Abstract
Although discovered long ago, posttranslational phosphorylation of histones has been in the spotlight only recently. Information is accumulating almost daily on phosphorylation of histones and their roles in cellular physiology and human diseases. An extensive cross talk exists between phosphorylation and other posttranslational modifications, which together regulate various biological processes, including gene transcription, DNA repair, and cell cycle progression. Recent research on histone phosphorylation has demonstrated that nearly all histone types are phosphorylated at specific residues and that these modifications act as a critical intermediate step in chromosome condensation during cell division, transcriptional regulation, and DNA damage repair. As with all young fields, apparently conflicting and sometimes controversial observations about histone phosphorylations and their true functions in different species are found in the literature. Accumulating evidence suggests that instead of functioning strictly as part of a general code, histone phosphorylation probably functions by establishing cross talk with other histone modifications and serving as a platform for recruitment or release of effector proteins, leading to a downstream cascade of events. Here we extensively review published information on the complexities of histone phosphorylation, the roles of proteins recognizing these modifications and the resuting physiological outcome, and, importantly, future challenges and opportunities in this fast-moving field.
INTRODUCTION
To fit the enormous length of the eukaryotic genome into the nucleus of a tiny cell requires some very sophisticated packaging. But this packaging must also be highly flexible and malleable so that the genome can be correctly replicated, transcribed, and finally translated. To meet these requirements, DNA is organized in a higher-order nucleoprotein complex known as chromatin. The basic unit of chromatin is the nucleosome, which is essentially DNA wrapped around a core of histone proteins. Each nucleosome is made up of an octamer of core histones (two each of H2A, H2B, H3, and H4), and around this histone core, the DNA is wrapped in two superhelical turns of 147 bp (67). Nucleosomes are spaced at intervals and linked by 20 to 60 bp of linker DNA to form an approximately 10-nm “beads on a string” structure, with H1 linker histones contacting the exit and entry points of the DNA strand that is spooled onto each nucleosome (141). Structurally, histones can be divided into the core domain, which makes up approximately 75% of the protein and is composed of histone fold motifs that physically interact with themselves to form the H2A/2B and H3/4 heterodimers, and the flexible tail domain, which makes up the remaining 25% of the protein. The tail domain is structurally undefined but has been found to be highly conserved. The tail domains are located at the amino termini of the four histones and the carboxyl terminus of H2A and are generally defined by their sensitivity to proteases.
Nucleosomes are believed to be very dynamic, which allows DNA-regulatory factors to access parts of the DNA. The dynamicity of the chromatin structure is afforded by various covalent posttranslational modifications which could be described as marks on the core histone tails established by modifying enzymes and subsequent changes in the association of modified histones with DNA and with other effector proteins. Recent mass spectrometry data have revealed that the histone globular domains are also modified and that many residues which had previously been implicated in gene expression through genetic screens in yeast (71, 98) are targets for posttranslational modifications. There is plenty of evidence that the core histones can be modified by methylation, acetylation, phosphorylation, ubiquitylation, sumoylation, and ADP-ribosylation. Histones can be simultaneously modified in more ways than one, at various amino acid residues. This leads to a wide range of unique modifications and combinations of modifications, each of which is expected to have functional consequences. Recent studies have revealed that the histone posttranslational modifications are involved in diverse functions, from chromatin packaging and DNA condensation during mitosis and meiosis to gene transcription and DNA damage response. As an example, acetylation and methylation of histones H3 and H4 have been linked to transcriptional activation or repression of certain genes, whereas phosphorylation of these histones during mitosis and meiosis has been implicated in chromosome condensation, while the same modification in interphase cells regulates gene transcription. What might the roles of these marks on histones be? There are several possibilities: first, these marks could directly and independently control any given physiological function by altering histone-DNA interaction. Alternatively, there may be an indirect signaling role of histone residues and their marks, allowing or causing release of effector proteins recognizing these residues and modifications and thus modulating a specific physiological function that is dependent on both modification and recruitment/release of effector proteins. In this context, it is conceivable that a particular modification may be both necessary and sufficient for regulating a physiological pathway. Alternatively, the same modification may be necessary but not sufficient for regulating another physiological pathway. This “necessary and sufficient” paradigm may in part help explain some of the apparently conflicting and controversial observations in the field.
While histone acetylation and methylation have been studied extensively over the years (40, 65, 72, 103, 108), histone phosphorylation, although discovered early on, has only recently become a topic of vigorous investigation, with information about new cellular functions of phosphorylated histones continually being reported in the literature. Histone phosphorylation has been associated with a variety of cellular processes, including transcriptional regulation, apoptosis, cell cycle progression, DNA repair, chromosome condensation, and developmental gene regulation (20, 25, 53, 60, 69, 81). In most cases, phosphorylation of serine and threonine residues of the histone tails appears to be involved in chromatin condensation during mitosis and meiosis; for example, C-terminal phosphorylation of Thr119 in histone H2A is linked to regulation of chromatin structure and function during mitosis (25), and H3 Ser10 (H3S10) phosphorylation is related to chromatin compaction during mitosis. Yet H3S10 phosphorylation has also been shown to play a role in transcriptional activation of NF-κB pathway genes and immediate early genes like c-jun and c-fos (69). With regard to DNA repair, phosphorylation of histone H2AX at Ser139 (γ-H2AX) has been identified as one of the early events after a DNA double-strand break that helps recruit DNA damage repair proteins to the site (101). The list of histone phosphorylations—which often occur in combination with other histone modifications—and their cellular effects, which is sometimes controversial and conflicting and reflects the youth of an evolving and dynamic field, is long and continues to grow. In this review, we attempt to present a comprehensive picture of the complexities of histone phosphorylation, focusing on recent discoveries while revisiting some key older observations with the aim of revealing just how widespread the effects are in cellular physiology.
HISTONE H2A
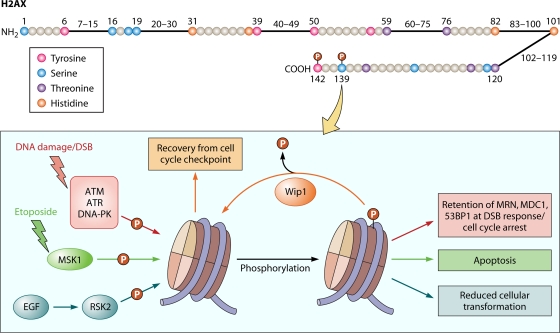
Histone H2A, especially the H2AX (Fig. 1 and Table 1) variant, has been widely studied and implicated mostly in DNA damage response and genomic instability pathways. In mammalian cells, one of the early responses to double-strand DNA breaks is the phosphorylation of histone H2AX at Ser139. Several kinases are known to be involved in DNA damage-induced H2AX phosphorylation, including members of the PI3 kinase family (like ATR [activated upon replication stress] [129] and the kinase ATM [activated by DNA damage] [14]. Other studies have shown that DNA-PK is activated by nucleosomes and in turn phosphorylates H2AX at Ser139 in an acetylation-dependent manner (97). The trigger for H2AX phosphorylation extends along the DNA up to two megabases from the site of the double-strand break and is required for the accumulation and retention of DNA damage response proteins such as MRN, MDC1, and 53BP1 (66, 101, 119, 122). Following repair of the damaged DNA, the cell must recover from the cell cycle checkpoint; to achieve this, a chromatin-associated phosphatase, Wip1, dephosphorylates H2AX at Ser139 and allows cells to enter the next cell division cycle (85). It is tempting to speculate that phosphorylation and dephosphorylation of Ser139 may dictate the direct recruitment and release of double-strand-break-sensing proteins in response to DNA damage, thereby affecting subsequent recovery and cell cycle progression.
Fig. 1.
(Top) Cartoon of the human H2AX histone showing all the serine, threonine, tyrosine, and histidine residues. Residues that have been reported to be phosphorylated are shown with a red “P.” (Bottom) Schematic of some of the events occurring at S139 in mammalian cells. Events that occur in yeast cells at the corresponding S129 are summarized in Table 1. (For details, see Table 1 and the text.)
Table 1.
Histone residues that are phosphorylateda
| Histone | Residue | Organism | Kinase(s) | Protein(s) recognizing (bound or released) the modification | Function(s) |
|---|---|---|---|---|---|
| H2A | Serine 139 (H2AX) | Mammals | ATR, ATM, DNA-PK, RSK2, MSK1 | DSB-sensing proteins (MRN, MDC1, P53BP1), AP1 | DNA repair, decreased EGF-mediated cellular transformation, apoptosis |
| Tyrosine 142 (H2AX) | Mammals | WSTF | ND | Decision between cell survival and apoptosis | |
| Serine 1 | Mammals | MSK | ND | Inhibition of transcription | |
| Threonine 119 | Mammals | NHK-1, Aurora B | PTB domain-containing protein Fe65 | Regulation of chromatin structure and function during mitosis | |
| Serine 121 | Fission yeast | Bub1 | Shugoshin | Maintaining chromosomal homeostasis by recruiting shugoshin at sister kinetochore | |
| Serine 129 | Yeast | ATM-related kinase Mec1 and Tel1 | NuA4, SWR1, INO80, cohesin | DSB repair | |
| H2B | Serine 10 | Yeast | Ste20 | ND | Apoptosis |
| Serine 14 | Various vertebrates | MstI | ND | Apoptosis | |
| Serine 32 | Mammals | Protein kinase C | ND | Possible involvement in apoptosis-related nucleosomal DNA fragmentation | |
| Serine 33 | Drosophila | CTK-TAF1 | ND | Transcriptional regulation | |
| Serine 36 | Mammals | AMPK | ND | Direct transcriptional and chromatin regulatory pathways leading to cellular response to stress | |
| H3 | Threonine 3 | Mammals | Haspin | Survivin | Correct localization of the CPC at the centromere |
| Threonine 6 | Mammals | PKCβ1 | ND | Androgen-dependent H3T6 phosphorylation prevents LSD1-mediated H3K4 demethylation, maintaining hormone-dependent gene activation | |
| Serine 10 | Yeast, mammals | Snf1, IpL1 (yeast), Aurora B (mammals), MSK1/2, IKKα, PKB/Akt, Rsk2, PIM1 | HP1, SRp20, ASF/SF2, 14-3-3 | Help in chromosome condensation during mitosis and meiosis; roles in transcription of certain genes | |
| Threonine 11 | Mammals | Chk1, PRK1, Dlk/Zip kinase | GCN5 | Transcriptional activation of certain genes; probable role at centromeres during mitosis | |
| Serine 28 | Mammals | Aurora B, MSK1/2 | Polycomb silencing complex | Help in chromosome condensation during mitosis and meiosis; roles in transcription of certain genes | |
| Tyrosine 41 | Mammals | JAK2 | HP1α | Role in differentiation related to hematopoiesis | |
| Threonine 45 | Mammals, budding yeast | Protein kinase C, S-phase kinase Cdc7-Dbf4 | ND | Apoptosis, role in DNA-damaged cells when the DNA is nicked, replication of DNA | |
| H4 | Serine 1 | Yeast, mammals | CK II | ND | DNA damage repair, mitosis and chromatin assembly |
| Histidines 18 and 75 | Mammals | Unknown | ND | Facilitation of DNA replication by destabilizing histone octamer |
DSB, double-strand break; ND, not determined.
Histone H2AX S139 phosphorylation has also been associated with apoptosis. In addition to double-strand-DNA-break repair in the survival pathway, phosphorylation of Ser139 on histone H2AX has also been implicated in apoptosis induced by death receptor activation (116). Etoposide-induced apoptotic chromatin condensation has been shown to be mediated by mammalian STE20-like kinase 1 (MSK1) through increased phosphorylation of H2AX S139 in Jurkat cells (132). In these two cases, the same modification (S139P) established by different kinases has completely opposite cellular outcomes. These results suggest that S139P is necessary but not sufficient to dictate both cellular outcomes and that other unique modifications or events that function in conjunction with S139P must occur in a pathway-specific manner.
A recent study by Zhu et al. investigated the role of H2AX Ser139 in epidermal growth factor (EGF)-mediated transformation of cells (140). They observed that EGF treatment resulted in phosphorylation of Ser139 and Ser16 of H2AX; they also identified ribosomal S6 kinase 2 (RSK2) as the enzyme responsible for these modifications (140). Their study revealed that EGF-dependent phosphorylation of Ser139 and Ser16 results in stabilization of H2AX and a decrease in AP1 transactivation. These together with a decreased H3S10 phosphorylation result in reduced cellular transformation.
Histone H2AX is the only H2A isoform present in yeast which is found to be phosphorylated in a DNA damage-dependent manner at serine 129. Consistent with a role for mammalian H2AX in DNA damage repair, yeast H2AX, in particular the C-terminal amino acids 129 to 132 with the SQE motif, is not necessary for cell cycle or transcriptional response to DNA damage but is important for double-strand DNA break repair by nonhomologous-end joining (31). Downs et al. have further shown that replacement of this serine with alanine impairs the double-strand-break-repair mechanism and makes the cells sensitive to the DNA damage-inducing agent methyl methanesulfonate (MMS) to the same extent as deletion of the SQEL motif (which is the consensus sequence for recognition of the PIKK kinase) (31). However, mutation of serine 129 with glutamate did not perturb the growth of the cells in media containing MMS, indicating that glutamic acid 129 in part mimics constitutive phosphorylation of serine 129 (31). Downs et al. proposed that mechanistically, this phosphorylation event upon DNA damage altered local chromatin structure, probably to facilitate DNA damage repair. This view was challenged by the work of Fink et al. (37), who suggested that histone H2A serine 129 phosphorylation functions not by alteration of chromatin structures but by recruitment of DNA repair proteins to the sites of lesions. At this point, several chromatin remodeling factors, like INO80, NuA4, and SWR1, have been found to be recruited in a DNA damage-induced and H2A phosphorylation-dependent manner (30, 91, 125). Continuing on the same theme, Unal et al. demonstrated that the yeast DNA damage checkpoint kinases Mec1p and Tel1p phosphorylated S129, generating a chromatin domain permissive to Mre11- and Scc2-dependent cohesion binding (124). Recent work from the Peterson laboratory has shown that recruitment of INO80 is involved in the ability of the cell to escape a prolonged cell cycle checkpoint arrest (96). To some extent, this effect can be physiologically compared to that of the mammalian phosphatase Wip1, which dephosphorylates DNA damage-induced H2AX at S139P and helps the cells to recover from cell cycle arrest. It is highly possible that serine 129 phosphorylation causes local and transient chromatin alteration at DNA double-strand breaks, allowing enhanced recruitment of DNA repair machinery.
The importance of this histone has also been analyzed in animal studies. First, deletion of H2AX in mice causes growth retardation and infertility in males, indicating that it is not essential for viability. Although no irradiation-induced cell cycle checkpoint was impaired, H2AX-null mice nonetheless are radiation sensitive, show increased chromosomal instability, and are immune deficient, implying a role for H2AX in DNA repair pathways. Interestingly, it has been seen that low-level gamma irradiation of mouse embryo fibroblasts (MEFs) does not influence cell cycle checkpoints, but a high (7-Gy) dose of gamma irradiation results in 100% mortality of H2A-null mice within 11 days, compared to a 20% death rate for unexposed littermates, indicating that irradiation-induced DNA damage does have a profound effect in H2AX null animals (16). Additional studies show that haploinsufficiency of histone H2AX also causes genomic instability, which in turn enhances the risk of cancer development in the absence of normal p53 function (8, 15). Double-strand-break repair can be mediated by homologous recombination (HR) or by nonhomologous-end joining (NHEJ). Bassing et al. and Celeste et al. observed that H2AX-deficient cells are impaired in HR but maintain functional NHEJ (8, 16). These results are in contrast to what has been observed in yeast, where histone H2A (and S129 phosphorylation) has been shown to be important for NHEJ and not homologous recombination (31). Interestingly, however, the phenotype of the early appearance of nondividing cells after passage 1 of H2AX-null MEFs is very similar to phenotypes observed in mice with mutations in the nonhomologous-end-joining proteins Ku70, Ku80, and ATM (7, 36, 46, 95). Additional and probably more site-specific DNA recombination studies must be carried out to firmly determine the necessary role of histone H2AX in double-strand-break repair by HR and/or NHEJ. Considering the critical role of DNA repair in physiology and the involvement of H2AX in the process, it is somewhat surprising that deletion of H2AX does not have a lethal phenotype and that some phenotypes, including impairment of DNA repair, are mild, indicating that H2AX is not necessary for overall viability and that in its absence, DNA damage repair pathways are affected but not completely eliminated. Similarly, mutational analysis in yeast indicated that serine 129 phosphorylation may not be critical for homologous recombination. Nonetheless, both mammalian cell- and yeast cell-based and animal studies clearly show that H2AX is important for DNA damage response. It remains to be seen if mutation of S139 will recapitulate all phenotypes seen in H2AX deletion mice. Development of histone H2AX-S139 mutant knock-in mice will facilitate future studies on the importance of phosphorylation of this residue in mouse physiology.
Although tyrosine phosphorylation is a major event in cellular cascades of kinase activation and signal transduction, this modification was only recently reported for histones (56, 115). A newly identified phosphorylation site in histone H2AX is Tyr142, which is phosphorylated by WSTF, a subunit of the WICH chromatin remodeling complex. A study from the Allis laboratory identified a novel, previously uncharacterized domain of WSTF that binds to and phosphorylates histone H2AX at Tyr142 upon DNA damage (134). WSTF-mediated phosphorylation of H2AX appears to prolong the retention of DNA damage response proteins at the site of damage. The mechanism is still not well understood, but it has been speculated that phosphorylation of Tyr142 may affect the kinetics of phosphorylation and dephosphorylation of H2AX such that in the absence of Tyr142 phosphorylation, the phosphorylation of Ser139 is affected and foci of DNA damage response proteins disassemble in less than 4 h (134). Cook et al. observed that the site of DNA damage-induced phosphorylated form of Tyr142 acts as a binding site for the PTB domain-containing adaptor protein Fe65 (24). Binding of Fe65 in turn facilitates docking of the proapoptotic transcription factor JNK1, thus triggering a DNA damage-dependent apoptotic pathway. Thus, it appears that the Tyr142 modification of histone H2AX drives a cell to either a repair-and-survival pathway or apoptosis (24, 134), though the cue that makes this crucial life-or-death decision is still unknown. These studies now set the stage for future investigations on this interesting modification and on Ser139 and Tyr142 cross talk. Since WSTF-WICH is an ATP-dependent chromatin remodeling complex, investigation of its role in H2AX Tyr142 phosphorylation and its subsequent effect on transcriptional regulation of genes involved in DNA damage response pathways would be of immense interest and significance. It also remains to be determined whether Tyr142 phosphorylation plays any role in regulating chromatin remodeling functions of the WSTF-WICH complex, particularly at DNA damage sites. It will be necessary to determine whether other ATP-dependent chromatin remodeling complexes have histone kinase activity on either Ser/Thr or Tyr residues.
The C terminus of histone H2A is also phosphorylated at residue Thr119. This modification is thought to regulate chromatin structure and function during mitosis. Histone H2A Thr119 phosphorylation is specific to the S phase and is caused primarily by Aurora B kinase and to a lesser extent by NHK-1 (nucleosomal histone kinase 1) (12). It has been shown that polo kinase can suppress the phosphorylation of Thr119 by NHK-1; however, the mechanism and the function of this are not yet known (12).
A recent report showed that the mitotic checkpoint protein Bub1 phosphorylates Ser121 of histone H2A in fission yeast, which in turn localizes the protein shugoshin. Shugoshin facilitates the recruitment of Aurora B and protein phosphatase 2A to the centromere, which senses anomalies in sister kinetochore spindle attachment and prevents chromosomal instability. These observations suggest a critical involvement of H2A phosphorylation in maintaining the chromosomal homeostasis and possibly prevention of tumorigenesis (64). As for other sites, Zhang et al. reported that the kinase MSK1 phosphorylated histone H2A at serine 1, thereby inhibiting transcription (137). Furthermore, their study revealed that acetylation of histone H3 by p300 and PCAF could suppress MSK-mediated transcription repression. This observation gave us an insight into histone cross talk and how histones coordinate to bring about cellular outcomes (137).
More recently, an interesting report by Kotova et al. showed that phosphorylation of the Drosophila homologue (H2Av) of the mammalian histone variants H2z/H2Ax directs the binding of poly(ADP-ribose) polymerase 1 (PARP1) to chromatin and also its enzymatic activation in a signal-dependent manner to activate downstream genes (68).
Future studies will need to determine whether—similar to the cytoplasmic Tyr phosphorylation signaling cascade—SH2-SH3 domain-containing nuclear proteins or proteins with a novel binding motif can associate with phosphorylated tyrosine residues of histones. In parallel, it will also be important to identify novel phosphotyrosine histone-binding motifs within nuclear proteins. Crystallographic, molecular modeling, and/or biochemical purification approaches may be critical in these investigations.
HISTONE H2B
Compared with histones H2A and H3 (discussed below), there are comparatively fewer reports of histone H2B phosphorylation (Fig. 2 and Table 1). In fact, there have been no reports of phosphorylation sites within the core or the C terminus of histone H2B. However, several N-terminal H2B phosphorylation sites are believed to play significant roles in cellular physiology and outcomes. In one study, caspase 3-activated mammalian sterile 20 (MstI) kinase was shown to phosphorylate H2B at Ser14 (21). Ser14-phosphorylated H2B has been implicated in apoptosis in a wide range of organisms, from frogs to mammals, and it has been suggested that H2B phosphorylation at this site facilitates apoptosis-related chromatin condensation, DNA fragmentation, and cell death. Based on these possible effects, there has been speculation about the potential of H2B Ser14 as a target for cancer drugs (21). Further, the Allis laboratory extended this finding to unicellular eukaryotes, where they showed that serine 10 of H2B is specifically phosphorylated in Saccharomyces cerevisiae by Ste20 kinase, a homologue of mammalian MstI, when the cells are treated with hydrogen peroxide to induce apoptosis (1). Together, these findings point to a conserved mechanism existing in a wide range of organisms that functions to maintain cellular homeostasis.
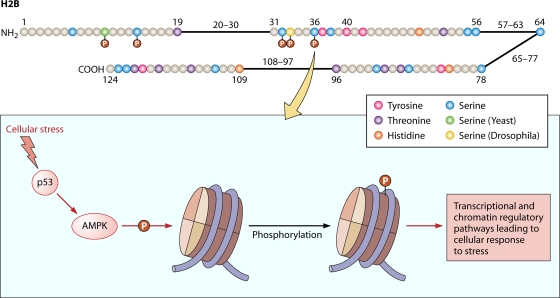
Fig. 2.
(Top) Cartoon of the human H2B histone showing all the serine, threonine, tyrosine, and histidine residues. Residues that have been reported to be phosphorylated are shown with a red “P.” Yeast has a serine at residue 10 (green), and Drosophila has a serine at residue 33 (yellow), both of which are also phosphorylated. (Bottom) Schematic of the events occurring at S36. (For details, see Table 1 and the text.)
Another study of histone phosphorylation profiles found that histone H2B was phosphorylated in almost all apoptotic cell lines studied (2). In leukemic HL-60 cells, drug-induced apoptosis triggers H2B phosphorylation, which in turn results in apoptosis-related nucleosomal DNA fragmentation. Protein kinase C, and to some extent protein kinase A, were identified as two potential kinases responsible for H2B phosphorylation in apoptosis. The study tentatively identified Ser32 as the phosphorylated residue responsible for this cellular function. More recently, the Berger laboratory reported that phosphorylation of H2B Ser36 occurs specifically in promoters and transcribed regions of genes that are upregulated in cells under stress (13). They showed that the kinase AMPK could phosphorylate the Ser36 residue in H2B. Furthermore, they identified a direct role for H2B Ser36 phosphorylation in transcriptional and chromatin regulatory pathways, leading to cellular responses to stress (Fig. 2). They showed that AMPK is recruited to specific promoters by the transcription factor p53—specifically to regions where p53 binds to proapoptotic genes, such as cptl1 and p21—and then subsequently phosphorylates the Ser36 residue of H2B associated with those transcribed regions. Additionally, AMPK recruitment may influence other stress-induced transcriptional activators and coactivators, such as Fox3a, PGC-1α, and TORC2. This mechanism partially explains one effect of histone phosphorylation in apoptosis. Moreover, Berger's group described an enrichment of Ser36-phosphorylated H2B along the length of the transcribed regions of stress-activated genes, supporting a role for phosphorylated H2B in transcriptional elongation by promoting RNA polymerase (Pol) II association at the promoter region (13). The authors concluded that H2B Ser36 phosphorylation may be a general cellular response to stress in which transcriptional regulation of certain genes occurs to support cell survival.
Since AMPK also senses the nutritional status of a cell, we further speculate that AMPK-mediated histone H2B Ser36 phosphorylation may link cellular energy status to genomic responses. The AMPK pathway is active in reactive oxygen species (ROS)-mediated cell survival/apoptosis decisions through the FOXO3-mediated transcriptional cascade (78), and AMPK is an important regulator of the circadian clock through CLOCK/BMAL1 target gene expression by controlling nutrient-dependent CRY1 stability within the cell (74). These findings lead us to hypothesize that H2B Ser36/Ser39 phosphorylation may also play regulatory roles in hormone signaling and circadian function, two active areas of current biomedical research.
A novel phosphorylation site on histone H2B was identified in Drosophila by Maile et al. (87). Their study showed that serine 33 was phosphorylated by the carboxyl-terminal kinase domain (CTK) of the Drosophila TFIID subunit TAFI. It was observed that phosphorylation of S33 at the promoters of the cell cycle regulatory gene string and the segmentation gene giant coincided with transcriptional activation (87). Since serine 33 of histone H2B is highly conserved, it would be interesting to see whether this residue is also phosphorylated in higher organisms and whether it plays any role in transcriptional activation of mammalian stress-responsive genes.
HISTONE H3
Among histones, histone H3 posttranslational modifications are probably the most studied. The first reports of histone H3 phosphorylation date back more than 30 years (113). Since then, there have been extensive reports on the various modifications of histone H3, including phosphorylation of Ser10, Ser28, and Thr11. Even though studies indicate that these modifications in yeast do not have profound physiological effects, many recent findings with both mammalian and yeast systems point to the fact that the modifications have cellular consequences (Fig. 3 and Table 1).
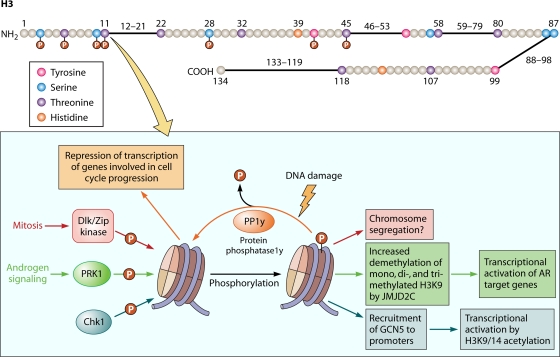
Fig. 3.
(Top) Cartoon of the human H3 histone showing all the serine, threonine, tyrosine, and histidine residues. Residues that have been reported to be phosphorylated are shown with a red “P.” (Bottom) Schematic of some of the events occurring at T11. (For details, see Table 1 and the text.)
Phosphorylation of Ser10 on histone H3 (H3S10P) has been well characterized. H3S10P during mitosis is conserved in eukaryotes, and many studies have characterized this modification throughout the cell cycle (49, 52, 130). In fact, H3S10P is used as a marker for mitotic cells. The kinases IpL1 and Snf-1 phosphorylate H3S10 in yeast (42, 79), whereas Aurora B, IKK, Rsk2, and AKT have been implicated in mammals (100, 105, 121, 135). The phosphorylation of H3S10 is first visible in the pericentromeric heterochromatin during late G2 phase. The modification subsequently spreads over the chromosomal arms, is completed by prophase, and remains visible during metaphase (52). Immunofluorescence studies clearly demonstrate the temporal and spatial relationship between chromosome condensation and histone H3S10 phosphorylation (43, 44). Dephosphorylation at this residue starts during anaphase and is completed within telophase, even before traces of chromosome decondensation become detectable (52, 81). These observations suggest that H3S10 phosphorylation is important for chromosome condensation and segregation; however, there have been studies showing that H3S10 phosphorylation is necessary only to initiate the condensation of the chromosomes, not to maintain it (126). Chromatin condensation/decondensation can be influenced by nucleosome remodeling. Interestingly, in their biochemical work, Shogren-Knaak et al. found that H3S10 phosphorylation does not directly affect SWI/SNF-dependent chromosome remodeling (114). In that study, the researchers used nucleosomal arrays which had H3S10P-modified histone ligated to them and compared the accessibility of these arrays to the restriction enzyme SalI in the presence of the ATP-dependent yeast remodeling factor SWI/SNF with that of unmodified arrays. It was found that the H3S10P modification does not contribute to the remodeling event (114). It is important to note here that in spite of evidence correlating H3S10 phosphorylation to chromosome condensation in mammalian cells, this phosphorylation does not have any appreciable physiological effect in yeast. It has been shown that in yeast, H3S10 phosphorylation peaks during mitosis, when chromosomes are maximally condensed and during the pachytene stage of meiosis (32, 54); however, the contribution of this histone modification to yeast cellular physiology has been questioned because mutation of serine 10 to alanine did not lead to any major defect. Budding yeast cells with S10A, S28A, and dual mutations (S10A S28A) were able to properly transmit mitotic or meiotic chromosomes during cell division (54). The sporulation time and cell cycle timing were also identical to those of the isogenic types, indicating a mechanism by which the cells bypass the S10 phosphorylation requirement for proper chromosome segregation, in contrast to various other organisms which require this phosphorylation mark. In this regard, it has been suggested by Hsu et al. that residues in the amino terminus of histone H2B have a milieu similar to that of H3S10 and that phosphorylation at these sites might be able to compensate for the loss of H3S10 phosphorylation in budding yeast (54). Hence, it seems that the need for H3S10 phosphorylation in chromosome condensation and segregation is not universal, which opens up an area for further investigations into other modifications which might act synergistically or redundantly in this process.
One open question in chromatin research is what the role of cell cycle-specific H3S10 phosphorylation might be; we wonder whether cell cycle-specific phosphorylation of the residue in conjunction with unique protein partners could participate in defined pathways, independent of the observed global spread of H3S10 phosphorylation over the chromatin. A recent study by the Allis laboratory reported that depletion or inhibition of Aurora B kinase, which phosphorylates H3 at Ser10, causes retention of heterochromatin protein 1 (HP1) on mitotic chromosomes; the authors suggested that H3S10P may mediate dissociation of HP1 from mitotic chromosomes, thereby facilitating their condensation and/or segregation (38). It is interesting that trimethylation of a lysine residue (Lys9) of H3 is responsible for recruitment of HP1 to discrete chromatin sites, but during M phase, the release of HP1 occurs without changes in levels of H3 Lys9 trimethylation. This same study showed that phosphorylation of H3 Ser10 was responsible for the ejection of HP1 from the chromosomes during M phase. Thus, phosphorylation of Ser10 disrupts the interaction between the HP1 chromodomain and the trimethylated Lys9, thereby shifting the equilibrium to the HP1 unbound state. This work revealed the presence of a more complex regulatory mechanism within the cell that relies on the combinatorial readout of interaction between a more stable modification (H3 Lys9 trimethylation) and a more dynamic modification (H3 Ser10 phosphorylation) (38). This histone modification-based switch may act as a part of a ‘histone readout,’ marking regions within the genome that need to be permanently silenced in the next generation. Use of such a binary switch to trigger HP1 ejection from mitotic chromatin may also be necessary for proper chromosome segregation.
In our laboratory, Loomis et al. took a biochemical approach to determine the possible interactions and functions of histone H3S10P (81). Interestingly, the authors showed that H3 Lys9 trimethylated peptides could bind the splicing factor SRp20 in addition to HP1 protein. Moreover, HP1 and SR proteins (SRp20/ASF/SF2) failed to bind the chromatin in the presence of the phosphorylation mark. In vitro studies further demonstrated that H3S10P is the major determinant that directs the loading and unloading of these proteins from the chromatin. These observations raised the question of whether release of HP1 and SRp20/ASF/SF2 from mitotic chromatin occurs independently or as a coupled phenomenon. Loomis et al. subsequently demonstrated that the dissociation of HP1 is dependent on the RNA-splicing proteins SRp20 and ASF/SF2, the binding of which is also H3S10 phosphorylation sensitive (81). Additionally, they showed that SRp20 and ASF/SF2 bind to chromatin in a cyclic manner, remaining bound during interphase and in the postmitotic chromosome and then dissociating during mitosis. The dissociation of the proteins from mitotic chromosomes coincides with H3S10 phosphorylation. In other experiments, knocking down ASF/SF2 expression affected the dissociation of HP1 from the chromatin despite H3S10 phosphorylation, indicating a possible role of the SR protein in ejection of HP1 during mitosis. Previous studies on ASF/SF2 had indicated its involvement in cell cycle progression; these studies were confirmed by the observation that knockdown of AFS/SF2 inhibits G2/M progression and delays G0/G1 entry (81). Another interesting finding by Loomis et al. was that the phosphorylation of the SR proteins influences binding to H3S10P. Phosphorylated forms of both SRp20 and ASF/SF2 failed to bind H3S10P, and the association required dephosphorylation of H3S10 (81). Thus, there appears to be an interplay of cellular kinase activity for histone marks (H3S10P) and proteins that recognize and are sensitive to these modifications for histone binding (SRp20, ASF/SF2), which introduces yet another layer of mechanistic cross talk between cellular and chromatin signaling pathways.
The cyclic patterns of binding, retention, and release of SRp20 and ASF/AF2 (and HP1) onto interphase, postmitotic, and mitotic chromosomes, as well as their roles in cell cycle progression, have been well documented, yet there is little evidence to indicate whether direct binding of ASF/SF2 and HP1 to the chromosome and its dissociation from mitotic chromatin are critical for chromosome condensation and cell cycle progression. This is primarily because of the limitations of the experimental system. Nonetheless, use of permanently chromosome-tethered recombinant ASF/SF2 containing chromo- or bromodomain fusions may facilitate an initial investigation of this important issue. Alternatively, use of H3S10A replacement histones (143) in mammalian cell-based assays would help address, at least in part, the question of whether phosphorylation-sensitive binding and release of these proteins are indeed necessary for cell cycle progression. Other possibilities are that ASF/SF2 and SRp20 are necessary for splicing of key cell cycle regulatory mRNAs and that physical non-chromatin-mediated association of SR proteins with components of cell cycle regulators is important for their effect on cell cycle progression. With regard to a plausible role for SR protein release, we speculate that such an event might prevent premature binding of mitosis-specific proteins to chromatin and that release of SR proteins allows binding of yet-to-be-discovered proteins. A biochemical affinity purification scheme may help in the identification of such factors.
The Ser28 residue of the histone H3 N-terminal tail is also phosphorylated and known to play a role in mitosis. Like H3S10, H3S28 is also phosphorylated during mitosis and meiosis, albeit to a lesser extent (100). The distribution of H3S28 phosphorylation is also very similar to that of H3S10P, initiating during prophase and then being maintained until early anaphase (43). H3S28 phosphorylation has also been shown to coincide with chromosome condensation during mitosis (44). The dynamics of H3 Ser28 phosphorylation is a consequence of the interplay between the kinase Aurora B and its phosphatase counterpart, PP1. PP1 is inactivated by Cdc2 kinase-mediated phosphorylation during mitosis, thereby allowing the appearance of the Ser28 phosphorylation mark on H3 (44). After chromosome segregation, Aurora B no longer remains associated with chromatin, resulting in decrease in H3 Ser28 phosphorylation. The biological significance of H3S28 phosphorylation is not well understood, but there are data indicating that it may be involved in chromosome condensation and subsequent mitosis. It will be interesting to investigate whether phosphorylation of H3S28, similar to H3S10, assists in recruitment or ejection of certain factors from the chromatin in a cell cycle-specific manner to assist cell cycle progression. In this context, we note that overexpression of a kinase-dead mutant of Aurora B abolishes phosphorylation of both histones H3 Ser10 and Ser28, accompanied by incomplete chromosome condensation and misalignment of chromosomes on the metaphase plate (44). These findings establish important roles for both H3 Ser10 and Ser28 phosphorylations as well as the kinases involved in chromosome dynamics. Taken together, these data highlight the importance of H3 Ser10 and Ser28 phosphorylation during mitosis. It will be necessary to determine whether Ser10 and Ser28 phosphorylation cross talk dictates, at least in part, chromatin dynamics during cell cycle progression.
A small fraction of nucleosomes also show phosphorylation of the Ser10 and Ser28 residues of histone H3 during interphase, and these modifications appear to be related to transcriptional activities (17, 44). The first link between gene expression induction and H3 phosphorylation was made in a study in which mammalian cells were treated with phorbol esters and growth factors and induction of immediate-early genes was accompanied by phosphorylation of H3 serine residues within the N-terminal tail (86). Further investigation using chromatin immunoprecipitation (ChIP)-based assays confirmed these findings (18, 123). H3 Ser10 and H3 Ser28 phosphorylations have been observed in distinct chromosomal regions during interphase in cells treated with various transcriptional inducers. Cells treated with the phorbol ester 12-O-tertadecanoylphorbol-13-acetate (TPA) show activated Ras/ERK/MAPK signaling, which in turn activates the mitogen- and stress-activated protein kinases 1 and 2 (MSK1 and MSK2) (33, 117). These kinases then phosphorylate Ser10 and Ser28 on H3. The same study also demonstrated the requirement of H3 Ser10 phosphorylation to activate c-fos in response to TPA treatment (117). Other studies have shown that H3S10 phosphorylation-dependent c-fos activation can be regulated by a variety of kinases depending on the nature of the stimulus: MSK1 and MSK2 phosphorylate H3S10 upon activation by ERK, as does tumor necrosis factor-α-induced IKK-α (4, 35, 117, 135). Additionally, PIM1 kinase has been shown to phosphorylate H3S10 in MYC-dependent transcriptional activation (142). Other kinases, such as PKB/Akt and Rsk2, have also been reported to phosphorylate H3S10 (20, 105).
A recent work from the Cheung laboratory showed that direct phosphorylation of H3S28 on the c-fos promoter by the kinase MSK1 is sufficient to activate transcription even in the absence of upstream signaling (76). Moreover, H3S28 phosphorylation at the α-globin promoter by MSK1 could abolish the transcriptional repression imposed by the Polycomb complex. This modification also triggered a methyl-acetylation switch in the adjacent K27 residue, thereby transactivating the gene (76, 77). This indicates that histone modifications are the points of convergence of signaling pathways and that this nodal event is crucial for gene expression.
Another role of H3S10 phosphorylation during interphase involves transcriptional activation via the 14-3-3 family of proteins. Macdonald et al. have shown that certain isoforms of 14-3-3 bind phosphorylated H3 Ser10 or Ser28 (84). It was subsequently shown that the binding affinity of 14-3-3 was stronger if the Lys14 residue was acetylated in addition to Ser10's being phosphorylated (127, 133). There is ample debate in this field as to whether transcriptional regulation by H3S10 phosphorylation is synergistic with or independent of adjacent lysine acetylation. The Berger laboratory showed in 2000 that H3S10 phosphorylation-mediated transcriptional activation of genes was functionally linked to acetylation of lysine 14 (80). Their work showed that phosphorylation of H3S10 increased acetylation of the adjacent lysine residue by the histone acetyltransferase (HAT) Gcn5 at the promoters of genes that are transcriptionally activated. They further showed that promoters requiring HAT activity of Gcn5 almost always required H3S10 phosphorylation for their activation and that H3S10 phosphorylation improved lysine 14 acetylation at these promoters. A year later, the Berger laboratory further characterized the nutrient- and energy-sensing kinase Snf1 as the kinase responsible for transcription-associated H3S10 phosphorylation in yeast (79). They studied the INO1 gene and concluded that Snf1-mediated phosphorylation of H3S10 resulted in Gcn5-mediated lysine 14 acetylation and subsequent transcriptional activation. Interestingly, however, Snf1 was not the kinase that promoted serine 10 phosphorylation, which preceded lysine 14 acetylation of the HO gene promoter during transcriptional activation, suggesting that additional yeast kinases exist and that these yet-to-be-identified kinases may be recruited in a gene- and signal-specific manner to promote HO gene transcription. Interestingly, however, more recent findings from the Berger laboratory have shown that the lysine 14 acetylation is not always preceded by serine 10 phosphorylation. At the promoter of the GAL1 gene, the Lys14 acetylation mark is present before the Ser10 phosphorylation mark; however, both the modifications are necessary for transcriptional activation of the gene (127).
There is controversy regarding the role of H3S10P in coupling S10P to histone acetylation. The Peterson laboratory used purified GCN5-containing SAGA complex and nucleosomal substrate to show that there is no increase in the binding affinity of the SAGA complex for phosphorylated H3S10 (39, 114). Genome-wide binding studies under various physiological conditions should resolve this issue. Additionally, as with mitotic H3S10P, transcription-linked H3S10P may trigger release of repressor proteins such as the INHAT (inhibitor of acetyltransferase) complex (73, 106, 109, 110), thereby linking histone phosphorylation, inhibitor/repressor release, and hyperacetylation. In vivo genome-wide binding studies will be necessary to test this possibility.
What additional mechanisms might be involved in H3S10P-mediated transcriptional activation? Work by Walter et al. revealed that the 14-3-3 family of proteins bind H3S10P-enriched promoters although they have higher affinity for the dual phosphorylation-acetylation mark, consistent with a synergistic cooperative model for histone modification cross talk in transcription even though the Lys14 acetyl mutant (K14R) showed only a 15% increased impairment in recruitment of the 14-3-3 protein Bmh1 to the promoter compared to the S10A strain (127). Also, in mammalian cells, 14-3-3 ζ proteins were recruited to c-fos and c-jun promoters associated with dually modified H3 (phospho-Ser10-acetyl-Lys14 H3) (84). 14-3-3 has also been reported to bind the phospho-Ser10 nucleosomes on the FOSL1 promoter to trigger transcriptional elongation through a series of downstream events involving H4K16 acetylation by enhanced recruitment of MOF histone acetyltransferase and subsequent recruitment of the bromodomain protein BRD4 (143). In Drosophila, H3 has been reported to be phosphorylated at Ser10 by the kinase Jil1, and as in mammalian cells, H3S10P-dependent recruitment of 14-3-3 proteins is necessary for early transcriptional elongation (63). This is another insight into the role of H3S10 phosphorylation in transcription.
The cooccurrence of H3S10 and -28P in mitotic and interphase cells, albeit at different sites, raises the question of how cells distinguish these marks in a cell cycle-specific manner. We wonder if, similar to interphasic 14-3-3 proteins, there is a mitosis-specific protein that recognizes H3S10 phosphorylation. It would be interesting to identify such proteins and characterize the distinguishing features of such proteins transducing mitotic and interphase H3S10 phosphorylation events. Additionally, identification of Snf1 in yeast opens up the possibility of the existence of a similar mammalian kinase whose function might be to contribute to regulated transcription of specific promoters. In this context we note the Berger laboratory recently showed that the mammalian homolog of yeast Snf1, the signaling kinase AMPK, activated stress-induced genes by phosphorylating histone H2B serine 36 (13). However, AMPK has not been shown to phosphorylate H3S10, and similarly, yeast Snf1 has yet to be shown to phosphorylate histone H2BS36, raising the possibility that the substrate specificity diverged for Snf1 kinase in yeast and mammals or that the mechanisms by which H3S10 and H2BS36 become phosphorylated and regulate transcription in yeast and mammals are different. In this context, we note that Zhang et al. observed EGF-induced phosphorylation of H3S10 at the promoters of Brf (transcription factor for RNA Pol III genes) and TBP genes. Also, H3 is phosphorylated at S28 by EGF directly on the promoters of tRNA, 5S rRNA, and 7SL RNA, thereby inducing transcription. Increased activation of these genes result heightened translation, leading to cellular transformation (136). In any case, generation of knock-in mice with site-specific mutations (S10A, S28A, and S10A-S28A) will be extremely valuable in demonstrating the role, if any, of these residues in animal viability and key physiological pathways. As with yeast, such mutant mice might give us some surprising results.
Currently, another residue of H3, Thr11, is being actively studied, and a possible significance in transcription is emerging (Fig. 3 and Table 1) (29). The Thr11 residue of H3 is phosphorylated during mitosis and interphase by an array of kinases, including the serine/threonine protein kinase Chk1, protein kinase C-related kinase 1 (PRK1), and Dlk/Zip kinases (90, 99, 112). Studies have shown that H3 Thr11 phosphorylation is predominant during mitosis and is concentrated at the centromeres. The Dlk/Zip kinase has been implicated in phosphorylating Thr11 of H3 preferentially at the centromeres, and Ser10 phosphorylation is replaced by Thr11 phosphorylation exclusively at the centromeres during mitosis (99). This observation supports some novel role of Thr11 phosphorylation at the centromeres during mitosis. In two separate studies, H3 Thr11 was found to be phosphorylated differentially at the promoters of certain genes during interphase, resulting in different transcriptional outcomes. Metzger et al. showed that during interphase, PRK1 phosphorylates H3 at Thr11 on certain androgen receptor-responsive genes (e.g., PSA and KLK2) and facilitates gene activation by accelerating demethylation of the mono-, di-, and trimethylated Lys9 repression mark by the demethylase JMJD2C (90). Furthermore, ligand-induced acetylation of H3 Lys9/Lys14 is facilitated following the demethylation event. Under such circumstances, the presence of serine 5-phosphorylated RNA Pol II can be detected at the target promoters, resulting in subsequent gene activation. It is interesting that phosphorylation of the adjacent Ser10 residue blocks demethylation of trimethylated H3 Lys9 by JMJD2A, which is a close homolog of JMJD2C (90, 93). These results indicate that the context and site specificity of the histone modifications are as important as the modification itself for such cross talk to occur. Interestingly, immunostaining of prostate cancer samples revealed increased levels of PRK1 and H3T11P, which correlated with high Gleason scores, indicating aggressive biology of the tumors. This finding suggests a role for PRK1 in prostate tumorigenesis, which probably occurs, in part, through transcriptional control of androgen receptor-responsive genes via H3 Thr11 phosphorylation (90). PRK1 is known to regulate other nuclear receptors, such as mineralocorticoid and progesterone receptors; it will be interesting to investigate the broader role, if any, of this modification and the kinase in transcriptional regulation by other nuclear receptors and inducible transcription factors such as NF-κB. Similarly, PRK1 recruitment by AR may cause phosphorylation of nonhistone substrates, such as components of the enhanceosome or transcription initiation complex, and these additional events may also contribute to the transcriptional output of AR target genes. A possible link between PRK1-mediated H3 Thr11 phosphorylation and tumorigenesis in ER+/− breast tumors would be a similarly compelling subject for investigation.
In other studies of H3 Thr11 phosphorylation, a structure-function analysis using phosphohistone peptides and S. cerevisiae mutants showed that Thr11 phosphorylation is necessary for the recruitment of the acetyltransferase GCN5 and is required for optimal transcription from GCN5-dependent promoters (23). It is interesting that GCN5 HAT activity is also required in H3S10-mediated transcriptional regulation of certain genes which need Lys14 acetylation for complete activation. We speculate here that a similar role for GCN5 might also exist in H3T11 phosphorylation-mediated gene activation. It will therefore be relevant to investigate the role of lysine 14 acetylation or acetylation of other adjacent lysines to see whether a broader histone modification cross talk exists. H3 Thr11 phosphorylation by Chk1 has also been related to the transcriptional repression of certain genes upon DNA damage (112). DNA damage causes Chk1 to dissociate rapidly from the chromatin, resulting in a loss of H3 Thr11 phosphorylation. This in turn causes reduced binding of the acetyltransferase GCN5 at promoters of important cell cycle genes, like cyclin B1 and Cdk1, and subsequent reduced acetylation of H3 Lys9. Deacetylation of H3 at these promoters results in transcriptional repression (112). It would be interesting to determine whether H3 Thr11 phosphorylation also acts as a recognition module for the docking of certain transcription factors or an unknown H3T11P-binding protein. It will also be important to resolve the question of the relative contributions of H3S10 and H3T11 phosphorylation in GCN5/SAGA recruitment and histone acetylation.
Recently, the Schule laboratory reported that threonine 6 of H3 is phosphorylated by protein kinase C beta 1 (PKCβ1) in an androgen-dependent manner (89). Here, an intricate cross talk between histone phosphorylation and methylation has been shown to exist to maintain the expression of certain genes in the presence of androgen hormone. The study revealed that androgen treatment results in colocalization of androgen receptor (AR), the lysine-specific histone demethylase LSD1, and PKCβ1 on target gene promoters. While LSD1 demethylates H3K4, resulting in gene repression, it was noted that in the presence of H3T6 phosphorylation, LSD1 could no longer demethylate residue K4, thereby allowing androgen-mediated gene expression. Moreover, it was observed that increased PKCβ1 levels correlated with more aggressive forms of prostate cancer and that knockdown of the kinase resulted in reduced cell proliferation. Together, these studies firmly established a role for threonine 6 phosphorylation in nuclear hormone action, cell proliferation, and cancer.
Recently, Shimada et al. reported that protein phosphatase 1γ (PP1γ) removes the phosphate group from H3 threonine 11 (111). They showed that PP1γ was able to dephosphorylate phosphorylated H3T11 in a DNA damage-dependent manner (111). This urges us to question the role of phosphorylation and dephosphorylation of H3T11 in DNA damage and opens up avenues for further work.
Thr3 of H3 has also been found to be phosphorylated. Wang et al. reported haspin to be the kinase responsible for the phosphorylation (128). During mitosis, phosphorylation of Thr3 by haspin helps in the precise positioning of the chromosome passenger complex (CPC) at the inner centromeres. The authors showed that survivin, a protein component of the CPC, binds directly to phosphorylated Thr3 and ensures proper localization of the CPC. Studies with mutants of survivin revealed that the BIR domain of survivin is involved in the direct interaction with the H3 Thr3 phosphorylated residue. This study opened up the possibility that the BIR domain is a module for binding phosphorylated residues. It was also observed that H3 Thr3 phosphorylation prevents H3 from inhibiting Aurora B by interfering with its autophosphorylation. The authors concluded that H3 Thr3 phosphorylation might have a dual role: first, in positioning the CPC properly at the centromeres, and second, in facilitating activation of Aurora B by autophosphorylation at the inner centromeres (128). Since H3 Thr3 and Ser10 have the same kinase, it would be interesting to investigate the existence of cross talk between the two modifications in various cellular contexts and signaling processes.
Finally, there are some lesser known C-terminal phosphorylations of H3 that appear to play roles in cellular physiology. H3 Thr45 is phosphorylated by protein kinase C, and this modification is involved in apoptosis of cells in which DNA is nicked (57). Thr45 has also been reported to be phosphorylated in budding yeast by the kinase Cdc7-Dbf4. This modification has been related to proper replication of DNA in budding yeast (5). Another significant C-terminal modification is H3 Tyr41 phosphorylation. A recent study has implicated this modification—driven by JAK2—in normal hematopoiesis as well as in leukemia (26). Inhibition of JAK2 results in decreased H3 Tyr41 phosphorylation at the Imo2 promoter, resulting in reduced expression of this hematopoietic oncogene. Interestingly, similar to H3S10P, Tyr41 phosphorylation is also involved in the eviction of HP1 from chromatin. HP1α, but not HP1β, binds the Tyr41 residue of H3 through its chromodomain. Phosphorylation of H3Tyr41 excluded HP1α binding to this region. Consequently, inhibition of the nonreceptor tyrosine kinase JAK2 results in decreased expression of Imo2 along with reduced H3 Tyr41 phosphorylation and subsequent HP1α recruitment (26). It remains to be seen whether H3 Tyr41 phosphorylation plays a role in a similar eviction of SR proteins from mitotic chromatin. While these results suggest that signal-specific modification of either Ser10 or Tyr41 might have identical physiological outcomes, it would be interesting to explore the possibility of novel signaling functions of H3Tyr41 through the recruitment of as-yet-undiscovered proteins.
HISTONE H4
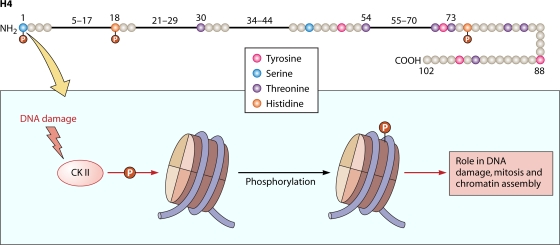
Phosphorylation at Ser1 of histone H4 is another well-documented modification (Fig. 4 and Table 1). This modification has been found in an array of organisms, including worms, flies, and mammals, suggesting its physiological importance. The localization and timing of this modification coincide with H3S10 phosphorylation. Therefore, H4 Ser1 phosphorylation is also regarded as a mitosis-specific modification. Although Ser1 phosphorylation correlates closely with chromatin condensation events during mitosis (6), the kinase responsible is unknown. H4 Ser1 phosphorylation has also been established as a DNA repair-linked histone modification, in which DNA damage triggers the phosphorylation of Ser1 by casein kinase II (CK II). ChIP studies have confirmed that histone H4 Ser1 phosphorylation occurs in regions close to double-strand breaks. Additionally, H4S1 phosphorylation plays a role in nonhomologous-end-joining repairs of double-strand breaks (22). We wonder whether H4 Ser1 and H2AX Ser139 phosphorylation may have complementary, overlapping, and/or redundant functions in DNA damage response. These results may also explain in part the mild phenotype of histone H2AX mutants in yeast and mammals. Finally, H4S1 has been reported to be involved in sporulation in yeasts and has been found to be conserved in both fly and mouse spermatogenesis (70).
Fig. 4.
Cartoon of the human H4 histone showing all the serine, threonine, tyrosine, and histidine residues. Residues that have been reported to be phosphorylated are shown with a red “P.” (Bottom) Schematic of the events occurring at S1. (For details, see Table 1 and the text.)
Apart from the Ser1 residue, there are a few uncommon residues that are phosphorylated in histone H4, including His18 and His75; however, the kinase responsible has not yet been identified. Two studies have reported a partial purification of cell fractions from various sources and the identification of candidate histidine kinases that phosphorylate histone H4 at His75 (55, 131). With regard to cellular function, it has been speculated that histone H4 His75 phosphorylation helps to stabilize the histone octamer through its imidazole ring (82, 83). His75 phosphorylation may lead to destabilization of the bond, thereby facilitating replication of its associated DNA during cellular proliferation. We anticipate that identification and characterization of histone H4 phosphorylation sites, the kinases and phosphatases involved, effector proteins recognizing phosphorylated histone H4 residues, and the associated cellular functions will be increasingly active areas of research.
HISTONE H1
Histone H1 is not part of the nucleosomal core; nevertheless, it plays an important role in nucleosomal stability and in maintaining higher-order chromatin structure. Histone H1 phosphorylation has been shown to be dependent on cell cycle progression in a variety of organisms. Histone H1 is found to be maximally phosphorylated during S phase and mitosis, followed by a drop to its lowest level during G1. During mitosis, there is a noticeable increase in histone H1 phosphorylation at the onset and through metaphase, followed by a sharp decrease at anaphase (3, 11, 47). The different subtypes of H1 histones, however, differ with regard to their degree of phosphorylation.
There are studies showing the involvement of histone H1 phosphorylation in chromatin condensation in a cell cycle-dependent manner; however, some data indicate that histone H1 phosphorylation might be more involved in chromatin decondensation rather than condensation (102). Hale et al. have shown that histone H1 binds through its C terminus to HP1 to maintain the higher-order compact chromatin structure but that CDK2-mediated phosphorylation of histone H1 disrupts this interaction and facilitates efficient cell cycle progression (51). Histone H1 phosphorylation apparently destabilizes chromatin structure, which in turn may permit access to the DNA by transcriptional factors and the replication machinery during G1 and S phase (19). There have also been reports that histone H1 phosphorylation is necessary for RNA polymerase I- and II-dependent transcription (138).
The N- and C-terminal ends of the histone H1 protein contain all the potential sites for phosphorylation. Two consensus sites (K[S/T]PXK and K[S/T]PK) are present on the tails; these sites are targeted by the CDC kinase family, and the serine and threonine residues are phosphorylated in a cell cycle-dependent manner (75). The levels of H1 phosphorylation appear to depend on the relative abundance of protein phosphatase 1 and CDC2/CDK2 kinases within the cell. It is generally believed that, unlike the other histones, it is the number of sites phosphorylated, as opposed to particular residues in histone H1, that is important in determining cellular effects (48). It is still not clear how histone H1 phosphorylation affects chromatin condensation and decondensation throughout the cell cycle. A recent study identified several sites on different histone H1 variants that are phosphorylated at different stages of the cell cycle. Sarg et al. showed that four cell cycle-specific kinases participate in this regulated phosphorylation event in Chinese hamster ovary (CHO) cells: the interphase-specific p33CDC2/cyclin A (kinase B), the late-interphase-specific p34CDC2/cyclin A (kinase A), the mitotic p34CDC2/cyclin B (kinase C), and the p34CDC2/unknown cyclin (kinase M) (104). Particular serine and threonine residues of the different variants are phosphorylated by these kinases at different stages of the cell cycle (104). Also, the Mizzen laboratory recently showed that specific sites in histone H1 variants H1.2 and H1.4 are phosphorylated exclusively in mitosis, whereas others are modified in both mitosis and interphase (138).
Further investigation revealed that interphase-specific H1.4 phosphorylation is concentrated at active 45S pre-rRNA gene promoters and is responsive to differential steroid hormone treatment at sites of steroid hormone response elements (138). This site-specific histone H1 phosphorylation during interphase assists RNA Pol I- and II-dependent transcription and has an as-yet-unexplained function in ribosome biogenesis and cell growth control.
CROSS TALK OF ENZYMES, MARKS, AND EFFECTOR PROTEINS
Given the abundance of different histone modifications, it is easy to appreciate the various levels of cross talk that exist between and within cellular and histone signaling pathways. There are certain modifications that work together toward the same outcome: histone H3 Ser10 and Ser28 phosphorylations both detected in condensed chromosomes during mitosis and meiosis. On the other hand, H3S10 phosphorylation blocks the methylation of H3K9 while dephosphorylation is not affected (34). It has been reported that ubiquitylation of histone H2A has an inhibitory effect on Aurora B, the primary kinase for H3 Ser10 and Ser28 phosphorylation. For proper chromosomal condensation during initiation of mitosis, histone H2A needs to be deubiquitylated by the enzyme Ubp-M to allow M-phase progression (61). Another study has shown that during mitosis, the levels of histone acetylation and histone methylation differ dramatically; specifically, the level of acetylated histone H4K16 decreases drastically, while the level of methylated histone H4K20 peaks (94). In a recent study, H2A serine 1 phosphorylation by MSK1 was shown to repress transcription, albeit in vitro; however, this repression can be removed by acetylation of H3 by p300 and PCAF (137). There are also several examples in which modification of one residue results in the exclusion of binding of a certain protein to an adjacent residue. A very good example of this phenomenon is the phosphorylation of H3S10, which affects the binding of the HP1 protein to methylated H3K9 during mitosis, while dephosphorylation of H3S10 augments the binding of HP1 back to methylated H3K9 (38). Thus, the phosphorylation of H3S10 and methylation of H3K9 dictate the binding of HP1 in and out of mitosis.
Several studies have demonstrated that additional factors may also interact with histone modifications in order to recruit or eject chromatin-modifying factors and bring about cellular changes. SRp20 and ASF/SF2 are two splicing factors that have been shown to bind interphase chromatin and are dissociated from mitotic chromatin. Knockdown of ASF/SF2 results in retention of HP1 on mitotic chromatin despite H3S10 phosphorylation (81). These observations hint at an intricate network of histone modifications, along with chromatin modifiers and other factors, which coordinate to allow proper cell cycle progression.
It is also important to note that the roles of the effector proteins are as important as the histone modifications themselves, and that the cross talk among the “effectors” provides yet another level of signaling complexity and control of cellular processes. These effector proteins have diverse roles, and like the SR proteins, they may participate in a number of overlapping pathways (139). It remains to be seen if these pathways communicate with each other through these key proteins.
Cross-regulation also exists at the level of functioning of histone-modifying enzymes. For example, isomerization of histone H3 Pro38 affects methylation of H3K36 by Set2 (92). In certain instances, an enzyme can recognize its substrate more efficiently in the presence of accompanying modifications; one example is that of GCN5 acetyltransferase, which can recognize H3 more efficiently when it is phosphorylated at Ser10 (23). Cross talk between histones can also occur when the modifications are present on different histone tails; for instance, ubiquitylation of histone H2B is required for trimethylation of H3K4.
With more advanced investigations being done on a global scale, it is possible to look at all the histone modifications in a particular histone at a given time (9, 41, 59). Bonenfant et al. did an extensive examination of different histone modifications throughout the cell cycle. Interestingly, phosphorylation of H3 and H4 occurred mainly during mitosis, whereas H2A modifications were constant through the cell cycle. G2/M-arrested cells displayed a general loss in acetylation of all the histones, whereas methylation patterns changed in a more complex manner, with levels of some methylated residues increasing while others decreased (9). Similarly, the work done by Garcia et al. with mitotic HeLa cells identified novel modification sites in histones; for example, the study revealed the presence of a novel phosphorylation mark on serine 31 of the histone H3 variants H3.2 and H3.3, along with dimethylation at lysine 36 (41). Such studies also have the potential to identify a larger number of novel modifications in different cellular contexts (here, cell cycle progression) and also give a complete picture of multiple histone modifications existing at a particular time. This information is critical in understanding the epigenetic signaling network within cells and their physiological relevance. Identification of the proteins recognizing these modifications and the enzymes controlling them (kinases and phosphatases) will be instrumental in future understanding at the molecular level of the roles of these novel modifications in organ and cellular physiology.
SUMMARY AND FUTURE DIRECTIONS
Given the diversity, multitude, and interaction of posttranslational histone modifications, it is a challenging task to assign specific biological effects to a single histone modification, especially when we must take into account all of the cross talk between and within the cellular and histone signaling pathways. The initial formulation of a histone code helped us understand this complex system more easily and stimulated research activities in the field (50, 58, 107, 118). However, these modifications cannot be easily interpreted, may function in a gene- and signal-specific manner, may not have universal coding capabilities, and can be interpreted only in combination with other modifications within a genomic, signaling, and regulatory context (107). A prime example of this conundrum is histone phosphorylation, which has been studied extensively in the recent past and is reviewed here to highlight major advances. However, because these histone modifications and their functions are not universal, the correct interpretation of the significance of these modifications also requires a detailed study in the context of evolutionarily divergent eukaryotes.
Major advances have been made in recent years regarding how histones are marked (modified), how marks are read by binding or effector proteins, and how such talks are interpreted into cellular outcome. However, much remains to be discovered, which makes future research in this discipline exciting, challenging, and thought provoking. Extensive studies of histone acetylation and methylation have led to the identification of certain cognate domains in proteins that facilitate interaction and binding with specific histone posttranslational modifications. For example, proteins bearing the 110-amino-acid bromodomain have been found to be more prone to bind proteins with acetylated lysines, including histones (10, 120). Several transcriptional coactivators, including HATs (10, 28), BET nuclear factors (BRD2, BRD4, and BDF1) (27, 88), and SWI/SNF remodeling factors (BRM and BRG1) (45, 62), have bromodomains through which they bind acetylated histones and assist in transcriptional activation of several genes. Similarly, there are distinct protein families that bind to methylated histone lysine residues through recognition modules like the chromodomain, the Tudor domain plant homeodomain finger, and the WD40 repeat domain (25). It is critical that such consensus sequences or domains be identified for the effector proteins that bind phosphorylated histone residues. Identification of such domains will help identify more interacting proteins and shed light on the various cellular pathways in which histone phosphorylation participates. In the context of cell cycle-dependent binding and release of cellular proteins onto chromatin, the major issue that remains unresolved is whether and how such cyclic binding and release of proteins are critical for cell cycle progression. Indirect evidence is strong, but direct experimental demonstration is lacking on this important issue. In this review, we provide some testable ideas on this matter. Increasing our understanding of the roles of different histone phosphorylation modifications in cellular signaling will help us decipher the complex web of histone and cellular signaling cross talk and its importance in cellular proliferation and response to stress. Ultimately, these efforts may reveal new therapeutic targets that will help us to target complex cellular processes and diseases, such as aging and cancer. To conclude, we hope readers will appreciate how vigorous studies in the chromatin field unveil the mechanisms that regulate histone phosphorylation under various cellular and environmental conditions and show how cells sense and utilize these modifications to carry out a diverse array of critical physiological functions. We understand that this is just a glimpse into the complex realm of histone phosphorylation, and more exciting information will continue to emerge as scientists around the world dig deeper into the secrets of chromatin.
ACKNOWLEDGMENTS
Work in our laboratory is supported in part by NIH grants RO1 CA 133755 and PO1 HD57877.
Due to space constraints, we were unable to include all contributions that have advanced the knowledge on this exciting topic.
D.C. dedicates this review to the memory of Jonathan Widom of Northwestern University, who suddenly passed away and was a major contributor in the field of chromatin research, as well as a friend and colleague.
We have nothing to declare.
Footnotes
Published ahead of print on 17 October 2011.
REFERENCES
- 1. Ahn S. H., et al. 2005. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell 120:25–36 [DOI] [PubMed] [Google Scholar]
- 2. Ajiro K. 2000. Histone H2B phosphorylation in mammalian apoptotic cells. An association with DNA fragmentation. J. Biol. Chem. 275:439–443 [DOI] [PubMed] [Google Scholar]
- 3. Ajiro K., Borun T. W., Cohen L. H. 1981. Phosphorylation states of different histone 1 subtypes and their relationship to chromatin functions during the HeLa S-3 cell cycle. Biochemistry 20:1445–1454 [DOI] [PubMed] [Google Scholar]
- 4. Anest V., Cogswell P. C., Baldwin A. S., Jr 2004. IκB kinase α and p65/RelA contribute to optimal epidermal growth factor-induced c-fos gene expression independent of IκBα degradation. J. Biol. Chem. 279:31183–31189 [DOI] [PubMed] [Google Scholar]
- 5. Baker S. P., et al. 2010. Histone H3 Thr 45 phosphorylation is a replication-associated post-translational modification in S. cerevisiae. Nat. Cell Biol. 12:294–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barber C. M., et al. 2004. The enhancement of histone H4 and H2A serine 1 phosphorylation during mitosis and S-phase is evolutionarily conserved. Chromosoma 112:360–371 [DOI] [PubMed] [Google Scholar]
- 7. Barlow C., et al. 1996. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell 86:159–171 [DOI] [PubMed] [Google Scholar]
- 8. Bassing C. H., et al. 2003. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell 114:359–370 [DOI] [PubMed] [Google Scholar]
- 9. Bonenfant D., et al. 2007. Analysis of dynamic changes in post-translational modifications of human histones during cell cycle by mass spectrometry. Mol. Cell. Proteomics 6:1917–1932 [DOI] [PubMed] [Google Scholar]
- 10. Bottomley M. J. 2004. Structures of protein domains that create or recognize histone modifications. EMBO Rep. 5:464–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bradbury E. M., Inglis R. J., Matthews H. R., Langan T. A. 1974. Molecular basis of control of mitotic cell division in eukaryotes. Nature 249:553–556 [DOI] [PubMed] [Google Scholar]
- 12. Brittle A. L., Nanba Y., Ito T., Ohkura H. 2007. Concerted action of Aurora B, Polo and NHK-1 kinases in centromere-specific histone 2A phosphorylation. Exp. Cell Res. 313:2780–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bungard D., et al. 2010. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science 329:1201–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Burma S., Chen B. P., Murphy M., Kurimasa A., Chen D. J. 2001. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 276:42462–42467 [DOI] [PubMed] [Google Scholar]
- 15. Celeste A., et al. 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5:675–679 [DOI] [PubMed] [Google Scholar]
- 16. Celeste A., et al. 2002. Genomic instability in mice lacking histone H2AX. Science 296:922–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cerutti H., Casas-Mollano J. A. 2009. Histone H3 phosphorylation: universal code or lineage specific dialects? Epigenetics 4:71–75 [DOI] [PubMed] [Google Scholar]
- 18. Chadee D. N., et al. 1999. Increased Ser-10 phosphorylation of histone H3 in mitogen-stimulated and oncogene-transformed mouse fibroblasts. J. Biol. Chem. 274:24914–24920 [DOI] [PubMed] [Google Scholar]
- 19. Chadee D. N., et al. 1995. Increased phosphorylation of histone H1 in mouse fibroblasts transformed with oncogenes or constitutively active mitogen-activated protein kinase kinase. J. Biol. Chem. 270:20098–20105 [DOI] [PubMed] [Google Scholar]
- 20. Cheung P., Allis C. D., Sassone-Corsi P. 2000. Signaling to chromatin through histone modifications. Cell 103:263–271 [DOI] [PubMed] [Google Scholar]
- 21. Cheung W. L., et al. 2003. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell 113:507–517 [DOI] [PubMed] [Google Scholar]
- 22. Cheung W. L., et al. 2005. Phosphorylation of histone H4 serine 1 during DNA damage requires casein kinase II in S. cerevisiae. Curr. Biol. 15:656–660 [DOI] [PubMed] [Google Scholar]
- 23. Clements A., et al. 2003. Structural basis for histone and phosphohistone binding by the GCN5 histone acetyltransferase. Mol. Cell 12:461–473 [DOI] [PubMed] [Google Scholar]
- 24. Cook P. J., et al. 2009. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature 458:591–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cruickshank M. N., Besant P., Ulgiati D. 2010. The impact of histone post-translational modifications on developmental gene regulation. Amino Acids 39:1087–1105 [DOI] [PubMed] [Google Scholar]
- 26. Dawson M. A., et al. 2009. JAK2 phosphorylates histone H3Y41 and excludes HP1α from chromatin. Nature 461:819–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dey A., Chitsaz F., Abbasi A., Misteli T., Ozato K. 2003. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. U. S. A. 100:8758–8763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dhalluin C., et al. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491–496 [DOI] [PubMed] [Google Scholar]
- 29. Di Croce L., Shiekhattar R. 2008. Thrilling transcription through threonine phosphorylation. Nat. Cell Biol. 10:5–6 [DOI] [PubMed] [Google Scholar]
- 30. Downs J. A., et al. 2004. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 16:979–990 [DOI] [PubMed] [Google Scholar]
- 31. Downs J. A., Lowndes N. F., Jackson S. P. 2000. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408:1001–1004 [DOI] [PubMed] [Google Scholar]
- 32. Dresser M. E., Giroux C. N. 1988. Meiotic chromosome behavior in spread preparations of yeast. J. Cell Biol. 106:567–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Drobic B., Dunn K. L., Espino P. S., Davie J. R. 2006. Abnormalities of chromatin in tumor cells. EXS 2006:25–47 [DOI] [PubMed] [Google Scholar]
- 34. Duan Q., Chen H., Costa M., Dai W. 2008. Phosphorylation of H3S10 blocks the access of H3K9 by specific antibodies and histone methyltransferase. Implication in regulating chromatin dynamics and epigenetic inheritance during mitosis. J. Biol. Chem. 283:33585–33590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Duncan E. A., Anest V., Cogswell P., Baldwin A. S. 2006. The kinases MSK1 and MSK2 are required for epidermal growth factor-induced, but not tumor necrosis factor-induced, histone H3 Ser10 phosphorylation. J. Biol. Chem. 281:12521–12525 [DOI] [PubMed] [Google Scholar]
- 36. Elson A., et al. 1996. Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 93:13084–13089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fink M., Imholz D., Thoma F. 2007. Contribution of the serine 129 of histone H2A to chromatin structure. Mol. Cell. Biol. 27:3589–3600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fischle W., et al. 2005. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438:1116–1122 [DOI] [PubMed] [Google Scholar]
- 39. Fry C. J., Shogren-Knaak M. A., Peterson C. L. 2004. Histone H3 amino-terminal tail phosphorylation and acetylation: synergistic or independent transcriptional regulatory marks? Cold Spring Harb. Symp. Quant. Biol. 69:219–226 [DOI] [PubMed] [Google Scholar]
- 40. Furumatsu T., Asahara H. 2010. Histone acetylation influences the activity of Sox9-related transcriptional complex. Acta Med. Okayama 64:351–357 [DOI] [PubMed] [Google Scholar]
- 41. Garcia B. A., et al. 2005. Modifications of human histone H3 variants during mitosis. Biochemistry 44:13202–13213 [DOI] [PubMed] [Google Scholar]
- 42. Glover D. M., Leibowitz M. H., McLean D. A., Parry H. 1995. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell 81:95–105 [DOI] [PubMed] [Google Scholar]
- 43. Goto H., et al. 1999. Identification of a novel phosphorylation site on histone H3 coupled with mitotic chromosome condensation. J. Biol. Chem. 274:25543–25549 [DOI] [PubMed] [Google Scholar]
- 44. Goto H., Yasui Y., Nigg E. A., Inagaki M. 2002. Aurora-B phosphorylates Histone H3 at serine28 with regard to the mitotic chromosome condensation. Genes Cells 7:11–17 [DOI] [PubMed] [Google Scholar]
- 45. Grune T., et al. 2003. Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol. Cell 12:449–460 [DOI] [PubMed] [Google Scholar]
- 46. Gu Y., et al. 1997. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity 7:653–665 [DOI] [PubMed] [Google Scholar]
- 47. Gurley L. R., D'Anna J. A., Barham S. S., Deaven L. L., Tobey R. A. 1978. Histone phosphorylation and chromatin structure during mitosis in Chinese hamster cells. Eur. J. Biochem. 84:1–15 [DOI] [PubMed] [Google Scholar]
- 48. Gurley L. R., Valdez J. G., Buchanan J. S. 1995. Characterization of the mitotic specific phosphorylation site of histone H1. Absence of a consensus sequence for the p34cdc2/cyclin B kinase. J. Biol. Chem. 270:27653–27660 [DOI] [PubMed] [Google Scholar]
- 49. Gurley L. R., Walters R. A., Barham S. S., Deaven L. L. 1978. Heterochromatin and histone phosphorylation. Exp. Cell Res. 111:373–383 [DOI] [PubMed] [Google Scholar]
- 50. Hake S. B., Allis C. D. 2006. Histone H3 variants and their potential role in indexing mammalian genomes: the “H3 barcode hypothesis.” Proc. Natl. Acad. Sci. U. S. A. 103:6428–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hale T. K., Contreras A., Morrison A. J., Herrera R. E. 2006. Phosphorylation of the linker histone H1 by CDK regulates its binding to HP1α. Mol. Cell 22:693–699 [DOI] [PubMed] [Google Scholar]
- 52. Hendzel M. J., et al. 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106:348–360 [DOI] [PubMed] [Google Scholar]
- 53. Houben A., et al. 2007. Phosphorylation of histone H3 in plants—a dynamic affair. Biochim. Biophys. Acta 1769:308–315 [DOI] [PubMed] [Google Scholar]
- 54. Hsu J. Y., et al. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102:279–291 [DOI] [PubMed] [Google Scholar]
- 55. Huang C. K., Laramee G. R. 1991. Stimulation of a histone H4 protein kinase in Triton X-100 lysates of rabbit peritoneal neutrophils pretreated with chemotactic factors: effect of leukotriene B4 and cytochalasin B. J. Leukoc. Biol. 49:158–162 [DOI] [PubMed] [Google Scholar]
- 56. Hunter T. 2009. Tyrosine phosphorylation: thirty years and counting. Curr. Opin. Cell Biol. 21:140–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hurd P. J., et al. 2009. Phosphorylation of histone H3 Thr-45 is linked to apoptosis. J. Biol. Chem. 284:16575–16583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jenuwein T., Allis C. D. 2001. Translating the histone code. Science 293:1074–1080 [DOI] [PubMed] [Google Scholar]
- 59. Jiang L., et al. 2007. Global assessment of combinatorial post-translational modification of core histones in yeast using contemporary mass spectrometry. LYS4 trimethylation correlates with degree of acetylation on the same H3 tail. J. Biol. Chem. 282:27923–27934 [DOI] [PubMed] [Google Scholar]
- 60. Johansen K. M., Johansen J. 2006. Regulation of chromatin structure by histone H3S10 phosphorylation. Chromosome Res. 14:393–404 [DOI] [PubMed] [Google Scholar]
- 61. Joo H. Y., et al. 2007. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature 449:1068–1072 [DOI] [PubMed] [Google Scholar]
- 62. Kadam S., Emerson B. M. 2003. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol. Cell 11:377–389 [DOI] [PubMed] [Google Scholar]
- 63. Karam C. S., Kellner W. A., Takenaka N., Clemmons A. W., Corces V. G. 2010. 14-3-3 mediates histone cross-talk during transcription elongation in Drosophila. PLoS Genet. 6:e1000975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kawashima S. A., Yamagishi Y., Honda T., Ishiguro K., Watanabe Y. 2010. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science 327:172–177 [DOI] [PubMed] [Google Scholar]
- 65. Khan S. N., Khan A. U. 2010. Role of histone acetylation in cell physiology and diseases: an update. Clin. Chim. Acta 411:1401–1411 [DOI] [PubMed] [Google Scholar]
- 66. Kobayashi S., Nakagawa S., Uehara N., Hamashima H., Tanaka A. 2002. DNA topology: topological transformation of DNA by reaction of cisplatin-DNA-core histone complexes with human DNA topoisomerase. Nucleic Acids Res. Suppl. 2002:283–284 [DOI] [PubMed] [Google Scholar]
- 67. Kornberg R. D. 1977. Structure of chromatin. Annu. Rev. Biochem. 46:931–954 [DOI] [PubMed] [Google Scholar]
- 68. Kotova E., et al. 2011. Drosophila histone H2A variant (H2Av) controls poly(ADP-ribose) polymerase 1 (PARP1) activation in chromatin. Proc. Natl. Acad. Sci. U. S. A. 108:6205–6210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kouzarides T. 2007. Chromatin modifications and their function. Cell 128:693–705 [DOI] [PubMed] [Google Scholar]
- 70. Krishnamoorthy T., et al. 2006. Phosphorylation of histone H4 Ser1 regulates sporulation in yeast and is conserved in fly and mouse spermatogenesis. Genes Dev. 20:2580–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kruger W., et al. 1995. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 9:2770–2779 [DOI] [PubMed] [Google Scholar]
- 72. Kulis M., Esteller M. 2010. DNA methylation and cancer. Adv. Genet. 70:27–56 [DOI] [PubMed] [Google Scholar]
- 73. Kutney S. N., Hong R., Macfarlan T., Chakravarti D. 2004. A signaling role of histone-binding proteins and INHAT subunits pp32 and Set/TAF-Iβ in integrating chromatin hypoacetylation and transcriptional repression. J. Biol. Chem. 279:30850–30855 [DOI] [PubMed] [Google Scholar]
- 74. Lamia K. A., et al. 2009. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 326:437–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Langan T. A., et al. 1989. Mammalian growth-associated H1 histone kinase: a homolog of cdc2+/CDC28 protein kinases controlling mitotic entry in yeast and frog cells. Mol. Cell. Biol. 9:3860–3868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lau P. N., Cheung P. 2011. Histone code pathway involving H3 S28 phosphorylation and K27 acetylation activates transcription and antagonizes polycomb silencing. Proc. Natl. Acad. Sci. U. S. A. 108:2801–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lau P. N., Cheung P. 2011. Unlocking polycomb silencing through histone H3 phosphorylation. Cell Cycle 10:1514–1515 [DOI] [PubMed] [Google Scholar]
- 78. Li X. N., et al. 2009. Activation of the AMPK-FOXO3 pathway reduces fatty acid-induced increase in intracellular reactive oxygen species by upregulating thioredoxin. Diabetes 58:2246–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lo W. S., et al. 2001. Snf1—a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 293:1142–1146 [DOI] [PubMed] [Google Scholar]
- 80. Lo W. S., et al. 2000. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell 5:917–926 [DOI] [PubMed] [Google Scholar]
- 81. Loomis R. J., et al. 2009. Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol. Cell 33:450–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Luger K., Mader A. W., Richmond R. K., Sargent D. F., Richmond T. J. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251–260 [DOI] [PubMed] [Google Scholar]
- 83. Luger K., Rechsteiner T. J., Flaus A. J., Waye M. M., Richmond T. J. 1997. Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol. 272:301–311 [DOI] [PubMed] [Google Scholar]
- 84. Macdonald N., et al. 2005. Molecular basis for the recognition of phosphorylated and phosphoacetylated histone h3 by 14-3-3. Mol. Cell 20:199–211 [DOI] [PubMed] [Google Scholar]
- 85. Macurek L., et al. 2010. Wip1 phosphatase is associated with chromatin and dephosphorylates gammaH2AX to promote checkpoint inhibition. Oncogene 29:2281–2291 [DOI] [PubMed] [Google Scholar]
- 86. Mahadevan L. C., Willis A. C., Barratt M. J. 1991. Rapid histone H3 phosphorylation in response to growth factors, phorbol esters, okadaic acid, and protein synthesis inhibitors. Cell 65:775–783 [DOI] [PubMed] [Google Scholar]
- 87. Maile T., Kwoczynski S., Katzenberger R. J., Wassarman D. A., Sauer F. 2004. TAF1 activates transcription by phosphorylation of serine 33 in histone H2B. Science 304:1010–1014 [DOI] [PubMed] [Google Scholar]
- 88. Maruyama T., et al. 2002. A mammalian bromodomain protein, brd4, interacts with replication factor C and inhibits progression to S phase. Mol. Cell. Biol. 22:6509–6520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Metzger E., et al. 2010. Phosphorylation of histone H3T6 by PKCβ(I) controls demethylation at histone H3K4. Nature 464:792–796 [DOI] [PubMed] [Google Scholar]
- 90. Metzger E., et al. 2008. Phosphorylation of histone H3 at threonine 11 establishes a novel chromatin mark for transcriptional regulation. Nat. Cell Biol. 10:53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Morrison A. J., et al. 2004. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119:767–775 [DOI] [PubMed] [Google Scholar]
- 92. Nelson C. J., Santos-Rosa H., Kouzarides T. 2006. Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell 126:905–916 [DOI] [PubMed] [Google Scholar]
- 93. Ng S. S., et al. 2007. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature 448:87–91 [DOI] [PubMed] [Google Scholar]
- 94. Nishioka K., et al. 2002. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell 9:1201–1213 [DOI] [PubMed] [Google Scholar]
- 95. Nussenzweig A., et al. 1996. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature 382:551–555 [DOI] [PubMed] [Google Scholar]
- 96. Papamichos-Chronakis M., Krebs J. E., Peterson C. L. 2006. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 20:2437–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Park E. J., Chan D. W., Park J. H., Oettinger M. A., Kwon J. 2003. DNA-PK is activated by nucleosomes and phosphorylates H2AX within the nucleosomes in an acetylation-dependent manner. Nucleic Acids Res. 31:6819–6827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Park J. H., Cosgrove M. S., Youngman E., Wolberger C., Boeke J. D. 2002. A core nucleosome surface crucial for transcriptional silencing. Nat. Genet. 32:273–279 [DOI] [PubMed] [Google Scholar]
- 99. Preuss U., Landsberg G., Scheidtmann K. H. 2003. Novel mitosis-specific phosphorylation of histone H3 at Thr11 mediated by Dlk/ZIP kinase. Nucleic Acids Res. 31:878–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Prigent C., Dimitrov S. 2003. Phosphorylation of serine 10 in histone H3, what for? J. Cell Sci. 116:3677–3685 [DOI] [PubMed] [Google Scholar]
- 101. Rogakou E. P., Pilch D. R., Orr A. H., Ivanova V. S., Bonner W. M. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858–5868 [DOI] [PubMed] [Google Scholar]
- 102. Roth S. Y., Allis C. D. 1992. Chromatin condensation: does histone H1 dephosphorylation play a role? Trends Biochem. Sci. 17:93–98 [DOI] [PubMed] [Google Scholar]
- 103. Sahin M., Sahin E., Gumuslu S., Erdogan A., Gultekin M. 2010. DNA methylation or histone modification status in metastasis and angiogenesis-related genes: a new hypothesis on usage of DNMT inhibitors and S-adenosylmethionine for genome stability. Cancer Metastasis Rev. 29:655–676 [DOI] [PubMed] [Google Scholar]
- 104. Sarg B., Helliger W., Talasz H., Forg B., Lindner H. H. 2006. Histone H1 phosphorylation occurs site-specifically during interphase and mitosis: identification of a novel phosphorylation site on histone H1. J. Biol. Chem. 281:6573–6580 [DOI] [PubMed] [Google Scholar]
- 105. Sassone-Corsi P., et al. 1999. Requirement of Rsk-2 for epidermal growth factor-activated phosphorylation of histone H3. Science 285:886–891 [DOI] [PubMed] [Google Scholar]
- 106. Schneider R., Bannister A. J., Weise C., Kouzarides T. 2004. Direct binding of INHAT to H3 tails disrupted by modifications. J. Biol. Chem. 279:23859–23862 [DOI] [PubMed] [Google Scholar]
- 107. Schreiber S. L., Bernstein B. E. 2002. Signaling network model of chromatin. Cell 111:771–778 [DOI] [PubMed] [Google Scholar]
- 108. Selvi B. R., Cassel J. C., Kundu T. K., Boutillier A. L. 2010. Tuning acetylation levels with HAT activators: therapeutic strategy in neurodegenerative diseases. Biochim. Biophys. Acta 1799:840–853 [DOI] [PubMed] [Google Scholar]
- 109. Seo S. B., et al. 2002. Regulation of histone acetylation and transcription by nuclear protein pp32, a subunit of the INHAT complex. J. Biol. Chem. 277:14005–14010 [DOI] [PubMed] [Google Scholar]
- 110. Seo S. B., et al. 2001. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell 104:119–130 [DOI] [PubMed] [Google Scholar]
- 111. Shimada M., Haruta M., Niida H., Sawamoto K., Nakanishi M. 2010. Protein phosphatase 1gamma is responsible for dephosphorylation of histone H3 at Thr 11 after DNA damage. EMBO Rep. 11:883–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Shimada M., et al. 2008. Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell 132:221–232 [DOI] [PubMed] [Google Scholar]
- 113. Shoemaker C. B., Chalkley R. 1978. An H3 histone-specific kinase isolated from bovine thymus chromatin. J. Biol. Chem. 253:5802–5807 [PubMed] [Google Scholar]
- 114. Shogren-Knaak M. A., Fry C. J., Peterson C. L. 2003. A native peptide ligation strategy for deciphering nucleosomal histone modifications. J. Biol. Chem. 278:15744–15748 [DOI] [PubMed] [Google Scholar]
- 115. Singh R. K., Gunjan A. 2011. Histone tyrosine phosphorylation comes of age. Epigenetics 6:153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Solier S., Pommier Y. 2009. The apoptotic ring: a novel entity with phosphorylated histones H2AX and H2B and activated DNA damage response kinases. Cell Cycle. 8:1853–1859 [DOI] [PubMed] [Google Scholar]
- 117. Soloaga A., et al. 2003. MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J. 22:2788–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Strahl B. D., Allis C. D. 2000. The language of covalent histone modifications. Nature 403:41–45 [DOI] [PubMed] [Google Scholar]
- 119. Stucki M., Jackson S. P. 2006. gammaH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amsterdam) 5:534–543 [DOI] [PubMed] [Google Scholar]
- 120. Taverna S. D., Li H., Ruthenburg A. J., Allis C. D., Patel D. J. 2007. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 14:1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Teperek-Tkacz M., Meglicki M., Pasternak M., Kubiak J. Z., Borsuk E. 2010. Phosphorylation of histone H3 serine 10 in early mouse embryos: active phosphorylation at late S phase and differential effects of ZM447439 on first two embryonic mitoses. Cell Cycle 9:4674–4687 [DOI] [PubMed] [Google Scholar]
- 122. Thiriet C., Hayes J. J. 2005. Replication-independent core histone dynamics at transcriptionally active loci in vivo. Genes Dev. 19:677–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Thomson S., Clayton A. L., Mahadevan L. C. 2001. Independent dynamic regulation of histone phosphorylation and acetylation during immediate-early gene induction. Mol. Cell 8:1231–1241 [DOI] [PubMed] [Google Scholar]
- 124. Unal E., et al. 2004. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell 16:991–1002 [DOI] [PubMed] [Google Scholar]
- 125. van Attikum H., Fritsch O., Hohn B., Gasser S. M. 2004. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119:777–788 [DOI] [PubMed] [Google Scholar]
- 126. Van Hooser A., Goodrich D. W., Allis C. D., Brinkley B. R., Mancini M. A. 1998. Histone H3 phosphorylation is required for the initiation, but not maintenance, of mammalian chromosome condensation. J. Cell Sci. 111(Pt. 23):3497–3506 [DOI] [PubMed] [Google Scholar]
- 127. Walter W., et al. 2008. 14-3-3 interaction with histone H3 involves a dual modification pattern of phosphoacetylation. Mol. Cell. Biol. 28:2840–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Wang F., et al. 2010. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science 330:231–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Ward I. M., Chen J. 2001. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276:47759–47762 [DOI] [PubMed] [Google Scholar]
- 130. Wei Y., Yu L., Bowen J., Gorovsky M. A., Allis C. D. 1999. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell 97:99–109 [DOI] [PubMed] [Google Scholar]
- 131. Wei Y. F., Matthews H. R. 1990. A filter-based protein kinase assay selective for alkali-stable protein phosphorylation and suitable for acid-labile protein phosphorylation. Anal. Biochem. 190:188–192 [DOI] [PubMed] [Google Scholar]
- 132. Wen W., et al. 2010. MST1 promotes apoptosis through phosphorylation of histone H2AX. J. Biol. Chem. 285:39108–39116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Winter S., et al. 2008. 14-3-3 proteins recognize a histone code at histone H3 and are required for transcriptional activation. EMBO J. 27:88–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Xiao A., et al. 2009. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature 457:57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Yamamoto Y., Verma U. N., Prajapati S., Kwak Y. T., Gaynor R. B. 2003. Histone H3 phosphorylation by IKK-α is critical for cytokine-induced gene expression. Nature 423:655–659 [DOI] [PubMed] [Google Scholar]
- 136. Zhang Q., Zhong Q., Evans A. G., Levy D., Zhong S. 2011. Phosphorylation of histone H3 serine 28 modulates RNA polymerase III-dependent transcription. Oncogene 30:3705–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Zhang Y., Griffin K., Mondal N., Parvin J. D. 2004. Phosphorylation of histone H2A inhibits transcription on chromatin templates. J. Biol. Chem. 279:21866–21872 [DOI] [PubMed] [Google Scholar]
- 138. Zheng Y., et al. 2010. Histone H1 phosphorylation is associated with transcription by RNA polymerases I and II. J. Cell Biol. 189:407–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Zhong X. Y., Wang P., Han J., Rosenfeld M. G., Fu X. D. 2009. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol. Cell 35:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Zhu F., et al. 2011. Phosphorylation of H2AX at Ser139 and a new phosphorylation site Ser16 by RSK2 decreases H2AX ubiquitination and inhibits cell transformation. Cancer Res. 71:393–403 [DOI] [PubMed] [Google Scholar]
- 141. Zhu Q., Wani A. A. 2010. Histone modifications: crucial elements for damage response and chromatin restoration. J. Cell Physiol. 223:283–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Zippo A., De Robertis A., Serafini R., Oliviero S. 2007. PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nat. Cell Biol. 9:932–944 [DOI] [PubMed] [Google Scholar]
- 143. Zippo A., et al. 2009. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 138:1122–1136 [DOI] [PubMed] [Google Scholar]