Abstract
MDM2 is a major regulator of p53 by acting as a ubiquitin E3 ligase. The central acidic domain and C-terminal RING domain of MDM2 are both indispensable for ubiquitination of p53. Our previous study suggested that ATM phosphorylation of MDM2 near the C terminus inhibits RING domain oligomerization, resulting in p53 stabilization after DNA damage. We present here evidence that these modifications allosterically regulate the functions of both acidic domain and RING domain of MDM2. Using chemical cross-linking, we show that the MDM2 RING domain forms oligomers including dimer and higher-order complexes in vivo. RING domain dimerization efficiency is negatively regulated by upstream sequence. ATM-mediated phosphorylation of the upstream sequence further inhibits RING dimerization. Forced oligomerization of MDM2 partially overcomes the inhibitory effect of phosphorylation and stimulates p53 ubiquitination. Furthermore, the ability of MDM2 acidic domain to bind p53 core domain and induce p53 misfolding are also suppressed by the same C-terminal ATM sites after DNA damage. These results suggest that the acidic domain and RING domain of MDM2 are both allosterically coupled to the intervening ATM sites, which enables the same modification to regulate multiple MDM2 functions critical for p53 ubiquitination.
INTRODUCTION
The p53 pathway is critical for maintenance of genomic stability and preventing development of cancer. The most notable feature of p53 is its stabilization after exposure to a wide range of stress signals such as hyperproliferation, nucleotide depletion, and DNA damage. These responses may be essential for its function as a tumor suppressor (14). In normal cells, p53 is present at very low levels due to rapid degradation through the ubiquitin-dependent proteasome pathway. p53 turnover is regulated by MDM2, which binds p53 and functions as an ubiquitin E3 ligase to promote p53 degradation by the proteasome (15, 16, 21). Additional E3 ligases such as Pirh2 and Cop1 have also been implicated as regulators of p53 turn over (12, 24). However, their physiological significance in p53 regulation and stress response still remain to be established. Current evidence suggests that MDM2 is a major and indispensable regulator of p53 level (18, 32).
MDM2 and p53 interact through their N-terminal domains in a high-affinity binding and through their central domains in a weak binding (48). Upon complex formation, the MDM2 C-terminal RING domain recruits ubiquitin-conjugating enzyme E2 that performs the transfer of activated ubiquitin to p53 lysine residues. To date, the understanding of molecular mechanisms that lead to p53 stabilization by different pathways remain incomplete. An important group of MDM2 regulators are proteins that bind to the central acidic domain, such as ARF, L5, L11, and L23 (37, 50). These basic proteins are important for mediating mitogenic stress and ribosomal stress signals to p53. They have been shown to inhibit MDM2-mediated p53 ubiquitination, but little is known about the mechanisms. They may act by neutralizing activities of the MDM2 acidic domain, which is critical for efficient p53 ubiquitination through unknown mechanisms (19, 31).
p53-MDM2 binding is essential for specific targeting of p53 for degradation and has been extensively studied as a target of regulation by DNA damage signaling. Several studies showed that DNA double-strand breaks induce phosphorylation of p53 S15 by DNA-PK and ATM (1, 30, 35, 39). ATM also activates Chk2, which in turn phosphorylates p53 on S20, which is part of the MDM2 binding site (1, 7, 38, 39). These findings suggested that p53 phosphorylation on the N terminus disrupts MDM2 binding and results in p53 stabilization. However, studies in mouse models showed that blocking mouse p53 phosphorylation on S18 or S23 (counterparts of human S15 and S20) only partially reduced p53 stabilization after DNA damage (6, 27). Single site mutation of S18 had no significant effect on p53 stabilization or tumor suppression function (5). These results indicate that robust p53 stabilization involves mechanisms besides p53 phosphorylation.
An alternative target for DNA damage stabilization of p53 is MDM2. MDM2 is phosphorylated by ATM, ATR, and C-Abl on sites near the C terminus (29, 40, 41). DNA damage also induces dephosphorylation of several serine residues in the acidic domain of MDM2 (3). We recently identified several novel phosphorylation sites near the RING domain of MDM2 and showed that blocking phosphorylation of all sites produced a constitutively active MDM2 that can prevent p53 stabilization after DNA damage. Therefore, phosphorylation of MDM2 appears to be a key step in p53 stabilization by DNA damage signals. MDM2 phosphorylation after DNA damage correlates with reduced oligomerization of the RING domain, suggesting that high-order complex formation is needed for efficient polyubiquitination of p53 (9). However, the functional significance of MDM2 oligomerization and its regulation by DNA damage in vivo still lack direct experimental support. The mechanism by which phosphorylation regulates MDM2 oligomerization has not been determined. It is also not clear whether phosphorylation by ATM regulates MDM2 through additional mechanisms.
In the present study, we further examined the effects of DNA damage on MDM2 E3 ligase function and its biochemical properties. The results show that sequence near the MDM2 RING domain has a role in negatively regulating RING dimerization and oligomerization, which is further potentiated by ATM-mediated phosphorylation. Artificially induced oligomerization of MDM2 significantly increases p53 ubiquitination. The ATM phosphorylation sites near the RING domain also regulate the p53 binding and misfolding functions of the acidic domain. These findings suggest that the ATM sites regulate multiple MDM2 domains to achieve efficient inhibition of p53 ubiquitination after DNA damage.
MATERIALS AND METHODS
Cell lines and plasmids.
MDM2 point mutants were generated by site-directed mutagenesis using a QuikChange kit (Stratagene). All MDM2 constructs used in the present study were human cDNA clones. MDM2-Praja fusion construct was provided by Allan Weissman (13). U2OS cells with stable expression of MDM2 mutants were generated by transfection of cytomegalovirus-driven MDM2 plasmids, followed by G418 selection and isolation of clonal cell lines. Induced oligomerization of MDM2 was achieved by using the dimerization kit provided by ARIAD. Three tandem copies of the FKBP ligand bind domain were fused to the N terminus of MDM2 by PCR cloning. DI-p53 was constructed by PCR subcloning, converting seven residues (underlined) in the MDM2 binding site of full-length wild-type p53 (1-MEEPQSDPSVEPPLSQETFSDLWKLLPENNVLSPLP-36) to a high-affinity MDM2 binding site in DI-p53 (1-MEEPQSDPSVEPPLSQETFEHWWSQLLSNNVLSPLP-36). The mutations destroy the epitope for DO-1 antibody but do not affect transcriptional activity.
Protein analysis.
To detect proteins by Western blot, cells were lysed in lysis buffer (50 mM Tris-HCl [pH 8.0], 5 mM EDTA, 150 mM NaCl, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride [PMSF], 50 mM NaF) and centrifuged for 5 min at 10,000 × g. Cell lysate (10 to 50 μg of protein) was fractionated by SDS-PAGE using gradient gel and transferred to Immobilon P filters (Millipore). The filter was blocked for 1 h with phosphate-buffered saline (PBS) containing 5% nonfat dry milk-0.1% Tween 20 and incubated with primary antibodies. The filter was developed using ECL-Plus reagent (Amersham). The MDM2 monoclonal antibodies 2A9, 3G9, 4B2, and 4B11 were used (8). DO-1 and Pab1801 were used for the detection of p53. For immunoprecipitation with conformation-specific p53 antibodies, the lysate was divided into equal halves, and each was immunoprecipitated for 18 h at 4°C with wild-type conformation-specific antibody (Pab1620) or mutant conformation-specific antibody (Pab240). The precipitated p53 was detected by using rabbit polyclonal antibody FL393 (Santa Cruz Biotechnology).
Ubiquitination assays. (i) In vivo assay.
H1299 cells in 10-cm plates were transfected with 5 μg of Myc-ubiquitin, 1 to 2 μg of MDM2 and 1 μg of p53 expression plasmids using calcium phosphate precipitation method. Thirty-two hr after transfection, cells were precipitated using p53 antibody Pab1801 in the presence of 10 mM iodoacetamide, and probed with anti-Myc antibody by Western blotting.
(ii) In vitro assay.
SJSA cells were treated with 10 Gy of ionizing radiation (IR) in the presence of 30 μM MG132 for 2 h. MDM2 was immunoprecipitated with 2A9 antibody. The substrate p53 was produced by in vitro translation in rabbit reticulocyte lysate by using the TNT system (Promega) in the presence of [35S]methionine. Portions (15 μl) of packed protein A-beads loaded with MDM2 from a 15-cm plate of SJSA cells were treated with 1 U of calf intestinal phosphatase (CIP) for 0.5 h at 37°C when indicated, washed with lysis buffer and reaction buffer (50 mM Tris [pH 7.5], 2.5 mM MgCl2, 15 mM KCl, 1 mM dithiothreitol, 0.01% Triton X-100, 1% glycerol), incubated with 5 μl of in vitro-translated p53 for 2 h at 4°C, and washed with reaction buffer. The beads loaded with MDM2-p53 complex were incubated with 100 ng of purified human ubiquitin-activating enzyme His6-E1, 250 ng of His6-UbcH5b (E2), 5 μg of ubiquitin or K0-ubiquitin (Biomol), and 20 μl of reaction buffer in the presence of 4 mM ATP. The mixture was incubated at 37°C for 2 h with shaking, boiled in SDS sample buffer, and fractionated by SDS-PAGE. The gel was dried, and p53 was detected by autoradiography.
GST pulldown assay.
MDM2 C-terminal fragments were produced by in vitro translation in rabbit reticulocyte lysate using the TNT system (Promega) in the presence of [35S]methionine. Bacterial lysate expressing glutathione S-transferase (GST) and GST-MDM2-294-491 were applied to glutathione-agarose beads according to the manufacturer's protocol (Pierce). The beads loaded with GST and GST-MDM2-294-491 were incubated with in vitro translated MDM2 fragments in buffer containing 20 mM HEPES (pH 7.4), 150 mM NaCl, 0.1% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 10% glycerol, and 0.5 mg of bovine serum albumin/ml at 4°C for 1 h. The beads were washed in RIPA buffer (50 mM Tris-Cl [pH 7.4], 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate), boiled in SDS sample buffer and fractionated by SDS-PAGE. The gel was dried and bound MDM2 was detected by autoradiography.
Protease sensitivity assay.
SJSA cells were treated with 10 Gy of IR for 2 h. The cells were lysed in lysis buffer, and the extract was preanalyzed for MDM2 level by Western blotting. Cell extract containing identical amount of MDM2 was mixed with MDM2-null murine embryonic fibroblast (MEF) lysate to prepare digestion substrates with identical total protein levels (20 μg) and identical MDM2 levels. The mixtures were incubated with trypsin (0.05 ng) for the indicated time points and analyzed by Western blotting with C terminal-specific antibody 4B11.
Chemical cross-linking.
H1299 cells were transfected with indicated plasmids for 32 h and lysed in lysis buffer (50 mM Tris-HCl [pH 8.0], 5 mM EDTA, 150 mM NaCl, 0.5% NP-40, 1 mM PMSF, 50 mM NaF). Cell lysate containing 20 μg of protein was incubated with glutaraldehyde for 20 min at 4°C or disuccinimidyl suberate (DSS; Pierce) for 30 min at 25°C at the indicated concentrations. The reaction was stopped by adding 2× Laemmli SDS sample buffer, and the samples were boiled and analyzed by Western blotting. For in vivo cross-linking, H1299 cells were transfected for 32 h. The cells were suspended in PBS and washed with ice-old PBS. DSS (1 to 2 mM) was added to the cells. The mixture was incubated for 30 min at 25°C and the reaction was quenched with 20 mM Tris-HCl (pH 7.5). The samples were boiled after the addition of Laemmli SDS sample buffer and analyzed by Western blotting.
BiFC assay.
The MDM2 RING domain (positions 362 to 491) was fused to YFP-1-154 (YN) and YFP-155-238 (YC). U2OS cells were cotransfected with YN and YC fusion plasmids using Lipofectamine reagents for 24 h. Cells were then cultured at 30°C for 10 h (to allow maturation of fluorophore) in the absence or presence of 50 nM neocarzinostatin (NCS) and fixed for 5 min with 4% formaldehyde. The cells were then stained using MDM2 antibody 4B11 and rhodamine-conjugated secondary antibody. Multiple photographs with red and green double exposures were taken and the fraction of MDM2-positive cells (∼300) that have visible YFP fluorescence was then counted in a blinded fashion for different samples.
RESULTS
DNA damage inhibits p53 ubiquitination.
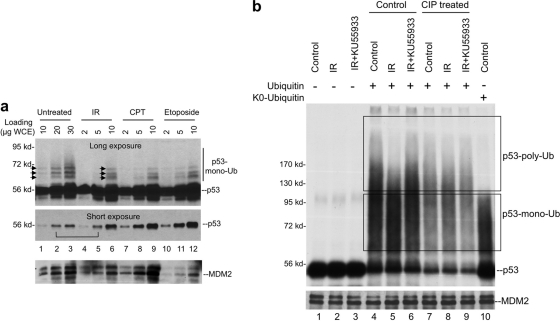
DNA damage treatments such as IR cause stabilization of p53. Whether p53 ubiquitination level is reduced after DNA damage is controversial. An early study showed that the total level of ubiquitinated p53 increases after IR, but it is not clear whether IR changes the fraction of p53 that is ubiquitinated (28). We analyzed the level of endogenous p53-ubiquitin conjugates by rapidly boiling cells in Laemmli SDS sample buffer containing iodoacetamide to preserve ubiquitinated p53. By titrating sample loading to obtain similar levels of unmodified p53, the fraction of ubiquitinated p53 can be compared. The results showed that IR significantly reduced the fraction of p53 that is ubiquitinated (Fig. 1a, compare lanes 2 and 5). As reported previously (28), the total amount of ubiquitinated p53 was higher in IR-treated cells (Fig. 1a, compare lanes 1 and 6), but it was likely due to a 5-fold increase in p53 level. IR treatment also caused subtle changes in the pattern of p53-ubiquitin bands, increasing the ratio of a low MW band relative to two higher-molecular-weight bands (Fig. 1a, arrows). Other DNA-damaging agents (camptothecin and etoposide) also induced similar changes. Therefore, DNA damage significantly reduces the fraction of p53 conjugated to ubiquitin and may also reduce the number of ubiquitin conjugated to each p53 molecule.
Fig. 1.
DNA damage inhibits p53 ubiquitination. (a) U2OS cells were treated with 10 Gy of IR for 4 h, 0.5 μM CPT for 8 h, or 50 μM etoposide for 8 h. Cells were suspended in Laemmli SDS-PAGE sample buffer with 10 mM iodoacetamide and immediately boiled. Whole-cell lysate was loaded at the indicated amounts and analyzed by p53 Western blotting. Long and short exposures were obtained to determine the levels of p53-ubiquitin conjugate and unmodified p53. (b) SJSA cells were treated with 10 Gy of IR for 2 h in the presence or absence of ATM inhibitor KU55933. MDM2 was immunoprecipitated by using 2A9 antibody. The MDM2-loaded beads were then used to capture in vitro-translated p53, followed by incubation with E1, E2, and ubiquitin. p53 ubiquitination was detected by autofluorography. Dephosphorylation treatment using CIP was performed on MDM2 beads prior to the capture of p53. p53 monoubiquitination products were identified by the use of K0-ubiquitin in the ubiquitination reaction.
The in vivo p53-ubiquitin level detected above reflects the combined effects of changes in MDM2 E3 ligase activity, MDM2-p53 binding, and p53 deubiquitination. To further determine whether DNA damage induces changes in MDM2 E3 ligase activity, MDM2 was immunopurified from cells and used to capture and ubiquitinate in vitro translated p53. This assay allows better control of several variables and specifically detects changes in ubiquitin chain formation. The result showed that MDM2 from untreated cells was highly active in promoting p53 ubiquitination in vitro. Use of lysine-free ubiquitin mutant (K0-ubiquitin) revealed that products smaller than 130 kDa could not be distinguished from p53 monoubiquitinated on multiple sites (Fig. 1b, lane 10). Therefore, only products of >130 kDa can be confidently defined as p53 with polyubiquitin chains (Fig. 1b, upper box). Based on this information, IR clearly reduced p53 polyubiquitination by MDM2 (Fig. 1b, compare lanes 4 and 5). The irradiated MDM2 had similar ability to initiate p53 ubiquitination as revealed by similar degree of substrate depletion (Fig. 1b, lanes 1 and 4 and lanes 2 and 5), but the products appeared to accumulate in the low-molecular-weight range (Fig. 1b, compare lanes 4 and 5).
Treatment of cells using ATM-specific inhibitor KU55933 abrogated the effect of IR, restoring the yield of high-molecular-weight p53-ubiquitin conjugates (Fig. 1b, compare lanes 5 and 6). Therefore, the suppression of p53 poly ubiquitination by IR in vitro involves ATM-mediated phosphorylation of MDM2. Furthermore, dephosphorylation of irradiated MDM2 with CIP in vitro changed its p53 ubiquitination activity to a profile similar to nonirradiated MDM2 (Fig. 1b, compare lanes 7 and 8 [note that the p53 input levels for lanes 7, 8, and 9 were lower due to CIP treatment, which is not shown on this gel]). These results indicate that the ability of MDM2 to promote p53 polyubiquitination is inhibited by ATM-mediated phosphorylation after DNA damage. The reduction of low-molecular-weight endogenous p53-ubiquitin conjugates in damaged cells suggests that p53 monoubiquitination is also inhibited by phosphorylation in vivo (Fig. 1a), although this effect was not recapitulated in vitro (Fig. 1b).
DNA damage promotes MDM2 phosphorylation near the RING domain.
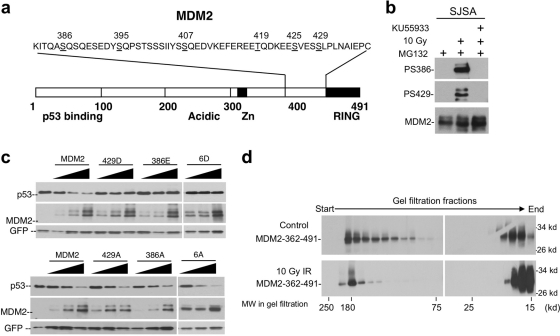
We recently reported the identification of multiple phosphorylation sites near the MDM2 RING domain (S386, S395, S407, T419, S425, and S429) (Fig. 2a) (9). Phosphorylation antibody analysis of selected sites suggest that they are rapidly phosphorylated by ATM after IR (Fig. 2b). Mutational analysis showed that they are critical for p53 stabilization. A mutant with alanine substitution of all six sites (MDM2-6A) is hyperactive in p53 ubiquitination and prevents p53 accumulation after IR. These published findings are confirmed in subsequent experiments (Fig. 2c). Additional experiments also revealed that of the six sites on the MDM2 C terminus, phosphorylation of four sites can each mediate significant response to DNA damage, demonstrating that the sites act in a redundant and additive fashion (data not shown). Our previous study also revealed that the phosphorylated MDM2 RING domain showed reduced ability to form high-molecular-weight oligomers in gel filtration chromatography (9). However, gel filtration did not detect RING dimers suggested by other studies to be important for MDM2 E3 ligase function (Fig. 2d), possibly due to limited resolution. To gain further insight on the mechanism of p53 stabilization, we decided to use several different assays to examine the effects of phosphorylation on MDM2 biochemical functions.
Fig. 2.
Multiple DNA damage-inducible phosphorylation sites regulate MDM2. (a) Diagram of MDM2 and relative positions of phosphorylation sites (underlined) targeted by ATM after DNA damage. (b) SJSA cells were treated with 10 Gy of IR, ATM inhibitor KU55933, and MG132 (to equalize MDM2 levels). At 4 h postirradiation, MDM2 was immunoprecipitated and probed by using phosphorylation-specific antibodies. The membrane was stripped and reprobed using MDM2 antibody. (c) Phosphorylation mimic mutations inactivate MDM2, whereas alanine substitutions increase MDM2 activity. H1299 cells were transfected with p53 and MDM2 mutants for 48 h, and p53 degradation was determined by Western blotting. MDM2-6A and MDM2-6D represent mutants with alanine or aspartic acid substitutions of all six phosphorylation sites (386, 395, 407, 419, 425, and 429). (d) SJSA cells were treated with 10 Gy of IR. MDM2 was immunoprecipitated with 2A9 after 2 h and cleaved with caspase 3. The C-terminal fragment (residues 362 to 491) containing the RING domain and phosphorylation sites was fractionated by gel filtration chromatography and detected by Western blotting using 4B11 antibody. IR treatment reduced the ratio of high-molecular-weight RING complexes.
Full-length MDM2 forms oligomers.
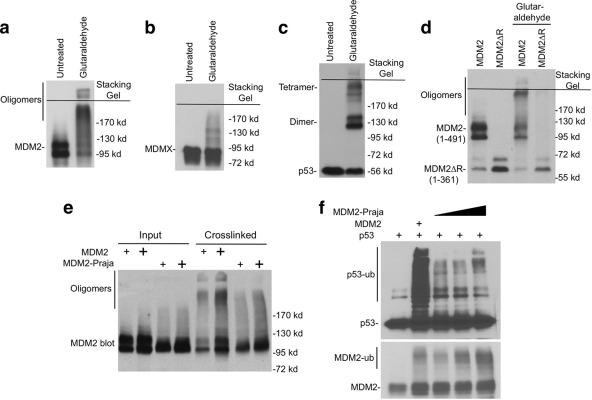
Recent evidence suggest that dimerization of E3 is often necessary for ubiquitin ligase function (2, 25, 43). MDM2 RING domain has been shown to form a homodimer (or oligomer) in a yeast two-hybrid assay (44). To test whether MDM2 expressed in human cells forms dimer or oligomer, we performed chemical cross-linking of MDM2 in cell extract using glutaraldehyde and DSS. The result showed that a significant fraction of full-length MDM2 was cross-linked into high-molecular-weight species (Fig. 3a), whereas MDMX oligomerization was not significant (Fig. 3b). Under the same condition, p53 was cross-linked into dimers and tetramers (Fig. 3c). The absence of distinct MDM2 dimer suggests that some of the MDM2 dimers further interact to form oligomeric complexes in vivo. MDM2 interaction with other cellular proteins such as ribosomal proteins may also lead to variation in the cross-linked products. Deletion of C-terminal region containing the RING domain (MDM2-1-361) significantly reduced oligomer formation, suggesting that oligomerization was mediated mainly by the RING domain (Fig. 3d). These results suggest that MDM2 exists as oligomeric complexes in vivo.
Fig. 3.
MDM2 forms oligomers. (a, b, and c) H1299 cells transiently transfected with MDM2, MDMX, and p53 were analyzed for oligomerization status by glutaraldehyde (0.02%) cross-linking and Western blotting. (d) H1299 cells were transfected with wild-type and mutant MDM2. The cell lysate was cross-linked using glutaraldehyde and analyzed by Western blotting with MDM2 antibody. (e) H1299 cells were transfected with MDM2 and MDM2-Praja, followed by glutaraldehyde cross-linking and Western blotting with MDM2 antibody. (f) H1299 cells were transfected with Myc-ubiquitin, p53, MDM2, and MDM2-Praja. p53 ubiquitination was determined by p53 immunoprecipitation, followed by anti-Myc Western blotting. MDM2 auto-ubiquitination was determined by Western blotting of whole-cell extract using MDM2 antibody.
To begin testing the functional significance of RING domain oligomerization, we compared MDM2 with the MDM2-Praja hybrid protein. In MDM2-Praja, the MDM2 RING (positions 437 to 491) was replaced with the RING of the Praja E3 ligase (13). MDM2-Praja failed to form high-molecular-weight products after chemical cross-linking, indicating that the Praja RING has low oligomerization efficiency (Fig. 3e). Compared to MDM2, MDM2-Praja was a poor E3 ligase for p53 (Fig. 3f). However, MDM2-Praja auto ubiquitination was as efficient as MDM2 (Fig. 3f), a finding consistent with a previous report that MDM2-Praja was able to degrade itself but not p53 (13). Therefore, RING domain oligomerization is not needed for auto-ubiquitination but appears to correlate with efficient trans-ubiquitination of p53.
Sequence adjacent to MDM2 RING domain inhibits oligomerization.
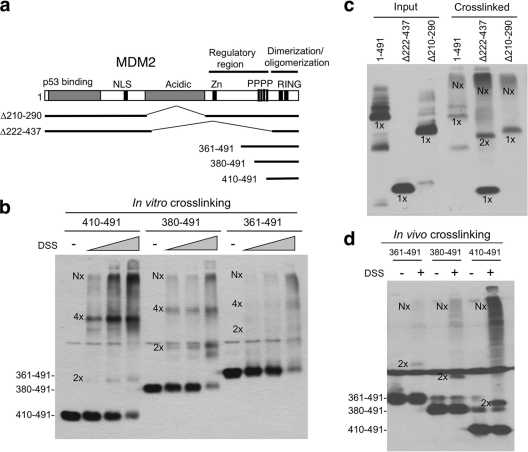
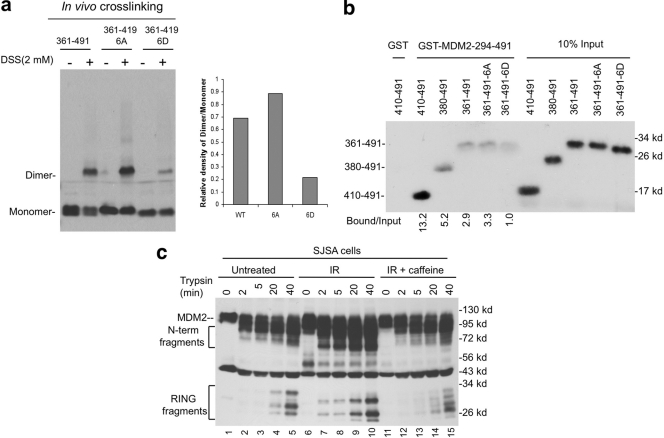
The MDM2 RING domain (positions 429 to 491) purified from bacteria has been shown to form homodimers in solution (20), but it is not clear whether the RING has the structure needed to form higher-order oligomers. We generated three progressively shortened RING domain constructs (361-491, 380-491, and 410-491) (Fig. 4a) that express proteins detectable by the 4B11 antibody. In vitro chemical cross-linking of these proteins in transfected H1299 cell extract showed that the shortest fragment (residues 410 to 491) produced significant amount of dimer, tetramer and high-molecular-weight oligomeric species, whereas the longest fragment (residues 361 to 491) produced much less dimer and tetramer intermediates (Fig. 4b). In vivo cross-linking by treating cells with DSS (cell permeable) before lysis also showed strong dimer and oligomer formation by the shortest RING fragment, although the tetramer band was not distinct (Fig. 4d). Therefore, both in vitro and in vivo cross-linking results suggest that sequence upstream of the RING domain has a role in suppressing dimerization and oligomerization.
Fig. 4.
An internal MDM2 region inhibits RING domain oligomerization. (a) Diagram of MDM2 deletion mutants. (b) Lysate of H1299 cells transfected with MDM2 C-terminal fragments were analyzed by DSS cross-linking in vitro (50, 100, and 200 nM) and Western blotting with 4B11 antibody. Dimers, tetramers, and oligomers are indicated by 2x, 4x, and Nx, respectively. (c) H1299 cells transfected with wild-type MDM2 and internal deletion mutants were analyzed by glutaraldehyde cross-linking and Western blotting with 4B11 antibody. (d) H1299 cells transfected with MDM2 RING fragments were treated with 1 mM DSS for 30 min before cell lysis and analyzed by Western blotting with 4B11.
Consistent with this notion, chemical cross-linking of MDM2 internal deletion mutant Δ222-437 (all sequence upstream of the RING removed) produced significant amounts of dimers and oligomers, whereas mutant Δ210-290 did not produce a discernible dimer intermediate (Fig. 4c). Overall, these results suggest that the MDM2 RING domain (residues 410 to 491) has intrinsic ability to form dimers and higher-order oligomers. Sequence upstream of the RING (residues 290 to 410) limits these interactions, possibly has a role in regulating MDM2 activity and preventing aggregate formation.
DNA damage inhibits MDM2 RING domain dimerization.
To test whether phosphorylation of the upstream sequence regulates MDM2 RING domain dimerization, the 361-491 fragments with 6A and 6D mutations were analyzed by in vivo cross-linking in H1299 cells. In vivo dimerization of 6D (phospho mimic) was much weaker than 6A (phosphorylation resistant) (Fig. 5a). As expected, transiently transfected wild type (residues 361 to 491 [wt361-491]) was partially phosphorylated (forming a double band) and showed intermediate dimerization efficiency compared to 6A and 6D. To rule out 6A causing structural effects other than blocking phosphorylation, the 361-491 fragments were also produced by in vitro translation and tested for binding to GST-MDM2-294-491 (the concentration of in vitro-translated RING was too low to detect homodimerization by cross-linking [data not shown]). In the absence of phosphorylation, wt361-491 migrated as a single band and showed the same pulldown efficiency as 361-491-6A, whereas 361-491-6D continued to show weaker binding (Fig. 5b). In the GST pulldown assay, MDM2 RING domain alone (residues 410 to 491) also showed much stronger affinity in binding to GST-MDM2-294-491 than fragments containing longer upstream sequences (residues 380 to 491 and 361 to 491), a finding consistent with the data in Fig. 4. These results suggest that the region located upstream of the RING domain suppresses RING dimerization and that the inhibitory effect is further enhanced by phosphorylation.
Fig. 5.
DNA damage inhibits MDM2 RING domain dimerization. (a) H1299 transfected with MDM2 wild-type, 6A, and 6D RING fragments were cross-linked with 2 mM DSS for 30 min before cell lysis and analyzed by Western blotting with 4B11. The dimerization efficiency was determined by densitometry and shown in the bar graph. (b) MDM2 RING domain fragments produced by in vitro translation were incubated with GST-MDM2-294-491 loaded beads in a pulldown assay. Binding efficiency was determined by autoradiography and densitometry. (c) SJSA cells were treated with 10 Gy of IR for 2 h. Cell extracts with identical total protein and MDM2 levels were incubated with trypsin for the indicated time points and analyzed by Western blotting with 4B11.
DNA damage alters RING domain protease sensitivity.
Formation of dimer or oligomeric complexes is expected to change the protease sensitivity of the MDM2 RING domain region. To test this prediction, MDM2 in control and irradiated cell extracts was incubated with identical amounts of trypsin, followed by detection of C-terminal fragments using the 4B11 antibody. The results showed that IR treatment increased the trypsin sensitivity of the MDM2 C terminus, producing higher yields of both N and C-terminal cleavage products (Fig. 5c, lanes 2 and 3 versus lanes 7 and 8). The different mobilities of the C-terminal fragments after irradiation (compare lanes 5 and 10) also suggest conformational change or steric hindrance leading to the use of different cleavage sites. These changes were reversed after the ATM/ATR activity was inhibited using caffeine. These results are consistent with the interpretation that phosphorylation changes the conformation of the RING domain, altering access to trypsin cleavage sites at or near the RING domain.
MDM2 RING domain interaction and regulation in vivo.
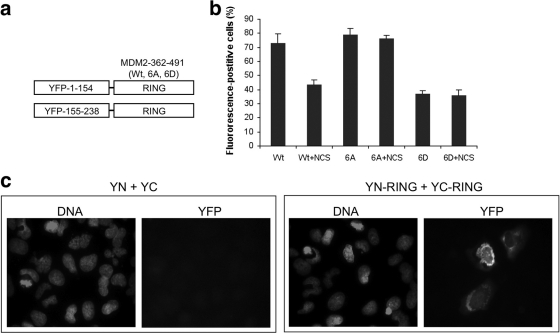
To directly visualize MDM2 RING domain interaction and regulation in vivo, a bimolecular fluorescence complementation assay (BiFC) was used (17). The C-terminal region (residues 362 to 491) of MDM2 was fused to the N (positions 1 to 154) and C (positions 155 to 238) terminal halves of yellow fluorescence protein (YFP) (Fig. 6 a). When coexpressed in cells, RING domain-mediated dimerization or oligomerization of two YFP fragments should reconstitute a fluorescent protein. As expected, cotransfection of YN-RING and YC-RING into U2OS cells produced strong fluorescence with diffused localization, whereas YN and YC cotransfection did not generate a signal (Fig. 6c). This result demonstrates significant intermolecular interaction mediated by the MDM2 RING domain in living cells.
Fig. 6.
DNA damage inhibits MDM2 dimerization in vivo. (a) The C-terminal region (residues 362 to 491) of wild-type MDM2, MDM2-6A, and MDM2-6D were fused to YFP N- and C-terminal fragments. (b) Each pair of YFP N- and C-terminal fusion proteins were cotransfected into U2OS cells, counterstained with 4B11, and analyzed for the ratio of YFP-positive to MDM2-positive cells. DNA damage was induced with NCS at 50 nM for 10 h. The data represent the averages of three experiments. (c) Representative photographs of YFP fluorescence signal in U2OS cells cotransfected with control YFP N/C fragments or YFP N/C fragments fused to the MDM2 RING domain.
Using this assay, MDM2 RING domain interaction in vivo was semiquantitatively analyzed after DNA damage by counting YFP-positive cells among cells stained positive by MDM2 RING domain antibody 4B11. Treatment of wild-type MDM2 RING fusion with DNA-damaging drug NCS caused ∼35% reduction in YFP-positive cells. Furthermore, MDM2-6D RING domain fusion produced only 50% YFP-positive cells compared to the wild type when expressed at similar levels (Fig. 6b). Both 6A and 6D fluorescence were not affected by DNA damage. Although this assay only produced a moderate phenotype from DNA damage (interaction of the two YFP moieties during chromophore maturation is likely to increase the stability of the RING complex), the results showed that in vivo interaction between MDM2 RING domain was inhibited by DNA damage and by phospho-mimic mutations. The result corroborates the inhibition of RING interaction by phospho-mimic mutations in cross-linking assay (Fig. 5a).
Assisted oligomerization of MDM2 stimulates p53 ubiquitination.
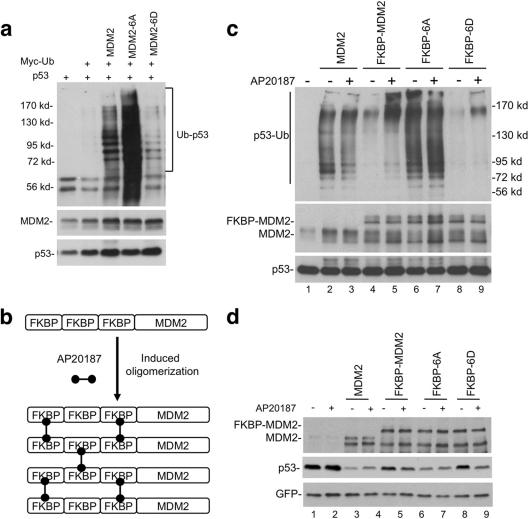
As we reported previously, MDM2-6D phospho-mimic mutant has significant deficiency in p53 degradation (Fig. 2c) and p53 ubiquitination (Fig. 7a), whereas MDM2-6A was more active than wild-type MDM2 due to resistance to basal phosphorylation. If MDM2 dimerization and oligomerization are important for ubiquitination of p53, artificially inducing MDM2 oligomerization should promote its activity. Therefore, we fused three tandem FKBP36V domains to the N terminus of MDM2. Treatment of cells with dimeric ligand AP20187 should promote dimerization and further oligomerization of MDM2 (Fig. 7b). The preferred option of fusing FKBP36V domains to MDM2 C terminus was ruled out due to a critical role of the extreme C terminus in E3 activity (45).
Fig. 7.
Forced oligomerization stimulates MDM2 activity. (a) p53 was cotransfected with MDM2 and Myc-ubiquitin into H1299 cells. Polyubiquitinated p53 was detected by p53 immunoprecipitation, followed by Myc Western blotting. (b) Three tandem copies of FKBP ligand binding domain were fused to the N terminus of MDM2 to facilitate induced oligomerization by treatment with dimeric ligand AP20187. (c) p53 was cotransfected with 3× FKBP-MDM2 and Myc-ubiquitin into H1299 cells. Cells were treated with 100 nM AP20187 for 4 h. Polyubiquitinated p53 was detected by p53 immunoprecipitation, followed by Myc Western blotting. (d) p53 was cotransfected with 3× FKBP-MDM2 fusion constructs into H1299 cells. Cells were treated with 100 nM AP20187 for 16 h. p53 degradation was detected by DO-1 Western blotting.
As expected, FKBP-MDM2 activity was significantly stimulated after treatment with the dimeric ligand, generating more high- and low-molecular-weight forms of p53-ubiquitin conjugates (Fig. 7c, compare lanes 4 and 5). Induced oligomerization of FKBP-MDM2-6D also enhanced its ability to induce p53 ubiquitination. FKBP-MDM2-6A was not stimulated by AP20187, probably because it already forms dimers and oligomers efficiently without the ligand (Fig. 7c). Consistent with the increase in p53 polyubiquitination, forced oligomerization also enhanced the ability of FKBP-MDM2 and FKBP-MDM2-6D to degrade p53 (Fig. 7d). In additional assays, AP20187 treatment did not cause significant difference in p53 binding to FKBP-MDM2 (data not shown). Overall, these results show that assisted oligomerization of MDM2 partially overcomes the inhibitory effect of phosphorylation and stimulates the ubiquitination of p53.
Stable MDM2-p53 binding does not substitute for MDM2 dimerization.
MDM2-p53 binding is critical for p53 ubiquitination and is also regulated by phosphorylation of p53 N-terminal sites. Stable MDM2-p53 binding is expected to also facilitate processive elongation of polyubiquitin chain on p53 by allowing repeated rounds of ubiquitin conjugation without MDM2 dissociation. Therefore, we tested whether stable MDM2-p53 binding eliminates the need for MDM2 dimerization.
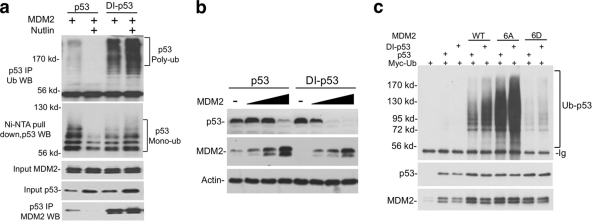
Using the information from our recent study (33), seven amino acid substitutions were introduced into p53 N terminus to convert its natural MDM2 binding site (17-ETFSDLWKLLPE-28, Kd = 160 nM) to an optimized high-affinity binding site (17-ETFEHWWSQLLS-28, Kd < 1 nM) (33). The resulting full-length DI-p53 showed increased binding to MDM2 (∼6-fold) in coimmunoprecipitation assay (Fig. 8 a, bottom panel) and, surprisingly, retained full transcriptional activity (data not shown). In an in vivo ubiquitination assay, DI-p53 ubiquitination by MDM2 produced more polyubiquitinated forms (Fig. 8a, top panel) but showed little increase in the yield of monoubiquitinated p53 (Fig. 8a, middle panel). Furthermore, inhibition of MDM2-p53 binding using Nutlin completely abrogated p53 polyubiquitination and significantly reduced monoubiquitination. DI-p53-MDM2 binding was resistant to disruption by Nutlin (Fig. 8a). As expected, DI-p53 was more efficiently degraded by MDM2 (Fig. 8b). However, high-affinity p53 binding only marginally enhanced ubiquitination by the phospho-mimic MDM2-6D (Fig. 8c). This result shows that MDM2-p53 binding is an important determinant of ubiquitination efficiency, but stable binding does not substitute for the unique function provided by MDM2 RING domain dimerization.
Fig. 8.
Stable MDM2-p53 binding does not substitute for MDM2 dimerization. (a) H1299 cells were transfected with DI-p53 containing high-affinity MDM2 binding site, His6-ubiquitin, Myc-ubiquitin, and MDM2. p53 ubiquitination was analyzed after Ni-NTA pulldown or p53 immunoprecipitation. MDM2 binding efficiency was determined by p53 immunoprecipitation and MDM2 Western blotting. (b) Wild-type p53 and DI-p53 were cotransfected with increasing doses of MDM2 into H1299 cells. p53 degradation was determined by Western blotting with Pab1801. (c) Wild-type p53 and DI-p53 were cotransfected with MDM2 mutants into H1299 cells. p53 ubiquitination was determined by immunoprecipitation with Pab1801 and anti-Myc Western blotting.
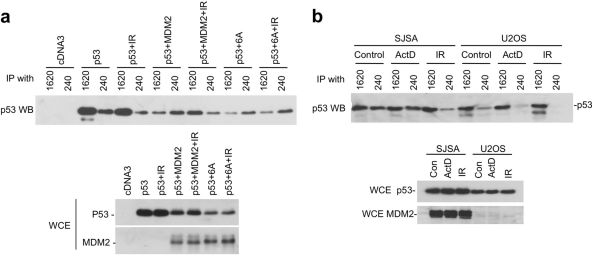
The p53 misfolding function of MDM2 is regulated by the ATM sites.
MDM2 binding to p53 induces a conformational change at the p53 DNA-binding domain (36). We recently found that the MDM2 acidic domain is responsible for the misfolding of p53 (10), providing a functional assay to examine the effect of C-terminal phosphorylation on the acidic domain. In the p53 conformation assay, cotransfection of MDM2 with wild-type p53 caused a switch of p53 antibody reactivity from Pab1620 positive (wild type specific) to Pab240 positive (mutant specific) if p53 degradation is blocked by MG132 (10, 36). We found that IR treatment prevented the conformational switch induced by MDM2 (Fig. 9a). However, IR failed to revert the conformational change induced by MDM2-6A (Fig. 9a). The MDM2-1-290 fragment containing only the acidic domain also efficiently induced p53 misfolding, but this activity was not reverted by IR (data not shown). As expected, the MDM2-6D mutant was less efficient in misfolding p53 compared to wild-type MDM2 and was not responsive to IR (data not shown).
Fig. 9.
p53 misfolding function of MDM2 is regulated by the ATM sites. (a) H1299 cells were transfected with p53 and MDM2 for 24 h and treated with 30 μM MG132 for 5 h to inhibit p53 degradation. Cells were also treated with 7 Gy of IR, as indicated, 4 h before harvesting. Cell lysates were immunoprecipitated with wild-type conformation-specific (Pab1620) or mutant conformation-specific (Pab240) antibodies. The precipitated p53 was detected by Western blotting with FL393 antibody. Whole-cell extracts (WCE) were analyzed for protein levels. (b) SJSA (amplified MDM2) and U2OS cells were treated with 30 μM MG132 for 5 h to inhibit p53 degradation. Cells were also treated with 7 Gy of IR as indicated 4 h before harvest or with 5 nM actinomycin D for 16 h. Cell lysates were immunoprecipitated with Pab1620 or Pab240. The precipitated p53 was detected by Western blotting with FL393. WCE were analyzed for protein levels.
Depending on endogenous MDM2 expression level, a small (U2OS) to moderate (SJSA) fraction of endogenous p53 displayed the mutant conformation after blocking proteasome-mediated degradation. IR treatment (but not ribosomal stress by actinomycin D) also efficiently reverted endogenous p53 to Pab1620-positive state (Fig. 9b). Therefore, the MDM2 C-terminal ATM sites are not needed for inducing p53 misfolding but are capable of regulating the function of the acidic domain after DNA damage.
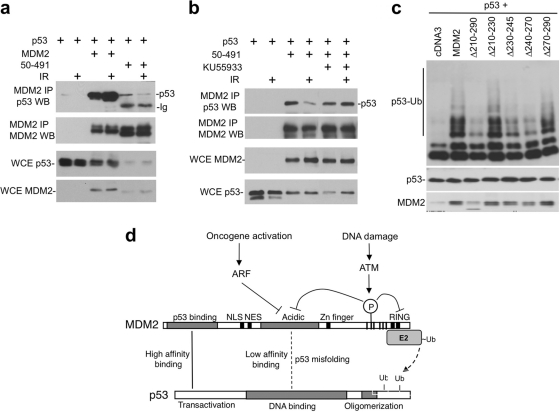
ATM regulates acidic domain-p53 core domain binding.
The MDM2 acidic domain (residues 200 to 300) binds to the p53 DNA-binding domain with low affinity (48). This interaction is thought to position the RING domain for optimal ubiquitination of p53 C-terminal lysines (46). We found that MDM2-50-491 (without the high-affinity N-terminal domain) binding to p53 was inhibited by IR (Fig. 10a). Furthermore, the ATM inhibitor KU55933 prevented IR inhibition of MDM2-50-491 binding to p53 (Fig. 10b), suggesting that phosphorylation of the ATM sites regulates the interaction between MDM2 acidic domain and p53 core domain. Previous studies showed that the MDM2 acidic domain is critical for ubiquitination of p53. Our results are consistent with this notion. Internal deletion of the entire acidic region (Δ210-290) strongly inhibited p53 ubiquitination (Fig. 10c). Smaller internal deletions suggested that residues 230 to 270 were critical (Fig. 10c). Interestingly, this region overlaps with the binding site for the ARF protein (residues 235 to 259) (42), which is an efficient inhibitor of p53 ubiquitination. Overall, these results showed that ATM phosphorylation also regulates the interaction between the MDM2 acidic domain and p53 core domain, which is necessary for p53 conformational change and ubiquitination.
Fig. 10.
ATM inhibits MDM2 acidic domain binding to p53 core. (a) H1299 cells were transfected with p53, MDM2, and MDM2-50-491 for 36 h. Cells were treated with 7 Gy of IR and harvested 4 h later. MDM2 was immunoprecipitated with 2A9 antibody, and the coprecipitated p53 was detected by FL393 Western blotting. (b) The ATM inhibitor KU55933 was used to treat cells during and after IR in an assay similar to that in panel a, showing that the inhibition of MDM2-50-491 and p53 binding by IR was mediated by ATM. (c) H1299 cells were transfected with His6-ubiquitin, p53, and MDM2 acidic domain internal deletion mutants. p53 ubiquitination was analyzed by Ni-NTA pulldown and p53 Western blotting. (d) Model summarizing the mechanism of p53 regulation by ATM. MDM2 bindings to p53 through N-terminal high-affinity interaction, which facilitates a weak second-site interaction between the acidic domain and core domain that activates or positions the RING for ubiquitin transfer to p53. MDM2 RING domain dimerization and oligomerization are also critical for p53 ubiquitination. Phosphorylation by ATM inhibits acidic domain-core binding and RING dimerization, thus blocking two critical steps in p53 ubiquitination. ARF inhibits MDM2 acidic domain function during oncogenic stress, possibly acting through a mechanism similar to phosphorylation.
DISCUSSION
The results described in this study suggest that DNA damage causes p53 stabilization by inducing phosphorylation of MDM2 near the RING domain and preventing the synthesis of polyubiquitin chain on p53. The ATM phosphorylation sites regulate the dimerization of the adjacent RING domain, and they also regulate the p53-binding and misfolding functions of the central acidic domain. Both domains are critical for efficient ubiquitination of p53. Their functional disruption by the same phosphorylation sites suggest the presence of cross talk and allosteric interactions between the domains (Fig. 10d).
Our previous study provided initial evidence that ATM-mediated phosphorylation regulates the homo-oligomerization of the MDM2 RING domain. The results described here further demonstrate that phosphorylation inhibits MDM2 RING domain dimerization and higher-order oligomerization. Furthermore, we provide direct evidence that the oligomerization state of MDM2 affects p53 ubiquitination efficiency. RING dimer formation appears to be a prerequisite for further oligomerization, since MDM2 fragments that dimerize poorly also form less high-molecular-weight oligomers. Tetrameric species were detected under some conditions, suggesting that higher order complexes were formed through dimerization of dimers. Nuclear magnetic resonance and X-ray crystallography studies also suggest that MDM2 RING domain forms dimer in vitro (20, 26). A recent study using recombinant MDM2 RING domain revealed the formation of high-molecular-weight complexes (34). Therefore, MDM2 RING domain alone has strong dimer/tetramer formation propensity and substantial oligomerization activity.
In certain dimeric RING domain E3 ligases (such as Brca1-Bard1), the sequence adjacent to the RING helps to stabilize dimer formation by providing an additional binding interface (4). Our results suggest that sequence upstream of the MDM2 RING domain may function by antagonizing the intrinsic dimerization activity of the RING. We found that fusing the MDM2 sequence from residues 361 to 440 to another monomeric protein does not lead to dimer formation (unpublished observations). However, deletion of this sequence increases MDM2 dimerization efficiency. Therefore, this region may normally limit the interaction between MDM2 RING domains through steric or allosteric effects. ATM-mediated phosphorylation may act by further enhancing the ability of this regulatory region in blocking MDM2 dimerization. Biochemical studies showed that purified recombinant MDM2 RING domain is prone to forming large protein complexes in vitro. Although this property may be beneficial for the polyubiquitination of p53, uncontrolled oligomerization will lead to nonproductive aggregation. It has been observed that the purified MDM2 RING domain (residues 429 to 491) has a strong tendency to form a nonspecific aggregate (20). The sequence adjacent to the RING may have a key role in maintaining an appropriate level of MDM2 oligomerization under physiological conditions.
Dimer formation is important for the activity of several RING domain E3 ligases (4, 49). Although the RING dimerization interface is distant from the E2 binding site, dimerization conceals a large surface area and may induce long-range changes that affect the E2 binding site. RING dimerization may be necessary for recruitment of charged E2, catalyzing ubiquitin transfer from E2 to substrate lysines, or rapid exchange of spent and charged E2. Furthermore, dimeric E3 may allow alternate reloading of charged E2 and more efficient synthesis of poly ubiquitin chains. Since phosphorylation of MDM2 C-terminal sites inhibits RING dimerization, it provides an effective mechanism for regulating p53 ubiquitination and degradation. Our results also suggest that MDM2 RING domain forms higher-order oligomers beyond dimers in vivo, and phosphorylation of MDM2 strongly suppresses oligomerization. Therefore, MDM2 may function most efficiently when multiple dimers assemble into oligomers, recruiting multiple E2 to p53 and enhancing the processivity of chain elongation. As expected from this model, induced oligomerization of MDM2 using the FKBP domain increases the yield of high-molecular-weight p53-ubiquitin conjugates.
In addition to regulating RING oligomerization, our results showed that the ATM sites also regulate the MDM2 acidic domain, revealing a second mechanism of p53 stabilization. The function of the MDM2 acidic domain in p53 ubiquitination is well established, although the mechanism remains speculative (19, 31). An acidic domain peptide has been shown to bind to the DNA-binding surface of p53 through charge interactions (48). Phosphorylation of the acidic domain by GSK3 stimulates ubiquitination of p53 (22, 23). We showed that the acidic domain also causes conformational changes in the p53 core and that the MDM2-p53 complex does not bind DNA (10). The MDM2 acidic domain-p53 core interaction may serve to position the C-terminal RING for efficient ubiquitination of p53 C-terminal lysines or expose the target lysines in the p53 core domain. It is also possible that acidic domain-p53 core binding is needed to allosterically activate the RING for ubiquitin transfer to p53. Phosphorylation of the MDM2 C terminus abrogates the p53 binding and misfolding function of the acidic domain, thus blocking a critical step in the ubiquitination of p53.
Although there is currently no structural information for the entire MDM2 molecule, our results provide indirect evidence that the acidic domain, ATM sites, and C-terminal RING domain are allosterically coupled. Phosphorylation of the ATM sites alters the conformation and function of both domains (Fig. 10d). The amino acid sequence upstream of the MDM2 RING domain is predicted to be intrinsically unstructured. This feature enables a peptide region to adopt different conformations upon interaction with other structures (possibly the adjacent RING domain and acidic domain). Phosphorylation of ATM sites in this region should alter the conformational flexibility and charge characteristics, providing a basis for allosteric regulation.
Our results also add to the growing evidence that different domains of MDM2 are functionally and allosterically connected. In such a model, ligand binding or modification of one region allosterically alters the structure and function of other domains. It has been suggested that p53 binding to MDM2 N terminus stimulates the acidic domain binding to p53 core. This second-site interaction is critical for p53 ubiquitination (46). Our results suggest that the acidic domain and C-terminal RING domain are also conformationally linked through the intervening region containing the ATM sites. These findings predict that the N-terminal p53 binding site and C-terminal RING domain may communicate directly or indirectly. In fact, a point mutation in the RING domain has been shown to change the conformation of the acidic domain and increase p53 binding by the N-terminal domain (47). There is also biochemical evidence that the RING domain of MDM2 binds to the acidic domain (11), suggesting the presence of an intramolecular interaction. Given this information, it is not entirely surprising that the C-terminal phosphorylation sites regulate multiple MDM2 domains.
The ARF tumor suppressor induces p53 stabilization through ATM-independent mechanisms. A major ARF binding site is located between residues 235 and 259 in the MDM2 acidic domain. Our results showed that the structural integrity of this region is critical for p53 ubiquitination. We recently also showed that ARF inhibits the ability of MDM2 acidic domain to induce p53 misfolding (10). Therefore, ATM phosphorylation of MDM2 and ARF binding appear to exert similar effects on the acidic domain function (Fig. 10d). These results suggest that different signaling pathways ultimately target the same MDM2 functional domains and use similar mechanisms to inhibit p53 ubiquitination.
ACKNOWLEDGMENTS
We thank ARIAD for providing the induced dimerization system, the Moffitt Molecular Biology Core for DNA sequence analyses, Hong-Gang Wang for providing BiFC vectors, and Alvaro Monteiro for providing GST-BRCA1.
This study was supported in part by grants from the National Institutes of Health to J.C. (CA141244 and CA109636).
Footnotes
Published ahead of print on 10 October 2011.
REFERENCES
- 1. Banin S., et al. 1998. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281:1674–1677 [DOI] [PubMed] [Google Scholar]
- 2. Barbash O., et al. 2008. Mutations in Fbx4 inhibit dimerization of the SCF(Fbx4) ligase and contribute to cyclin D1 overexpression in human cancer. Cancer Cell 14:68–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blattner C., Hay T., Meek D. W., Lane D. P. 2002. Hypophosphorylation of Mdm2 augments p53 stability. Mol. Cell. Biol. 22:6170–6182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brzovic P. S., Rajagopal P., Hoyt D. W., King M. C., Klevit R. E. 2001. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat. Struct. Biol. 8:833–837 [DOI] [PubMed] [Google Scholar]
- 5. Chao C., et al. 2003. Cell type- and promoter-specific roles of Ser18 phosphorylation in regulating p53 responses. J. Biol. Chem. 278:41028–41033 [DOI] [PubMed] [Google Scholar]
- 6. Chao C., Herr D., Chun J., Xu Y. 2006. Ser18 and 23 phosphorylation is required for p53-dependent apoptosis and tumor suppression. EMBO J. 25:2615–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chehab N. H., Malikzay A., Appel M., Halazonetis T. D. 2000. Chk2/hCds1 functions as a DNA damage checkpoint in G1 by stabilizing p53. Genes Dev. 14:278–288 [PMC free article] [PubMed] [Google Scholar]
- 8. Chen J., Marechal V., Levine A. J. 1993. Mapping of the p53 and mdm-2 interaction domains. Mol. Cell. Biol. 13:4107–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng Q., Chen L., Li Z., Lane W. S., Chen J. 2009. ATM activates p53 by regulating MDM2 oligomerization and E3 processivity. EMBO J. 28:3857–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cross B., et al. 2011. Inhibition of p53 DNA binding function by the MDM2 protein acidic domain. J. Biol. Chem. 286:16018–16029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dang J., et al. 2002. The RING domain of Mdm2 can inhibit cell proliferation. Cancer Res. 62:1222–1230 [PubMed] [Google Scholar]
- 12. Dornan D., et al. 2004. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 429:86–92 [DOI] [PubMed] [Google Scholar]
- 13. Fang S., Jensen J. P., Ludwig R. L., Vousden K. H., Weissman A. M. 2000. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 275:8945–8951 [DOI] [PubMed] [Google Scholar]
- 14. Harris S. L., Levine A. J. 2005. The p53 pathway: positive and negative feedback loops. Oncogene 24:2899–2908 [DOI] [PubMed] [Google Scholar]
- 15. Haupt Y., Maya R., Kazaz A., Oren M. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296–299 [DOI] [PubMed] [Google Scholar]
- 16. Honda R., Tanaka H., Yasuda H. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420:25–27 [DOI] [PubMed] [Google Scholar]
- 17. Hu C. D., Chinenov Y., Kerppola T. K. 2002. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9:789–798 [DOI] [PubMed] [Google Scholar]
- 18. Jones S. N., Roe A. E., Donehower L. A., Bradley A. 1995. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378:206–208 [DOI] [PubMed] [Google Scholar]
- 19. Kawai H., Wiederschain D., Yuan Z. M. 2003. Critical contribution of the MDM2 acidic domain to p53 ubiquitination. Mol. Cell. Biol. 23:4939–4947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kostic M., Matt T., Martinez-Yamout M. A., Dyson H. J., Wright P. E. 2006. Solution structure of the Hdm2 C2H2C4 RING, a domain critical for ubiquitination of p53. J. Mol. Biol. 363:433–450 [DOI] [PubMed] [Google Scholar]
- 21. Kubbutat M. H., Jones S. N., Vousden K. H. 1997. Regulation of p53 stability by Mdm2. Nature 387:299–303 [DOI] [PubMed] [Google Scholar]
- 22. Kulikov R., Boehme K. A., Blattner C. 2005. Glycogen synthase kinase 3-dependent phosphorylation of Mdm2 regulates p53 abundance. Mol. Cell. Biol. 25:7170–7180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kulikov R., Winter M., Blattner C. 2006. Binding of p53 to the central domain of Mdm2 is regulated by phosphorylation. J. Biol. Chem. 281:28575–28583 [DOI] [PubMed] [Google Scholar]
- 24. Leng R. P., et al. 2003. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 112:779–791 [DOI] [PubMed] [Google Scholar]
- 25. Li W., et al. 2009. Mechanistic insights into active site-associated polyubiquitination by the ubiquitin-conjugating enzyme Ube2g2. Proc. Natl. Acad. Sci. U. S. A. 106:3722–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Linke K., et al. 2008. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 15:841–848 [DOI] [PubMed] [Google Scholar]
- 27. MacPherson D., et al. 2004. Defective apoptosis and B-cell lymphomas in mice with p53 point mutation at Ser 23. EMBO J. 23:3689–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maki C. G., Howley P. M. 1997. Ubiquitination of p53 and p21 is differentially affected by ionizing and UV radiation. Mol. Cell. Biol. 17:355–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maya R., et al. 2001. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 15:1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mayo L. D., Turchi J. J., Berberich S. J. 1997. Mdm-2 phosphorylation by DNA-dependent protein kinase prevents interaction with p53. Cancer Res. 57:5013–5016 [PubMed] [Google Scholar]
- 31. Meulmeester E., et al. 2003. Critical role for a central part of Mdm2 in the ubiquitylation of p53. Mol. Cell. Biol. 23:4929–4938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Montes de Oca Luna R., Wagner D. S., Lozano G. 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378:203–206 [DOI] [PubMed] [Google Scholar]
- 33. Phan J., et al. 2010. Structure-based design of high-affinity peptides inhibiting the interaction of p53 with MDM2 and MDMX. J. Biol. Chem. 285:2174–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Poyurovsky M. V., et al. 2007. The Mdm2 RING domain C terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J. 26:90–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Prives C., Hall P. A. 1999. The p53 pathway. J. Pathol. 187:112–126 [DOI] [PubMed] [Google Scholar]
- 36. Sasaki M., Nie L., Maki C. G. 2007. MDM2 binding induces a conformational change in p53 that is opposed by heat-shock protein 90 and precedes p53 proteasomal degradation. J. Biol. Chem. 282:14626–14634 [DOI] [PubMed] [Google Scholar]
- 37. Sherr C. J. 2006. Divorcing ARF and p53: an unsettled case. Nat. Rev. Cancer 6:663–673 [DOI] [PubMed] [Google Scholar]
- 38. Shieh S. Y., Ahn J., Tamai K., Taya Y., Prives C. 2000. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 14:289–300 [PMC free article] [PubMed] [Google Scholar]
- 39. Shieh S. Y., Ikeda M., Taya Y., Prives C. 1997. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91:325–334 [DOI] [PubMed] [Google Scholar]
- 40. Shinozaki T., Nota A., Taya Y., Okamoto K. 2003. Functional role of Mdm2 phosphorylation by ATR in attenuation of p53 nuclear export. Oncogene 22:8870–8880 [DOI] [PubMed] [Google Scholar]
- 41. Sionov R. V., et al. 2001. c-Abl regulates p53 levels under normal and stress conditions by preventing its nuclear export and ubiquitination. Mol. Cell. Biol. 21:5869–5878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sivakolundu S. G., et al. 2008. Intrinsically unstructured domains of Arf and Hdm2 form bimolecular oligomeric structures in vitro and in vivo. J. Mol. Biol. 384:240–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tang X., et al. 2007. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell 129:1165–1176 [DOI] [PubMed] [Google Scholar]
- 44. Tanimura S., et al. 1999. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett. 447:5–9 [DOI] [PubMed] [Google Scholar]
- 45. Uldrijan S., Pannekoek W. J., Vousden K. H. 2007. An essential function of the extreme C terminus of MDM2 can be provided by MDMX. EMBO J. 26:102–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wallace M., Worrall E., Pettersson S., Hupp T. R., Ball K. L. 2006. Dual-site regulation of MDM2 E3-ubiquitin ligase activity. Mol. Cell 23:251–263 [DOI] [PubMed] [Google Scholar]
- 47. Wawrzynow B., et al. 2009. A function for the RING finger domain in the allosteric control of MDM2 conformation and activity. J. Biol. Chem. 284:11517–11530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yu G. W., et al. 2006. The central region of HDM2 provides a second binding site for p53. Proc. Natl. Acad. Sci. U. S. A. 103:1227–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang L., et al. 2011. The IDOL-UBE2D complex mediates sterol-dependent degradation of the LDL receptor. Genes Dev. 25:1262–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang Y., Lu H. 2009. Signaling to p53: ribosomal proteins find their way. Cancer Cell 16:369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]