Abstract
The transcriptional coactivator Cbp plays an important role in a wide range of cellular processes, including proliferation, differentiation, and apoptosis. Although studies have shown its requirement for hematopoietic stem cell (HSC) development, its role in adult HSC maintenance, as well as the cellular and molecular mechanisms underlying Cbp function, is not clear. Here, we demonstrate a gradual loss of phenotypic HSCs and differentiation defects following conditional ablation of Cbp during adult homeostasis. In addition, Cbp-deficient HSCs reconstituted hematopoiesis with lower efficiency than their wild-type counterparts, and this response was readily exhausted under replicative stress. This phenotype relates to an alteration in cellular fate decisions for HSCs, with Cbp loss leading to an increase in differentiation, quiescence, and apoptosis. Genome-wide analyses of Cbp occupancy and differential gene expression upon Cbp deletion identified HSC-specific genes regulated by Cbp, providing a molecular basis for the phenotype. Finally, Cbp binding significantly overlapped at genes combinatorially bound by 7 major hematopoietic transcriptional regulators, linking Cbp to a critical HSC transcriptional regulatory network. Our data demonstrate that Cbp plays a role in adult HSC homeostasis by maintaining the balance between different HSC fate decisions, and our findings identify a putative HSC-specific transcriptional network coordinated by Cbp.
INTRODUCTION
The cyclic AMP response element binding protein (CREB)-binding protein (CREBBP; here, CBP or Cbp) and its paralogue, p300, are large multidomain proteins with transcriptional coactivator properties that participate in numerous cellular processes during development and homeostasis (10). They modulate locus-specific transcription by a number of separate mechanisms, including their direct catalytic activity, which can acetylate both histone (2) and nonhistone (11) proteins, as well as through multiple protein-protein interactions with transcription factors, chromatin-remodeling complexes, and the basal transcriptional machinery (3). Furthermore, through interaction with multiple components within wider transcriptional networks, Cbp/p300 may further integrate and orchestrate whole transcriptional programs.
Precise temporal and spatial control of specific transcriptional programs is required for the development, maintenance, and differentiation of organ systems. Strong evidence has linked Cbp function with both normal and malignant hematopoiesis. Cbp and p300 have been demonstrated to have tumor suppressor roles in the development of hematological malignancies in mice (18, 23). Germ line mutations of CBP occur in the cancer predisposition syndrome Rubinstein-Taybi syndrome (35). CBP is also the direct target of chromosomal translocations associated with acute myeloid leukemia (AML), for which its C terminus is fused to the chromatin modifiers MLL (MLL1) and MOZ (MYST3) (7, 39). Previous studies have shown that Cbp and p300 functions are required for early embryonic development as well as hematopoietic stem cell (HSC) specification. Homozygous loss of either Cbp or p300 leads to embryonic lethality between embryonic day 9 (E9) and E11.5 due to defects in neuralation and other proliferative defects (23, 29, 30, 41, 47). Cbp−/− embryos exhibit pallor and demonstrate defects in primitive hematopoiesis (30). Furthermore, Cbp−/− embryonic stem (ES) cells contributed significantly to other tissues but failed to generate any hematopoietic changes in chimeric animals (34). Studies of Cbp and p300 heterozygous mice, where one allele of Cbp is already lost in developing HSCs and in other tissues, have suggested differential roles for the paralogues: for Cbp in HSC self-renewal and for p300 in hematopoietic differentiation during early development (34). Studies have shown that some transcription factors, such as Runx1, that are critical during early-stage hematopoiesis become dispensable in adult HSC maintenance (16). Although studies of adult mice have demonstrated the importance of Cbp and p300 for B- and T-cell development (19, 46), no studies have demonstrated the consequences of Cbp deficiency in normal adult HSC maintenance and function. A recent study also suggested a potential role for Cbp in the hematopoietic microenvironment (53). Despite all these studies, the cellular mechanisms as well as the hematopoietic transcriptional network by which Cbp regulates hematopoiesis and adult HSC function have not been addressed. Here, using functional studies of a Cbp conditional knockout mouse model together with integrated genomic analyses, we demonstrate that Cbp regulates HSC quiescence, apoptosis, and differentiation, and we identify putative hematopoietic transcriptional networks coordinated by Cbp.
MATERIALS AND METHODS
Mice.
Cbp conditional knockout mice (18) were bred with Mx-Cre transgenic mice (22), both on the C57BL/6 background. To induce Cre-mediated recombination, 6- to 10-week-old mice were administered 5 doses of poly(I)·poly(C) (300 μg per dose; Sigma) by intraperitoneal injection every other day over 10 days. Deletion efficiency was determined by PCR of genomic DNA extracted from various tissues. Peripheral blood (PB) samples were taken from the lateral saphenous vein and collected in EDTA-treated tubes (Sarstedt, Germany). Automated total and differential blood cells were counted using a Vet abc automated counter (Scil Animal Care, Viernheim, Germany). All mice were housed in a pathogen-free animal facility. Experiments were conducted under UK Home Office regulations.
Flow cytometry analysis.
Single-cell suspensions of bone marrow (BM) cells were prepared by flushing both femurs and tibiae and by obtaining spleen cells, with cell populations homogenized with Dulbecco's phosphate-buffered saline (DPBS; Invitrogen) supplemented with 2% fetal bovine serum (FBS; Sigma). Cells were filtered through a 70-μm nylon cell strainer (BD Biosciences). All BM, spleen, and PB samples were treated with red blood cell lysis buffer (5 PRIME, Germany) prior to the subsequent experiments. Cell numbers were determined using a CASY cell counter (Schärfe System GmbH, Germany). All staining was performed in DPBS with 0.1% bovine serum albumin and 1 mM EDTA. Antibodies used were as follows: affinity-purified rat anti-mouse lineage markers CD4, CD45R, LY-6C/G, TER119, CD19, and CD8a (all from Invitrogen) and CD3 (17A2; BD Biosciences). Other surface markers were as follows: from BD Biosciences, c-Kit (2B8; phycoerythrin [PE] conjugated), CD45.1 (A20; PE conjugated), CD45.2 (104; fluorescein isothiocyanate [FITC] conjugated), and CD4 (RM4-5; PE-Cy7 conjugated); from eBioscience, CD127 (interleukin-7 receptor α [IL-7Rα]; A7R34; biotin conjugated), CD135 (Flk-2/flt-3; A2F10; biotin conjugated), CD16/32 (93; biotin conjugated), CD34 (RAM34; FITC conjugated), Ly-6G (Gr1; PE-Cy7 conjugated), CD3 (PE-Cy7 conjugated), CD11b (M1/70; allophycocyanin [APC] conjugated), and streptavidin–PE-Cy7 conjugated secondary antibody; from Invitrogen, Ly-6A/E (Sca-1, D7; Alexa Fluor 647 conjugated), CD45R (APC conjugated), and Pacific blue anti-mouse IgG F(AB′)2 fragments. Flow cytometry analysis was performed on a CyAn ADP flow cytometer (Dako). All data were analyzed with FlowJo software (Tree Star, Inc.).
Isolation of the LSK population by flow sorting.
BM cells were harvested from femurs and tibiae from mice with the same genotype at 4 weeks after poly(I)·poly(C) treatment. BM cells were first stained with affinity-purified rat anti-mouse lineage markers (CD4, CD3, CD45R, LY-6C/G, TER119, CD19, and CD8a [listed above]). Lineage-positive cells were depleted using Dynabeads (sheep anti-rat IgG; Invitrogen) according to the manufacturer's instructions. Lineage-depleted BM cells were labeled with c-Kit PE-conjugated (BD Biosciences) and Sca-1 Alexa Fluor 647-conjugated (Invitrogen) antibodies. 7-Aminoactinomycin D (7-AAD) was added to exclude dead cells. The LSK population (Lin− c-Kithi Sca1hi) was sorted using a MoFlo cell sorter (Beckman Coulter).
Cell cycle and apoptosis analyses.
Mice were injected intraperitoneally with 1 mg of bromodeoxyuridine (BrdU; BD Biosciences). BM cells were collected 24 h later and stained with surface markers as stated above prior to BrdU detection, following the manufacturer's instructions (BrdU flow kit; BD Biosciences). For 5-fluorouracil (5-FU) treatment, 1 dose of 5-FU was injected intraperitoneally at a dose of 150 mg/kg of body weight. BM cells were collected 10 days later for fluorescence-activated cell sorting (FACS) analysis. The Ki-67 staining procedure was similar to that for BrdU detection, except a Ki-67–FITC antibody (BD Biosciences) was used without DNase treatment. Annexin V-FITC staining (Miltenyi Biotec) was performed according to the manufacturer's instructions. DNA content was labeled with 7-AAD (BD Biosciences).
Bone marrow transplantation assays.
Unfractionated BM cells from 6- to 10-week-old Cbp Mx or Cbp wild-type (wt) littermates (CD45.2) were collected as donor cells 4 weeks after poly(I)·poly(C) treatment. For competitive transplantations, these cells were mixed with competitor BM cells (CD45.1) at different ratios (1:1 and 3:1). A total of 1 × 106 cells were injected intravenously into lethally irradiated (550 rads; 2 doses) C57BL/6 recipients (CD45.1/45.2). For the serial transplantation experiment, 1 × 106 donor cells (CD45.2) were injected intravenously into lethally irradiated (550 rads; 2 doses) C57BL/6 recipients (CD45.1). At 16 weeks after the primary transplantation, 1 × 106 unfractionated BM cells from the primary recipients were pooled and injected into the secondary recipients. The whole procedure was repeated for the tertiary transplantation. PB chimerism was monitored by flow cytometry analysis monthly. Reconstitution of BM cells in the primary, secondary, and tertiary recipients was assessed at 16 weeks posttransplantation.
Homing assay.
Whole BM cells were collected from Cbp wt or Cbp Mx mice 4 weeks after poly(I)·poly(C) treatment. Single cells were labeled using a Vybrant carboxyfluorescein diacetate succinimidyl ester (CFDA SE [CFSE]) cell tracer kit (Invitrogen) for 15 min at 37°C. BM cells were washed in DPBS (Invitrogen) to remove excess dye. A total of 5 million labeled BM cells were transplanted into lethally irradiated recipients (CD45.1). BM cells from recipients were collected 24 h after transplantation. CFSE-positive stem and progenitor populations were assessed by flow cytometry.
Microarray analysis.
Total RNA was extracted from pooled sorted LSK cells by using TRIzol reagent (Invitrogen) and a standard protocol. Five nanograms of RNA was amplified using the Ovation RNA amplification system V2 (NuGEN) and labeled with biotin. The biotin-labeled cDNA samples were then hybridized onto the Illumina mouse WG-6 v2 bead chip array, following the manufacturer's instructions. Probe selection was performed using the R package Lumi (9). Probes that were successfully detected over background (P values of <0.01 in Lumi) in at least one sample were selected for subsequent analyses. Data were transformed using variance stabilization followed by normalization using quantile normalization (25). The comparisons in expression levels between samples were performed using the R package Limma. Probes differentially expressed (P values of <0.05) were selected for subsequent analyses.
GSEA.
Gene set enrichment analysis (GSEA) software and all curated gene sets (c2.All.v3.0.symbols.gmt) were obtained from the GSEA website (http://www.broadinstitute.org/gsea/index.jsp) (37). Gene enrichment analyses were performed using the default settings.
Chromatin immunoprecipitation and sequencing assay (ChIP-seq) and data analysis.
HPC-BM cells (33) were maintained in Iscove's modified Dulbecco's medium (IMDM) supplemented with 10% FBS, monothiolglycerol (MTG; Sigma), 100 ng/ml of stem cell factor, and 10 ng/ml of human IL-6. Cells were cross-linked with 1% formaldehyde. Immunoprecipitation was performed using rabbit polyclonal Cbp antibody (ab2832; Abcam) and rabbit IgG antibody (Abcam) as a negative control as described previously (8). Each sample was amplified and sequenced using an Illumina Genome Analyzer IIx. All sequencing reads for each sample were converted into density maps (.wig files). Peak calling was performed using the peak-finding program MACS (model-based analysis of ChIP-Seq) (52) as described in reference 45 with the following modified command line parameters for MACS: mfold, 16; t size, 35; bw, 100; P = 1e−5; gsize, 1,870,000,000; no lambda. All peaks were standardized to be 400 bp wide. Data were mapped to mouse reference genome (mm9) and displayed as UCSC genome browser custom tracks. For gene mapping and peak distribution analysis, genes were mapped to peaks by examining a 100-kb region around each peak and linking the nearest 3′ and 5′ genes to that peak. If a gene had a transcription start site within 1,000 bp from the peak, then only that gene was considered. Peak coordinates were checked for overlap with genes. If a peak overlapped 1,000 bp upstream of a gene, it was assigned as a promoter peak. The remaining peaks were either assigned as intragenic, if overlapping a gene by at least 1 bp, or intergenic, if not overlapping a gene or promoter region. The motif-finding program MEME (1) was used to find overrepresented motifs in three groups of peaks with the following parameters: -dna -nmotifs, 20; -evt, 0.01; -maxsize, 1,000,000. The middle 100 bp was taken for all peak regions, and peaks were excluded from analysis if they had more than 40% of repetitive sequence. The motif comparison tool TOMTOM (12) was then used to identify the motifs found by MEME.
Quantitative real-time PCR.
For gene expression analysis, RNA from flow-sorted LSK cells was extracted using TRIzol reagent (Invitrogen) following the standard protocol. cDNA was synthesized using a SuperScript VILO cDNA synthesis kit (Invitrogen) following the manufacturer's instructions. Quantitative reverse transcription-PCR (qRT-PCR) was carried out with SYBR green PCR master mix in the ABI Prism 7000 system (Applied Biosystems). The expression level of RNA was calculated using the standard curve method, normalized to the expression of β-actin. Results were from 2 individual PCRs from 3 individual samples. For ChIP-PCR, genomic DNA immunoprecipitated with the Cbp or rabbit IgG antibody was subjected to real-time PCR as described above. Input DNA was used to generate a standard curve. Enrichment of binding at particular genomic loci is presented as the percentage of the input DNA. All PCR results were from two individual experiments. Primer sequences are available upon request.
Statistical analysis.
An unpaired Student's t test was used for all analyses, within the GraphPad Prism4 software (GraphPad Software, Inc.).
Microarray data accession number.
Microarray and ChIP-seq data have been deposited into the NCBI Gene Expression Omnibus portal under the accession number GSE25274.
RESULTS
Deletion of Cbp alters hematopoietic lineage differentiation.
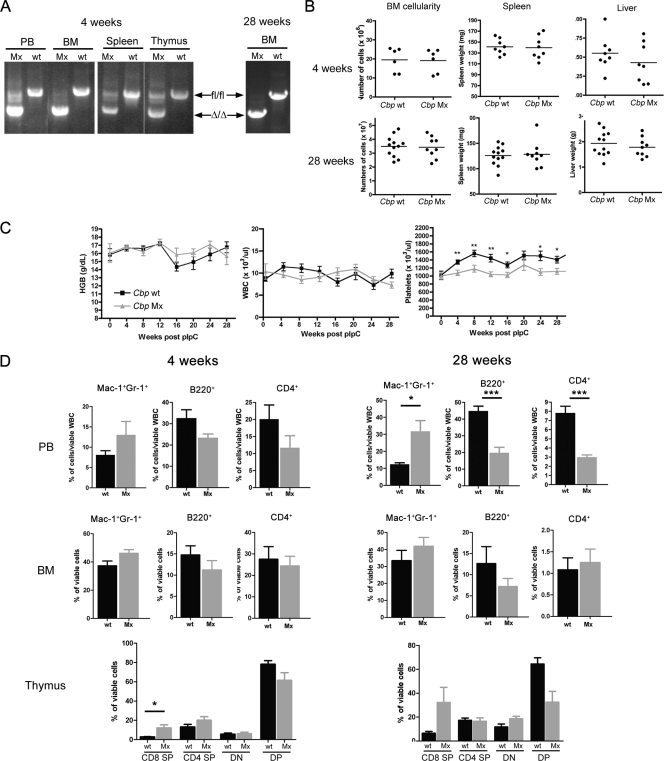
To study the role of Cbp in the adult hematopoietic system, we crossed Cbp conditional knockout mice (Cbp fl/fl) (18) with Mx-Cre transgenic mice (22), allowing Cre-mediated deletion of Cbp in the hematopoietic system upon injection of poly(I)·poly(C). Cre-mediated recombination at this genomic locus has been previously demonstrated to ablate Cbp, with no detectable Cbp protein expression (18, 19, 46). Efficient recombination was confirmed by PCR of genomic DNA at both 4 and 28 weeks post-poly(I)·poly(C) treatment (Fig. 1A). At both time points, no gross phenotypic abnormalities were observed in Cbp fl/fl; Mx-Cre+ mice (referred to here as Cbp Mx) and Cbp fl/fl; Mx-Cre− littermate control mice (referred to here as Cbp wt). BM cellularity and spleen and liver weights were similar between the two genotypes (Fig. 1B). In serial measures of PB, no differences were observed in total white blood cell counts or hemoglobin levels between genotypes. However, we observed a significant decrease in platelet numbers in Cbp Mx mice, which was evident from as early as 4 weeks post-poly(I)·poly(C) treatment (Fig. 1C). Furthermore, at 4 weeks after Cbp deletion, flow cytometry analysis of PB showed a decline in the frequencies of B-cell (B220+) and T-cell (CD4+) lineages in Cbp Mx mice, as has been previously reported with excision of Cbp in later lymphoid compartments (19, 46). This decrease was accompanied by a concomitant increase in the percentage of myeloid cells (Mac-1+ Gr-1+) (Fig. 1D). This differentiation bias toward the myeloid lineage became more marked over time, and by 28 weeks after poly(I)·poly(C) treatment the myeloid lineage was increased by around 3-fold in Cbp Mx mice, while both B- and T-cell lineages were decreased by 2.5-fold in comparison to Cbp wt mice (Fig. 1D). This pattern was recapitulated in BM cells at both time points, although it was less evident (Fig. 1D). It has previously been shown that deleting Cbp in thymocytes results in increased numbers of CD8 single-positive (SP) cells and decreased numbers of double-positive (DP) cells (18, 19). We observed a similar progressive phenotype at both time points. We and others have shown that poly(I)·poly(C)-induced Cre-mediated excision under the control of the Mx1 promoter is less efficient in thymocytes and spleen cells (Fig. 1A). Therefore, the initial phenotype we observed may have been diluted by the remaining Cbp fl/fl unexcised populations. Taken together, the above data suggest that deletion of Cbp in HSCs biases toward myeloid differentiation, resulting in thrombocytopenia, lymphopenia, and myeloid cell expansion over time.
Fig. 1.
Deletion of Cbp in adult bone marrow affects lineage differentiation. (A) Genotyping of genomic DNA extracted from different tissues collected from Cbp wt or Cbp Mx animals 4 and 28 weeks after poly(I)·poly(C) (pIpC) treatment demonstrated stable and nearly complete excision of Cbp in BM and PB. fl/fl and Δ/Δ represent Cbp floxed and null alleles, respectively. (B) Comparisons of BM cellularity (2 tibiae and 2 femurs) and spleen and liver weights between Cbp wt and Cbp Mx mice at 4 weeks and 28 weeks after poly(I)·poly(C) treatment. No differences were noted at either time point (n > 6 for each genotype). (C) PB counts of Cbp wt and Cbp Mx over 28 weeks after poly(I)·poly(C) treatment. Deletion of Cbp led to a significant decrease in platelet numbers, while white blood cell (WBC) and hemoglobin (HGB) counts were not affected. (D) FACS analyses of PB, BM, and thymus cells from both genotypes at 4 and 28 weeks after poly(I)·poly(C) treatment. The bias toward myeloid lineages at the expense of lymphoid lineages became more pronounced at the 28-week time point. Results are shown as means ± standard errors of the means (n > 15 for each genotype at 4 weeks; n = 8 for each genotype at 28 weeks). *, P < 0.05; **, P < 0.01; ***, P < 0.001. DN, CD4 and CD8 double-negative cells.
Loss of Cbp alters the composition of the hematopoietic stem and progenitor compartment.
The differentiation defects observed in Cbp Mx mice may reflect possible alterations in the hematopoietic stem and progenitor cell (HSPC) compartment. We therefore examined the effect of Cbp deletion on the composition of HSPCs. At 4 weeks post-poly(I)·poly(C) treatment, we observed a mild decrease in the frequency of the HSC-enriched LSK population (Lin− c-Kithi Sca1+) and a corresponding increased frequency of the myeloid progenitor compartment (Lin− IL-7R− c-Kithi Sca1−) (Fig. 2A and B). When the ratio of progenitors to LSK cells was compared (Fig. 2B), there was over a 2-fold relative increase in the production of progenitors in Cbp Mx compared to wt mice. However, there was no significant difference in composition within the heterogeneous LSK compartment, as determined by the frequencies of the long-term HSC (LT-HSC; L− K+ S+ CD34− Flt3−), short-term HSC (ST-HSC; L− K+ S+ CD34+ Flt−), and lymphoid-primed multipotent progenitor compartments (LMPP; L− K+ S+ CD34+ Flt3+) between Cbp Mx and wt mice (Fig. 2B). Within the myeloid progenitor compartment, no differences were noted in the frequency of the common myeloid progenitor (CMP) population (L− K+ S− CD34+ CD16/32int) between the two genotypes. However, a small increase in the granulocyte-monocyte progenitor (GMP; L− K+ S− CD34+ CD16/32hi) frequency, with a corresponding decrease in the megakaryocyte-erythroid progenitor cell (MEP; L− K+ S− CD34− CD16/32lo) frequency was noted in Cbp Mx mice. Although nonsignificant, this difference may have contributed to the thrombocytopenia and myeloid lineage expansion observed in the PB over time. The common lymphoid progenitor (CLP; Lin− IL-7R+ c-Kitint Sca1int) frequencies between the two genotypes were also similar.
Fig. 2.
Modest alterations in the BM stem and progenitor compartments occur following Cbp deletion. (A) Representative FACS plots of BM compartments of Cbp wt and Cbp Mx mice 4 weeks after poly(I)·poly(C) treatment. (B) Graphs showing frequencies of LSK and progenitors and their subpopulations 4 weeks after poly(I)·poly(C)-induced Cbp excision. The ratio of myeloid progenitor frequency to LSK frequency is shown (with the ratio for Cbp wt mice normalized to 1). Results shown are means ± standard errors of the means (SEM; n = 6). (C) Cbp-deficient LSK cells do not mobilize to the spleen. Representative FACS plots show the LSK and progenitor compartments (gates on Lin−) in spleens from Cbp wt and Cbp Mx mice 4 weeks after poly(I)·poly(C) treatment. Data are shown as means ± SEM (n = 4). (D) Frequencies of BM subpopulations analyzed at 28 weeks post-poly(I)·poly(C) treatment. Results are means ± SEM (n = 5). *, P < 0.05.
The changes in the HSPC compartment in Cbp Mx mice were progressive, and at 28 weeks post-poly(I)·poly(C) treatment, the frequency of LSK in Cbp Mx mice was significantly reduced in comparison to Cbp wt mice (Fig. 2D). This reduction in LSK compartment size was not due to increased mobilization or redistribution of stem and progenitor cells to the spleen or PB (Fig. 2C and data not shown). Although the size of the Cbp Mx LSK population was reduced, the ratio of progenitor to LSK in Cbp Mx mice remained stable and was still greater than twice that of Cbp wt mice (Fig. 2D). This suggests that a bias directing HSC differentiation over self-renewal is maintained over time following Cbp deletion, which may, in turn, contribute to the gradual reduction of phenotypic HSCs.
Cbp-deficient HSCs are capable of multilineage bone marrow reconstitution but engraft with decreased efficiency.
We next investigated if loss of Cbp also affected normal HSC function. We first assessed the long-term multilineage bone marrow reconstitution ability of Cbp-deficient BM cells in a noncompetitive transplantation experiment. One million unfractionated BM cells from either Cbp Mx or Cbp wt mice (CD45.2) were transplanted into lethally irradiated wild-type recipients (congenic CD45.1). Donor contribution to recipient PB reconstitution was monitored by flow cytometry at four weekly intervals for 16 weeks after transplantation. As shown in Fig. 3A, the degree of PB chimerism was similar between mice transplanted with Cbp Mx and Cbp wt BM cells. In addition, the frequencies of donor-derived B- and T-cell lineages in the PB of recipients transplanted with Cbp Mx cells were reduced. This reduction in the lymphoid lineage resembled the phenotype observed after deleting Cbp during homeostasis, suggesting that the differentiation defects caused by Cbp deficiency are predominantly cell autonomous (Fig. 3B). In keeping with the normal reconstitution ability of Cbp Mx cells in primary transplantation, no significant differences were observed in the homing abilities of progenitor or LSK cells between Cbp wt and Cbp Mx cells (Fig. 3C).
Fig. 3.
Cbp-deficient BM cells are capable of BM reconstitution with reduced efficiency. (A) Noncompetitive transplantation. Representative FACS plots of PB cells from recipients transplanted with either Cbp wt or Cbp Mx cells (left panel). The right panel shows the contributions of Cbp wt and Mx cells to the recipient PB cells over 16 weeks following transplantation. Results are means ± standard errors of the means (SEM; n = 10 for each genotype). (B) Graph demonstrating the percentages of donor-derived B-cell (B220+), T-cell (CD3+), and granulocytic (Mac-1+ Gr-1+) lineages in the PB of recipients at 16 weeks after the noncompetitive transplantation shown in panel A. (C) Cbp-deficient BM cells show no obvious defect in homing. Representative FACS plots are shown for BM cells from recipients transplanted with either Cbp wt or Cbp Mx BM cells that were labeled with CFSE prior to transplantation. The graphs on the right show percentages of total CFSE+, CFSE+ progenitor, and CFSE+ LSK cells of two genotypes detected in the recipient BM cells 24 h after transplantation. Data are means ± SEM (n = 6 for Cbp wt; n = 8 for Cbp Mx). (D) Competitive transplantation. Representative FACS analysis of PB cells from recipients transplanted with Cbp wt and Mx (test) and competitor cells at a 3:1 ratio (left panel). Graphs show the contributions of Cbp wt and Mx cells to the recipient PB reconstitution over a 22-week period following transplantation at a 1:1 ratio (middle panel) and 3:1 ratio (right panel). Results are means ± SEM (n = 5 for each genotype). *, P < 0.05; **, P < 0.01. (E) Deletion of Cbp in the BM niche does not significantly affect the reconstitution ability of wt donor BM cells. The graph shows the contributions of wt donor cells (45.1) to the PB cells of Cbp wt or Cbp Mx recipients over 20 weeks after transplantation (left panel). The right panel shows the percentages of different lineages derived from donor cells in the PB of these recipients at 20 weeks posttransplantation. Results are means ± SEM (n = 5 for each genotype).
We next examined the relative BM reconstitution efficiency of Cbp-deficient BM cells in a competitive setting. Cbp Mx or wt whole BM cells (test; CD45.2) were mixed with normal wt competitor BM cells (competitor; CD45.1) at a ratio of 1:1 or 3:1 (test:competitor ratio) and subsequently transplanted into recipient mice (CD45.1/45.2). Analyses of PB chimerism in recipients showed a reduced contribution from Cbp Mx cells, in comparison with Cbp wt cells, at both ratios (Fig. 3D). Taken together, these data demonstrate that Cbp-deficient BM cells are able to engraft and reconstitute lethally irradiated recipient mice; however, they do so with a reduced efficiency.
It has been reported that Cre expression under the Mx1 promoter is also induced in the HSC microenvironment (51). Moreover, it was shown recently that the Cbp+/− microenvironment fails to maintain normal HSC function (53). To examine if deletion of Cbp in the HSC microenvironment during homeostasis affects the reconstitution abilities of wt HSCs, we transplanted wt whole BM cells (CD45.1) into lethally irradiated Cbp wt or Cbp Mx recipients (CD45.2) 4 weeks after poly(I)·poly(C) treatment. No difference in the levels of PB chimerism or multilineage reconstitution could be detected over 20 weeks posttransplantation between recipients of the two genotypes (Fig. 3E). Although it is possible that excision under the control of the Mx1 promoter may not totally delete Cbp in all critical niche compartments, these data suggest that deletion of Cbp in the HSC niche during homeostasis has little effect on the engraftment and long-term multilineage reconstitution of donor cells.
Loss of Cbp leads to exhaustion of HSCs upon serial transplantation.
We further examined the effect of Cbp deficiency on long-term HSC self-renewal under the replicative stress of serial transplantation. Secondary transplantation was performed 16 weeks after the initial transplantation, whereby 106 total BM mononuclear cells were harvested from the primary recipients and transplanted into lethally irradiated secondary recipients. This process was iteratively repeated into tertiary recipients after another 16 weeks. As previously mentioned, Cbp Mx BM cells were able to engraft primary recipients to a similar level as Cbp wt cells (Fig. 3A). However, in the secondary recipients the contribution of Cbp Mx donor cells to total PB, as well as to individual lineages, was dramatically decreased compared to Cbp wt cells (Fig. 4A). In the tertiary transplant recipients, Cbp Mx cells were virtually undetectable, while Cbp wt cells still demonstrated a high level of engraftment.
Fig. 4.
Progressive exhaustion of Cbp-deficient HSCs under replicative stress. (A) The upper graphs show the contributions of Cbp wt and Mx cells to reconstitution of the recipient PB cells over 16 weeks following the secondary (2nd) and tertiary (3rd) transplantations. The Cbp Mx contribution was dramatically decreased in the secondary transplantation and extinguished in the tertiary. Contributions to different lineages of PB cells are also shown (lower panels). Results are means ± standard errors of the means (SEM; secondary transplantation, n = 10 for each genotype; tertiary transplantation, n = 6 for each genotype). **, P < 0.01; ***, P < 0.001. (B) FACS analyses of BM chimerism in the primary, secondary, and tertiary recipients. The upper panels show the contribution of donor-derived cells (CD45.2) to total BM cellularity. The middle panels show the contribution of CD45.2 cells to the LSK and progenitor compartments (HSPCs). The lower panel shows the frequencies of donor-derived LSK populations in the primary and secondary recipients of Cbp wt or Cbp Mx cells. Similar to the result for PB cells, the contribution of the CD45.2 Cbp Mx cells to BM cellularity and HSPC dramatically decreased with successive transplantations, indicating a severe defect in HSC self-renewal under replicative stress. All results are shown as means ± SEM (n ≥ 5 for each genotype). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Consistent with the results for PB, Cbp Mx cells were able to reconstitute total BM cells as well as the HSPCs in the primary recipients. In the secondary recipients, Cbp Mx donor cells showed decreased contributions to total BM cells and HSPCs (Fig. 4B, top and middle panels). Notably, we observed a significant reduction in the percentages of donor-derived LSK in both primary and secondary recipients of Cbp Mx compared to Cbp wt cells (Fig. 4B, lower panel). This recapitulates the reduced LSK frequency observed in Cbp Mx mice after poly(I)·poly(C) treatment (Fig. 2B and D), further indicating a cell-autonomous defect in stem cell self-renewal. In the tertiary recipients, virtually no evidence of Cbp Mx contribution to hematopoiesis was seen, while Cbp wt cells still significantly contributed to the total BM, LSK, and progenitor compartments (Fig. 4B). These data demonstrate that, under replicative stress, Cbp-deficient HSCs are exhausted, suggesting an inability to sufficiently maintain the HSC pool through impaired self-renewal.
Aberrant regulation of the cell cycle and increased apoptosis contribute to HSC exhaustion in Cbp-deficient mice.
To further investigate the cellular nature of our HSC exhaustion phenotype, we next assessed the cell cycle and apoptotic characteristics of Cbp-deficient HSPCs 4 weeks after poly(I)·poly(C) treatment. At 4 weeks after poly(I)·poly(C) administration, we detected a mild but statistically significant increase in apoptosis in Cbp Mx LSK cells, but not in the progenitor compartment, as assessed by staining with annexinV/7-AAD (Fig. 5A). In vivo BrdU incorporation showed no obvious differences in the proportions of either LSK or progenitors in S or G2/M phases in the absence of Cbp (Fig. 5B), suggesting that loss of Cbp has no effect on cycling of stem and progenitor cells. However, a noticeable increase in cells in G0/G1 in both LSK and progenitor compartments prompted us to further assess quiescent and cycling states, based on the intracellular expression of ki-67 and 7-AAD staining (Fig. 5C). Surprisingly, the LSK population from Cbp Mx mice demonstrated an increase in quiescent (G0) cells and a concomitant decrease in the G1 fraction. An increased proportion of quiescent cells was also demonstrated in the progenitor compartment of Cbp Mx mice. In consonance with the BrdU analysis, no change in the proportion of HSPCs in S/G2/M was observed between Cbp Mx and Cbp wt animals (Fig. 5C), confirming that deletion of Cbp had no obvious effect on stem and progenitor cells that were able to enter the cell cycle. Taken together, the above data demonstrated that deletion of Cbp leads to an accumulation of stem and progenitor cells in the quiescent state and an increase in apoptosis within the LSK compartment during adult homeostasis. These cellular characteristics further contribute to the HSC exhaustion phenotype we describe.
Fig. 5.
Cbp-deficient LSK and progenitor cells demonstrated alterations of quiescence and apoptosis. (A) Representative FACS plots of annexin V/7-AAD staining on LSK and progenitor cells from Cbp wt and Cbp Mx mice. Percentages of apoptotic LSK and progenitor cells (annexin V+ 7-AAD−) from Cbp wt and Cbp Mx animals show increased apoptosis in LSK but not progenitors from Cbp Mx mice. Results shown are means ± standard errors of the means (SEM; n = 8 for Cbp wt; n = 9 for Cbp Mx). *, P < 0.05. (B) Both Cbp wt and Mx mice were injected with 1 mg of BrdU 24 h before analysis. Representative FACS plots show cell cycle distributions of LSK and progenitors in Cbp wt and Mx mice. DNA content was stained with 7-AAD. Graphs show the percentages of cells in the different cell cycle phases. The proportions of LSK and progenitor cells in S or G2/M were similar between Cbp wt and Mx mice, suggesting that deletion of Cbp does not affect the cycling stem and progenitor cells. Data are mean ± SEM (n = 4 for Cbp wt; n = 5 for Cbp Mx). (C) The cell cycle status of LSK and progenitor cells was determined by the combined expression of the proliferation marker ki67 and the DNA-staining dye 7-AAD. Representative FACS plots from both Cbp wt and Cbp Mx mice are shown. Graphs show the proportions of LSK and progenitor cells at different stages of the cell cycle. Both Cbp Mx LSK and progenitors demonstrated an increase in quiescent (G0) fractions. Results are shown as means ± SEM (n = 11 for Cbp wt; n = 14 for Cbp Mx). *, P < 0.05; **, P < 0.01. (D) Analysis of the LSK compartment under stress conditions. Cbp wt or Mx mice were treated with one dose of 5-FU, 4 weeks after poly(I)·poly(C) treatment. Graphs show the frequencies of LSK and its subpopulations 10 days after 5-FU treatment (left and middle panels). The cell cycle status of the LSK compartment was determined based on the expression of Ki-67 and staining with the DNA dye 7-AAD (right panel). Results are means ± SEM (N > 6). *, P < 0.05; ***, P < 0.001.
The Cbp-deficient HSC phenotype is exacerbated by in vivo 5-FU treatment.
To gain further insights into the impaired self-renewal of Cbp-deficient HSCs under replicative stress, we challenged Cbp wt and Cbp Mx mice with one dose of 5-FU, 4 weeks after Cbp deletion, and monitored the recovery of the HSPC compartments as well as the cell cycle characteristics of these compartments. 5-FU treatment eliminated any cycling hematopoietic cells and therefore induced HSCs into the cell cycle to replenish the hematopoietic system. Ten days after 5-FU treatment, the LSK frequency in Cbp Mx mice was significantly lower than in Cbp wt mice (Fig. 5D). We also observed a marked reduction in the frequency of LT-HSCs and ST-HSCs, with a concomitant increase in MPP frequency in Cbp Mx mice, changes not apparent during homeostasis (Fig. 2B). Although total progenitor frequency was not altered, there was a significant increase in the proportion of GMP and a concomitant significant decrease in the proportion of MEP when Cbp Mx mice were compared to Cbp wt mice (see Fig. S1 in our supplemental material posted at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html). These data show an accentuation of the homeostatic phenotype and further demonstrate preferential differentiation and granulocyte/monocyte production (Fig. 5D). In addition, when we examined the cell cycle status of LSK cells after 5-FU treatment, there was a significant decrease in S/G2/M cycling LSK cells. These data together further suggest that under replicative stress, Cbp-excised LSK cells undergo increased differentiation and reduced cell cycle progression, which together lead to exhaustion of HSCs.
Gene expression analysis identified candidate HSC pathways regulated by Cbp.
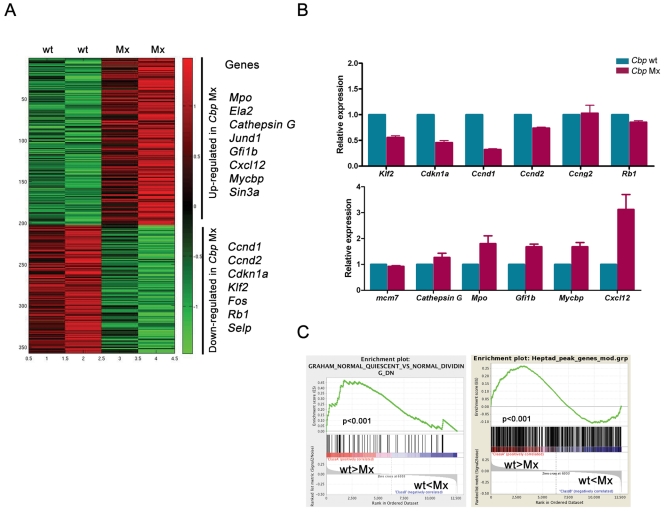
Cbp is a global transcriptional coactivator, interacting with numerous transcription factors, including important hematopoietic regulators like Gata-1, Gata-2, Runx2, and Rb1 (3). We therefore predicted that its loss would result in an alteration of multiple, rather than single, transcriptional programs. To identify alterations in global gene expression and the molecular mechanisms underlying the stem cell and cell cycle defects, we performed microarray analysis in the LSK populations of Cbp wt and Cbp Mx mice, a population enriched for hematopoietic stem cells. The analysis was performed at 4 weeks after Cbp deletion, the time point at which we had already documented changes in differentiation, apoptosis, and the cell cycle. Microarray analysis identified 341 unique transcripts (187 upregulated and 154 downregulated) differentially expressed (P < 0.05) between Cbp Mx and wt LSK for which the expression level varied by ±1.4-fold (log2, >±0.5) (Fig. 6A; see also our supplemental Table S1 at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html). The overall change in gene expression level was modest, with only around 30 genes showing greater-than-2-fold differences in expression level. However, consistent with our data, others have reported similar modest alterations in gene expression after deleting Cbp in mature T and B cells, suggesting a more subtle modulation of transcript levels upon Cbp loss (18, 46). Genes differentially expressed in LSK included genes implicated in the phenotypic abnormalities demonstrated in our Cbp Mx mice: abnormal cell cycle control [upregulation of Gfi1b and Cxcl12; downregulation of Ccnd1, Ccnd2, Cdkn1a(p21), Rb1, and Mcm7], and altered differentiation (upregulation of myeloid differentiation genes, including Mpo, Ela2, Cd16, and Ctsg; downregulation of lymphoid genes, including Rag1) (Fig. 6A). The differential expression of a subset of these genes was confirmed by qRT-PCR (Fig. 6B).
Fig. 6.
Microarray analysis identified candidate genes regulated by Cbp. (A) Heat map showing probes differentially expressed more than 1.4-fold (up- or downregulated) between Cbp wt and Cbp Mx LSK cells (P < 0.05). Selected genes, either up- or downregulated and proposed to contribute to our cellular phenotypes, are listed to the right of the heat map. (B) qRT-PCR analysis for selected genes confirmed differential expression levels for the majority of genes between Cbp wt and Mx LSK cells. Results are from 3 individual samples from each genotype and are shown as means ± standard errors of the means. Cbp wt samples were averaged and normalized to 1. (C) Representative GSEA, demonstrating that both Heptad-bound genes (right panel) and genes downregulated in quiescent versus dividing HSCs (left panel) are strongly enriched within genes downregulated in Cbp−/− LSK (nominal P value, <0.001).
In order to gain further insights into pathways and cellular processes altered by loss of Cbp, we next performed GSEA (http://www.broadinstitute.org/gsea) to compare our gene expression profile with all published curated gene sets (c2, version 3). Among 1,978 unselected gene sets analyzed, 22 gene sets were enriched in genes upregulated in Cbp−/− LSK cells, while 79 showed significant enrichment in genes downregulated in Cbp−/− LSK cells (nominal P value, <0.001). Interestingly, the latter group included gene sets with genes downregulated in quiescent versus dividing human CD34+ hematopoietic cells as well as genes related to the G1 cell cycle checkpoint (Fig. 6C; see also Table S2 at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html), in agreement with the cell cycle defect observed upon Cbp deletion. In addition, we recently identified 927 target genes bound by 7 major hematopoietic transcriptional regulators (Scl/Lyl1/Gata2/Runx1/Lmo2/Fli-1/Erg) in a hematopoietic stem and progenitor system (45). This Heptad target gene set is highly enriched for genes specifically expressed in HSCs. By GSEA, the Heptad gene set showed a strong correlation with genes downregulated in Cbp-deficient LSK cells (Fig. 6C; see also supplemental Table S3 at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html). These data suggest that loss of Cbp may alter HSC function through decreased expression of critical HSC-specific genes regulated by these core hematopoietic transcription factors.
Genome-wide analysis of Cbp occupancy identifies the HSC-specific transcriptional network coordinated by Cbp.
Cbp executes its function by interacting with a number of transcriptional regulators. The significant enrichment of Heptad target genes in genes downregulated after loss of Cbp suggests that Cbp may contribute to the regulatory network controlling normal hematopoiesis. We therefore performed ChIP-seq analysis in HPC-BM cells to identify the putative HSC transcriptional network coordinated by Cbp. The small available number of LSK cells precluded ChIP-seq analysis within this compartment. The immortalized HPC-BM cell line retains the HSC characteristics of self-renewal and multilineage potential in vivo without leukemic transformation (33), thus providing a surrogate HSPC transcriptional environment. Analyses of Cbp ChIP-seq data in comparison with control IgG data identified a total of 3,228 peaks associated with 2,395 genes, with 1,456 peaks (45.1%) located in the intergenic regions, 671 peaks (20.8%) in the promoter regions, and 1,101 peaks (34.1%) in the intragenic regions (Fig. 7A; see also supplemental Table S4 at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html). GSEA, using all genes bound by Cbp as the enquiry gene set, showed that these potential Cbp target genes were significantly enriched (P < 0.001) in genes downregulated, but not those upregulated, in Cbp-deficient cells, an observation in agreement with the well-recognized function of Cbp as a transcriptional coactivator (Fig. 7B; see also supplemental Table S5 at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html). Enrichment of binding at selected loci was confirmed by individual ChIP-qPCR (Fig. 7C). Among genes bound by Cbp were members of the Krüppel-like factor family (Klf2, Klf10, and Klf13), Runx1, Gfi1b, and Gata2 (Fig. 7A; see also supplemental Table S4 at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html). These genes have important functions in hematopoiesis, cell cycle regulation, and HSC self-renewal, further implicating Cbp function in the regulation of these cellular processes by coordinating critical transcription factors.
Fig. 7.
ChIP-seq analysis identified the potential hematopoietic transcription network coordinated by Cbp. (A) Representative ChIP-seq data, showing Cbp binding at the Klf2 and Runx1 loci. The scale of enrichment is shown to the left of the data. The relationships of a gene to the peaks and the genomic scale are shown below and above the individual tracks, respectively. (B) GSEA plot demonstrating that genes bound by Cbp are strongly correlated with genes downregulated in Cbp−/− LSK. (C) Real-time PCR verification of several genomic loci. The MyoD promoter region was used as a negative binding region. Results are from 2 individual PCR assays. Data are presented as means ± standard errors of the means. (D) Significant overlaps between genes bound by Cbp and the Heptad factors. The UCSC Genome Browser tracks demonstrate an example of Cbp binding at the Runx1 intragenic region, which is also bound by the Heptad factors (black box). (E) A de novo motif discovery identified Ets, Klf, and Creb1 motifs preferentially bound by Cbp.
Heptad target genes were strongly correlated with genes downregulated following Cbp deletion. We therefore investigated any common genes bound by both Cbp and the Heptad factors. When we compared 2,395 genes bound by Cbp with the 927 genes combinatorially bound by Heptad factors, there was a highly significant overlap of 324 genes (P = 6.428 e−085) (Fig. 7D; see also supplemental Table S6 at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html), implicating Cbp as a component of the hematopoietic transcriptional network regulated by the Heptad factors. Finally, de novo motif analysis of all peaks bound by Cbp demonstrated enrichment for Ets, Klf, and Creb1 consensus sequences (Fig. 7E). This suggests that the transcriptional activity of Ets (including the Heptad members Fli1 and Erg), Klf family members, and Creb1, which are all known to be involved in stem cell self-renewal, are coordinated by Cbp in the regulation of normal HSC function.
DISCUSSION
Hematopoietic stem cells have unique properties of long-term self-renewal and multilineage differentiation, which ensure the life-long production of all terminal hematopoietic lineages. To maintain the HSC pool and prevent HSC exhaustion, HSCs must precisely regulate often-antagonistic cellular processes, such as quiescence, proliferation, apoptosis, self-renewal, and differentiation, through tightly controlled transcriptional programs (32, 54). Based on our combination of cellular and molecular analyses, we suggest that the transcriptional coactivator Cbp regulates adult HSC homeostasis (Fig. 8) through the orchestration of cell fate decisions by the combinatorial actions of specific transcription factors.
Fig. 8.
Cbp regulates HSC fate decisions. The schematic diagram summarizes the role of Cbp in the maintenance of the HSC pool. The HSC pool is normally maintained through self-renewal, which reflects an ordered balance of proliferation and differentiation of HSCs with minimal cell death (left panel). In contrast, in Cbp−/− HSCs (right panel), these critical cell fate decisions are altered, leading to an increase in quiescence and apoptosis, to preferential differentiation, and ultimately HSC exhaustion through decreased self-renewal (red arrows). Based on our integrated genomic analyses, we propose that this occurs through the inability of Cbp−/− HSCs to properly coordinate the transcriptional programs necessary to regulate these cell fate decisions.
Following loss of Cbp in the adult HSC compartment, the size of the LSK compartment gradually decreased, with a concomitant increase in the myeloid progenitor compartment. This was accompanied by a fixed, over-2-fold increase in the progenitor-to-LSK ratio in Cbp Mx mice, indicating an altered balance between HSC self-renewal and myeloid differentiation with a bias toward the latter. In addition, gene expression analysis showed increased expression levels of myeloid differentiation-related transcripts, including Mpo, Ela2, and Cd16, and decreased lymphoid-related genes, including Rag1 in Cbp−/− LSK cells, thus providing molecular evidence for myeloid priming at the stem cell level following Cbp deletion. These data provide the first evidence that loss of Cbp in adult HSCs selectively increases myeloid over lymphoid lineage production. It is possible that Cbp deficiency in more-differentiated compartments also contributes to the differentiation defects observed, as has been demonstrated in other studies during lymphoid differentiation (19, 46).
Cbp has been shown to play a critical role during HSC specification and early embryonic development (23, 29, 30, 41, 47). Our phenotypic and functional data, in contrast, demonstrate a relatively milder defect upon loss of Cbp. Cbp-deficient HSCs were able to repopulate lethally irradiated recipients, albeit with reduced efficiency. These data are reminiscent of the relative subtle defects in HSC function after homeostatic loss of critical developmental regulators of hematopoiesis, such as Scl and Runx1 (16, 26). We speculate that loss of Cbp is compensated for by the cellular reserve of the adult HSC pool prior to deletion and a degree of functional redundancy with the paralogous coactivator p300 at the molecular level. Nevertheless, under the replicative stress of serial transplantation, and to a lesser extent following in vivo 5-FU treatment, Cbp-excised BM cells demonstrated an obvious inability to maintain the stem cell pool, indicating a marked and predominantly cell-autonomous defect in HSC self-renewal. The self-renewal potential is determined by the size of the stem cell pool, which is in turn influenced by the rate of loss of HSCs through apoptosis or differentiation and by the proliferative state of this pool. In addition to preferential differentiation of the HSC compartment, we demonstrated an increase in apoptosis and quiescence within the Cbp−/− LSK compartment, which cumulatively led to stem cell exhaustion (Fig. 8). These results provide the first detailed mechanistic data to account for the function of Cbp in adult HSC homeostasis and demonstrate that Cbp plays a role in the regulation of cellular fate decisions in adult HSCs.
Precise regulation of the cell cycle is critical for normal HSC function. Loss of function of many critical genes has been shown to cause a stem cell defect through an alteration of the balance between quiescence and cell cycle entry (14, 31). Previously, however, loss of stem cell function following deletion of a critical regulator has been almost uniformly accompanied by loss of quiescence and an increase in cell cycle progression. In contrast, our data demonstrated the opposite and corroborated a recent report whereby stabilization of Hif-1α through biallelic loss of the E3 ubiquitin ligase von Hippel-Lindau (VHL) protein also resulted in increased quiescence, a decrease in cell cycle entry, and a loss of HSC number and function (40). These data demonstrated that an increase in HSC quiescence may actually be detrimental to HSC function rather than protective, as has been seen following deletion of the Mef/Elf4 transcription factor (24), and the data further enforce the notion that exquisite control of the cell cycle is critical during adult HSC homeostasis. The mechanisms that regulate quiescence in stem cells are still poorly understood. However, a complex interplay between signals transduced from the stem cell niche through ligand-cell surface receptor modules, such as the Cxcl12-Cxcr4 (28) and Tpo-cMpl (49) axes, appear to control cell cycle entry and progression through intrinsic factors, such as Pten (48, 50), Gfi1/Gfi1b (15, 20), Cdkn1a/p21 (6), and Foxo transcription factors (27, 42). We identified altered expression levels of many genes implicated in cell cycle control following Cbp loss in the LSK population enriched for hematopoietic stem cells (Fig. 6; see also Supplemental Table S1 at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html), and it is likely that the cell cycle phenotype observed is a composite of these net transcriptional changes. For example, an alteration of the balance of cell cycle entry toward quiescence may be explained, at least in part, by an upregulation of Cxcl12 and Gfi1b, both positive regulators of quiescence, with Cxcl12 potentially forming an autocrine loop with Cxcr4 in HSCs. Although another positive regulator, Cdkn1a, was actually downregulated in this same analysis, these findings along with a loss of negative regulation of Foxo factors through Cbp-mediated acetylation (44) would be predicted to favor an increase of quiescence in HSCs.
In this study, we have provided the first genome-wide analysis of Cbp occupancy in the hematopoietic system and demonstrated that Cbp integrates into an HSC-specific transcriptional network (45). This “Heptad network” has been reconstructed from combinatorial genome-wide binding patterns in a stem cell model cell line and comprises seven transcription factors well known to regulate stem cell quiescence, self-renewal, and differentiation. Comparison of the combinatorial binding patterns of these Heptad factors revealed a highly significant overlap with genes downregulated upon Cbp loss in LSK cells and with our own Cbp genome-wide binding pattern. Furthermore, genome-wide motif analysis of Cbp-binding peaks revealed enrichment for consensus Ets binding sites (Fli1 and Erg are Ets family members). In addition, it is likely that Cbp coordinates additional transcriptional networks, as was evidenced by the motif enrichment for Klf and Creb1 consensus sequences at Cbp-bound regions. Krüppel-like factor family members have been implicated in ES cell self-renewal, and Klf4 is one of the four transcription factors required to reverse the highly differentiated state of somatic cells back to pluripotency (17, 38). Creb1/CREB1 has been demonstrated to be a critical regulator of both normal hematopoiesis and leukemogenesis (5, 36). We propose that Cbp may further regulate the function of these networks at a subset of loci through acetylation of histone and nonhistone proteins (including Heptad members, such as Runx1 and Gata2) (13, 21) and/or by providing a protein scaffold for nucleation of network members and other high-order transcriptional complex members.
Taken together, our data demonstrate that Cbp orchestrates multiple critical cell fate decisions in HSCs and suggest that this occurs through the ability of Cbp to integrate the transcriptional programs that mediate these decisions. In this respect, it behaves similarly to another epigenetic regulator, Dnmt1, to preserve HSC multipotency and self-renewal through control of critical transcriptional programs (4, 43), a finding that further underlines the role of epigenetic modulation as an important layer of control in HSC regulation.
ACKNOWLEDGMENTS
We thank Katrin Ottersbach and David Kent for helpful discussions.
Work in our lab is sponsored by CRUK, Medical Research Council (MRC; United Kingdom), Leukemia and Lymphoma Research (United Kingdom), The Wellcome Trust, the Leukemia and Lymphoma Society of America, and the NIHR Cambridge Biomedical Research Centre. B.H. is funded by an MRC (United Kingdom) senior clinical research fellowship, and M.A.D. is supported by a Wellcome-Beit intermediate clinical fellowship.
Footnotes
Published ahead of print on 17 October 2011.
REFERENCES
- 1. Bailey T. L., Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28–36 [PubMed] [Google Scholar]
- 2. Bannister A. J., Kouzarides T. 1996. The CBP activator is a histone acetyltransferase. Nature 384:641–643 [DOI] [PubMed] [Google Scholar]
- 3. Bedford D. C., Kasper L. H., Fukuyama T., Brindle P. K. 2010. Target gene context influences the transcriptional requirement for the KAT3 family of CBP and p300 histone acetyltransferases. Epigenetics 5:9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bröske A.-M., et al. 2009. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat. Genet. 41:1207–1215 [DOI] [PubMed] [Google Scholar]
- 5. Cheng J. C., et al. 2008. CREB is a critical regulator of normal hematopoiesis and leukemogenesis. Blood 111:1182–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheng T., et al. 2000. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science 287:1804–1808 [DOI] [PubMed] [Google Scholar]
- 7. Crowley J. A., Wang Y., Rapoport A. P., Ning Y. 2005. Detection of MOZ-CBP fusion in acute myeloid leukemia with 8;16 translocation. Leukemia 19:2344–2345 [DOI] [PubMed] [Google Scholar]
- 8. Dawson M. A., et al. 2009. JAK2 phosphorylates histone H3Y41 and excludes HP1α from chromatin. Nature 461:819–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Du P., Kibbe W. A., Lin S. M. 2008. lumi: a pipeline for processing Illumina microarray. Bioinformatics 24:1547–1548 [DOI] [PubMed] [Google Scholar]
- 10. Goodman R. H., Smolik S. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14:1553–1577 [PubMed] [Google Scholar]
- 11. Gu W., Roeder R. G. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595–606 [DOI] [PubMed] [Google Scholar]
- 12. Gupta S., Stamatoyannopoulos J. A., Bailey T. L., Noble W. S. 2007. Quantifying similarity between motifs. Genome Biol. 8:R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hayakawa F., et al. 2004. Functional regulation of GATA-2 by acetylation. J. Leukoc. Biol. 75:529–540 [DOI] [PubMed] [Google Scholar]
- 14. He S., Nakada D., Morrison S. J. 2009. Mechanisms of stem cell self-renewal. Annu. Rev. Cell Dev. Biol. 25:377–406 [DOI] [PubMed] [Google Scholar]
- 15. Hock H., et al. 2004. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature 431:1002–1007 [DOI] [PubMed] [Google Scholar]
- 16. Ichikawa M., et al. 2004. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat. Med. 10:299–304 [DOI] [PubMed] [Google Scholar]
- 17. Jiang J., et al. 2008. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat. Cell Biol. 10:353–360 [DOI] [PubMed] [Google Scholar]
- 18. Kang-Decker N., et al. 2004. Loss of CBP causes T cell lymphomagenesis in synergy with p27Kip1 insufficiency. Cancer Cell 5:177–189 [DOI] [PubMed] [Google Scholar]
- 19. Kasper L. H., et al. 2006. Conditional knockout mice reveal distinct functions for the global transcriptional coactivators CBP and p300 in T-cell development. Mol. Cell. Biol. 26:789–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Khandanpour C., et al. 2010. Evidence that growth factor independence 1b (Gfi1b) regulates dormancy and peripheral blood mobilization of hematopoietic stem cells. Blood 116:5149–5161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kitabayashi I., Aikawa Y., Nguyen L. A., Yokoyama A., Ohki M. 2001. Activation of AML1-mediated transcription by MOZ and inhibition by the MOZ-CBP fusion protein. EMBO J. 20:7184–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kuhn R., Schwenk F., Aguet M., Rajewsky K. 1995. Inducible gene targeting in mice. Science 269:1427. [DOI] [PubMed] [Google Scholar]
- 23. Kung A. L., et al. 2000. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 14:272–277 [PMC free article] [PubMed] [Google Scholar]
- 24. Lacorazza H. D., et al. 2006. The transcription factor MEF/ELF4 regulates the quiescence of primitive hematopoietic cells. Cancer Cell 9:175–187 [DOI] [PubMed] [Google Scholar]
- 25. Lin S. M., Du P., Huber W., Kibbe W. A. 2008. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 36:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mikkola H. K. A., et al. 2003. Haematopoietic stem cells retain long-term repopulating activity and multipotency in the absence of stem-cell leukaemia SCL/tal-1 gene. Nature 421:547–551 [DOI] [PubMed] [Google Scholar]
- 27. Miyamoto K., et al. 2007. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell 1:101–112 [DOI] [PubMed] [Google Scholar]
- 28. Nie Y., Han Y.-C., Zou Y.-R. 2008. CXCR4 is required for the quiescence of primitive hematopoietic cells. J. Exp. Med. 205:777–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oike Y., et al. 1999. Truncated CBP protein leads to classical Rubinstein-Taybi syndrome phenotypes in mice: implications for a dominant-negative mechanism. Hum. Mol. Genet. 8:387–396 [DOI] [PubMed] [Google Scholar]
- 30. Oike Y., et al. 1999. Mice homozygous for a truncated form of CREB-binding protein exhibit defects in hematopoiesis and vasculo-angiogenesis. Blood 93:2771–2779 [PubMed] [Google Scholar]
- 31. Orford K. W., Scadden D. T. 2008. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat. Rev. Genet. 9:115–128 [DOI] [PubMed] [Google Scholar]
- 32. Orkin S. H., Zon L. I. 2008. Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132:631–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pinto do Ó P., Richter K., Carlsson L. 2002. Hematopoietic progenitor/stem cells immortalized by Lhx2 generate functional hematopoietic cells in vivo. Blood 99:3939–3946 [DOI] [PubMed] [Google Scholar]
- 34. Rebel V. I., et al. 2002. Distinct roles for CREB-binding protein and p300 in hematopoietic stem cell self-renewal. Proc. Natl. Acad. Sci. U. S. A. 99:14789–14794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Roelfsema J. H., Peters D. J. M. 2007. Rubinstein-Taybi syndrome: clinical and molecular overview. Expert Rev. Mol. Med. 9:1–16 [DOI] [PubMed] [Google Scholar]
- 36. Shankar D. B., et al. 2005. The role of CREB as a proto-oncogene in hematopoiesis and in acute myeloid leukemia. Cancer Cell 7:351–362 [DOI] [PubMed] [Google Scholar]
- 37. Subramanian A., et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102:15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Takahashi K., Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676 [DOI] [PubMed] [Google Scholar]
- 39. Taki T., Sako M., Tsuchida M., Hayashi Y. 1997. The t(11;16)(q23;p13) translocation in myelodysplastic syndrome fuses the MLL gene to the CBP gene. Blood 89:3945–3950 [PubMed] [Google Scholar]
- 40. Takubo K., et al. 2010. Regulation of the HIF-1α level is essential for hematopoietic stem cells. Cell Stem Cell 7:391–402 [DOI] [PubMed] [Google Scholar]
- 41. Tanaka Y., et al. 2000. Extensive brain hemorrhage and embryonic lethality in a mouse null mutant of CREB-binding protein. Mech. Dev. 95:133–145 [DOI] [PubMed] [Google Scholar]
- 42. Tothova Z., et al. 2007. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 128:325–339 [DOI] [PubMed] [Google Scholar]
- 43. Trowbridge J. J., Snow J. W., Kim J., Orkin S. H. 2009. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell 5:442–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vogt P. K., Jiang H., Aoki M. 2005. Triple layer control: phosphorylation, acetylation and ubiquitination of FOXO proteins. Cell Cycle 4:908–913 [DOI] [PubMed] [Google Scholar]
- 45. Wilson N. K., et al. 2010. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell 7:532–544 [DOI] [PubMed] [Google Scholar]
- 46. Xu W., et al. 2006. Global transcriptional coactivators CREB-binding protein and p300 are highly essential collectively but not individually in peripheral B cells. Blood 107:4407–4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yao T. P., et al. 1998. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93:361–372 [DOI] [PubMed] [Google Scholar]
- 48. Yilmaz O. H., et al. 2006. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature 441:475–482 [DOI] [PubMed] [Google Scholar]
- 49. Yoshihara H., et al. 2007. Thrombopoietin/MPL signaling regulates hematopoietic stem cell quiescence and interaction with the osteoblastic niche. Cell Stem Cell 1:685–697 [DOI] [PubMed] [Google Scholar]
- 50. Zhang J., et al. 2006. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature 441:518–522 [DOI] [PubMed] [Google Scholar]
- 51. Zhang J., et al. 2003. Identification of the haematopoietic stem cell niche and control of the niche size. Nature 425:836–841 [DOI] [PubMed] [Google Scholar]
- 52. Zhang Y., et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zimmer S. N., et al. 2011. Crebbp haploinsufficiency in mice alters the bone marrow microenvironment leading to loss of stem cells and excessive myelopoiesis. Blood 118:69–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zon L. I. 2008. Intrinsic and extrinsic control of haematopoietic stem-cell self-renewal. Nature 453:306–313 [DOI] [PubMed] [Google Scholar]