of degradation-susceptible Clostridium difficile strains. J. Clin. Microbiol. 40:3546–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Garey K. W., Salazar M., Shah D., Rodrigue R., DuPont H. L. 2008. Rifamycin antibiotics for treatment of Clostridium difficile-associated diarrhea. Ann. Pharmacother. 42:827–835 [DOI] [PubMed] [Google Scholar]
- 9. Hecht D. W., et al. 2007. In vitro activities of 15 antimicrobial agents against 110 toxigenic Clostridium difficile clinical isolates collected from 1983 to 2004. Antimicrob. Agents Chemother. 51:2716–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jiang Z.-D., Ke S., Palazzini E., Riopel L., Dupont H. 2000. In vitro activity and fecal concentration of rifaximin after oral administration. Antimicrob. Agents Chemother. 44:2205–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a. Jiang Z.-D., DuPont H. L., La Rocco M., Garey K. W. 2010. In vitro susceptibility of Clostridium difficile to rifaximin and rifampin in 359 consecutive isolates at a university hospital in Houston, Texas. J. Clin. Pathol. 63:355–358 [DOI] [PubMed] [Google Scholar]
- 11. Koo H. L., Dupont H. L. 2010. Rifaximin: a unique gastrointestinal-selective antibiotic for enteric diseases. Curr. Opin. Gastroenterol. 26:17–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Loo V. G., et al. 2005. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N. Engl. J. Med. 353:2442–2449 [DOI] [PubMed] [Google Scholar]
- 13. Louie T., Miller M., Donskey M., Mullane K., Goldstein E. J. 2009. Clinical outcomes, safety, and pharmacokinetics of OPT-80 in a phase 2 trial with patients with Clostridium difficile infection. Antimicrob. Agents Chemother. 53:223–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Louie T., et al. 2011. Fidaxomicin versus vancomycin for Clostridium difficile infection. N. Engl. J. Med. 364:422–431 [DOI] [PubMed] [Google Scholar]
- 15. McDonald L. C., et al. 2005. An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 353:2433–2441 [DOI] [PubMed] [Google Scholar]
- 16. Murphy C. K., et al. 2006. In vitro activity of novel rifamycins against rifamycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 50:827–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Musher D. M., et al. 2005. Relatively poor outcome after treatment of Clostridium difficile colitis with metronidazole. Clin. Infect. Dis. 40:1586–1590 [DOI] [PubMed] [Google Scholar]
- 18. O'Connor J. R., et al. 2008. Rifampin and rifaximin resistance in clinical isolates of Clostridium difficile. Antimicrob. Agents Chemother. 52:2813–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peláez T. L., et al. 2002. Reassessment of Clostridium difficile susceptibility to metronidazole and vancomycin. Antimicrob. Agents Chemother. 46:1647–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pépin J., et al. 2005. Increasing risk of relapse after treatment of Clostridium difficile colitis in Quebec, Canada. Clin. Infect. Dis. 40:1591–1597 [DOI] [PubMed] [Google Scholar]
- 21. Salix Pharmaceuticals, Ltd 10 December 2003, posting date. Salix receives FDA notification that rifaximin amendment considered a complete response. Salix Pharmaceuticals, Raleigh, NC: http://www.businesswire.com/news/home/20031210005070/en/Salix-Receives-FDA-Notification-Rifamixin-Amendment-Considered [Google Scholar]
- 22. Salix Pharmaceuticals 2010. XIFAXAN (rifaximin) 550 mg tablets now available in U.S. pharmacies: new patient and physician assistance program (H.E.L.P.) launches to educate patients and assist with treatment adherence. Salix Pharmaceuticals, Raleigh, NC: http://www.salix.com/news-media/news/news-archive/10-05-24/xifaxan%C2%AE_rifaximin_550_mg_tablets_now_available_in_u_s_pharmacies.aspx [Google Scholar]
- 23. Scarpellini E., et al. 2007. High dosage rifaximin for the treatment of small intestinal bacterial overgrowth. Aliment. Pharmacol. Ther. 25:781–786 [DOI] [PubMed] [Google Scholar]
- 24. Scarpignato C., Pelosini I. 2005. I. Rifaximin, a poorly absorbed antibiotic: pharmacology and clinical potential. Chemotherapy 51(Suppl. 1):36–66 [DOI] [PubMed] [Google Scholar]
- 25. Tenover F. C., et al. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wilson V., et al. 2010. Predictors of death after Clostridium difficile infection: a report on 128 strain-typed cases from a teaching hospital in the United Kingdom. Clin. Infect. Dis. 50:e77–e81 [DOI] [PubMed] [Google Scholar]