Abstract
Chlamydia trachomatis infection has been suggested to induce host genome duplication and is linked to increased risks of cervical cancer. We describe here the mechanism by which Chlamydia causes a cleavage furrow defect that consistently results in the formation of multinucleated host cells, a phenomenon linked to tumorigenesis. Host signaling proteins essential for cleavage furrow initiation, ingression, and stabilization are displaced from one of the prospective furrowing cortices after Chlamydia infection. This protein displacement leads to the formation of a unique asymmetrical, unilateral cleavage furrow in infected human cells. The asymmetrical distribution of signaling proteins is caused by the physical presence of the Chlamydia inclusion at the cell equator. By using ingested latex beads, we demonstrate that the presence of a large vacuole at the cell equator is sufficient to cause furrow ingression failure and can lead to multinucleation. Interestingly, internalized latex beads of similar size do not localize to the cell equator as efficiently as Chlamydia inclusions; moreover, inhibition of bacterial protein synthesis with antibiotic reduces the frequency at which Chlamydia localizes to the cell equator. Together, these results suggest that Chlamydia effectors are involved in strategic positioning of the inclusion during cell division.
INTRODUCTION
Chlamydia trachomatis is an obligate intracellular bacterium that is the leading cause of bacterial sexually transmitted diseases worldwide (55). In recent epidemiological studies, Chlamydia infection has been linked to increased risk of cervical cancer (29, 43). It has been reported in various human and other mammalian cell lines that Chlamydia infection can lead to significant increases in host cell multinucleation (16), a phenotype well linked to tumorigenesis (14, 15, 36, 47). A defect in cytokinesis was theorized, but not proven, in causing multinucleation in Chlamydia-infected cells (16). As a result, the mechanism by which Chlamydia causes multinucleation remains unclear.
Numerous quality control mechanisms exist in eukaryotic cells to ensure the proper segregation of genetic material into two daughter cells during mitosis. In mammalian cells, cytoplasmic division at the end of mitosis is mediated by the formation of a bilateral cleavage furrow, which is under heavy spatiotemporal regulation (1). Cleavage furrow ingression is driven by force generated by the actomyosin filaments, consisting of actin and nonmuscle myosin II (1). The assembly of actomyosin filament is tightly controlled through phosphorylation of myosin II regulatory light chain, which in turn is regulated by RhoA through its effector proteins Rho kinase and citron kinase (30). In order to have continuous force generation throughout furrow ingression, Rho kinase and citron kinase localization is stabilized by anillin, a scaffolding protein recruited by RhoA (20). Ect2, a Rho guanine exchange factor, plays a critical role in defining the cleavage plane by recruiting and activating RhoA at the precise time and location (56). After degradation of the CDK1-cyclinB1 complex, centralspindlin complex assumes critical roles not only in the bundling of midzone microtubules (MTs) but also in the recruitment of Ect2 to the cell equator (38). The Chlamydia inclusion is a vacuolar structure in which the bacteria reside and replicate within the host cell, and it can continue to grow until it occupies most of the cell volume (48).
Chlamydia can exit the host cells by two distinct mechanisms: cell lysis and extrusion. The cell lysis pathway involves the activation of cysteine protease, which kills the host cell in the process (22). In the extrusion pathway, the Chlamydia inclusion pinches off as a vacuole, taking some or all of the bacteria from the current host, leaving the host cell intact (2, 22). In our study, we observed a unique asymmetry in cleavage furrow formation in human cells after Chlamydia infection, which often led to the formation of multinucleated cells. Signaling proteins essential for cleavage furrow initiation, ingression, and stabilization were displaced from regions of the cell cortex lacking the cleavage furrow. These observations provide evidence that Chlamydia contribute to host genome duplication by disrupting host cell cleavage furrow formation and ingression.
MATERIALS AND METHODS
Cell culture, transfection, reagents, and Chlamydia infection.
HeLa CCL-2 cells (American Type Culture Collection) and previously described CHO-IIA cells (49) were grown in Dulbecco modified Eagle medium (Wisent) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Wisent). All cell cultures were maintained at 37°C in 5% CO2 atmosphere. For the generation of a stable H2B cell line, HeLa cells were transfected with mCherry-H2B (46) using Fugene HD (Roche) for 3 days, followed by selection using 600 ng of puromycin (Invivogen)/ml for 2 weeks. For fluorescent live cell imaging, Cerulean-Lifeact (32, 41) was transiently transfected into the above-mentioned stable HeLa cell line using Fugene HD. Azithromycin, nocodazole, aphidicolin, and poly-l–lysine were purchased from Sigma and monastrol was purchased from Santa Cruz Biotechnology.
C. trachomatis L2 was propagated in HeLa cells and stored at −80°C. C. trachomatis was added to HeLa cells and centrifuged at 300 × g for 20 min to synchronize the infection.
Cell cycle synchronization.
Aphidicolin and nocodazole double synchronization protocol was adapted from Sauer et al. (44). HeLa cells were treated with 1.6 μg of aphidicolin/ml for 16 h to synchronize the cells at S phase without affecting bacterial replication (31). After aphidicolin washout, the cells were incubated with 40 ng of nocodazole/ml for 14 h to arrest cells in mitosis. Cells were infected at such a time point that it would be 24 h postinfection (hpi) by the time of mitotic cell collection. Mitotic cells were tapped off and washed twice. Released cells were plated onto poly-l-lysine-coated coverslips. Progression through mitosis was monitored by fixing cells with 4% paraformaldehyde (PFA) at the indicated time points. Mitotic stages were quantified by using previously described phenotypes (1, 4, 46, 51). In order to avoid potential artifacts caused by nocodazole, monastrol was used to synchronize HeLa cells for the study of signaling protein localization, differential interference contrast (DIC), and fluorescent live cell imaging. HeLa cells were incubated with 100 μM monastrol for 8 h before they were released for 55 min to obtain telophase cells (5). In order to obtain bead-containing telophase CHOIIA cells, the cells were allowed to phagocytose IgG-opsonized 15 μm beads overnight before they were treated with monastrol as described above.
Azithromycin was dissolved in ethanol and added to infected cells at 28 hpi, along with monastrol. For cytokinesis completion experiments, monastrol synchronized cells were released for 3 h with or without azithromycin treatment.
Immunofluorescence and antibodies.
To stain acetylated-α-tubulin, α-tubulin, actin and myosin II, HeLa cells grown on coverslips were fixed with 4% PFA in phosphate-buffered saline (PBS) for 15 min. To visualize RhoA and anillin, HeLa cells were fixed by ice-cold 10% trichloroacetic acid (TCA; BioShop) for 10 min. Cells fixed by PFA or TCA were permeabilized for 20 min using 0.1% Triton X-100 in PBS with 100 mM glycine. For Ect2, Mklp1, Plk1, and Prc1 staining, the cells were fixed and permeabilized using ice-cold 100% methanol for 10 min. Cells were blocked in 5% FBS overnight, followed by primary and fluorescent secondary antibody incubation for 1 h each. Actin and DNA were visualized by Alexa Fluor 488-phalloidin and DAPI (4′,6′-diamidino-2-phenylindole), respectively.
Primary antibodies against acetylated α-tubulin (T6793; 1:5,000), α-tubulin (T9026; 1:1,000), and myosin IIB heavy chain (M7939; 1:200) were purchased from Sigma-Aldrich. RhoA (26c4; 1:100), Mklp1 (N19; 1:100), Plk1 (F8; 1:100), Prc1 (H70; 1:100), and Chlamydia (Chlam III; 1:100) antibodies were purchased from Santa Cruz Technology. Ect2 antibody was a generous gift from Michael Glotzer. Anillin antibody was a gracious gift from William Trimble. Rabbit anti-Chlamydia antibody was kindly donated by David Hackstadt. Alexa Fluor 488-phalloidin (1:500) and DAPI (1:10,000) were obtained from Invitrogen. Donkey anti-mouse or donkey anti-rabbit Dylight secondary antibodies were purchased from Jackson Immunoresearch Laboratories and were used at recommended dilutions.
Phagocytosis.
Polystyrene beads (PolySciences) with a mean diameter of 15 μm were opsonized with 2 mg of human IgG (Sigma-Aldrich)/ml for 1 h, and unbound IgG was removed by one wash with PBS. The IgG-opsonized beads were then added to CHO-IIA cells overnight to allow internalization by phagocytosis. External beads were washed away before live cell DIC imaging.
Microscopy.
Epifluorescence images were acquired with an EC Plan-Neofluor ×40/1.3 oil objective and AxioCam MRm camera mounted on either an AxioVert 200M or Axio Observer Z1 microscope (Carl Zeiss). Structured illumination images were taken with the above mentioned objective and the images were captured using an AxioCam HRm camera with ApoTome module on AxioVert 200M microscope (Carl Zeiss). Spinning disk confocal images were taken with a Plan-Apochromat ×40/1.3 oil objective and CSU-X1 (Yokogawa). Images were captured with Cascade 1K EMCCD (Photometrics) on an Axio Observer Z1 (Carl Zeiss). All live cell imaging experiments were carried out with an Incubator XL-S1 with TempModule S1, CO2 module S1, heating insert P S1, and heating device humidity S1 connected to Axio Observer Z1 (Carl Zeiss). All live cell images were captured with an AxioCam MRm. Fluorescent live cell imaging was carried out with the same objective as epifluorescence images, while DIC live cell imaging was done with a ×20/0.8 Plan Apochromat air objective.
Quantifications.
For mitotic progression quantification, epifluorescence images of more than 100 control and 100 infected cells were quantified per time point per trial. The sizes and positions of Chlamydia inclusions during telophase were quantified based on epifluorescence images of 165 telophase cells at 1.5 h postrelease (hpr) and 94 telophase cells at 3 and 4 hpr from three independent experiments. Localization of actin, myosin II, anillin, RhoA, Ect2, Mklp1, Plk1, and Prc1 were quantified based on structured illumination images of more than 100 control and 100 infected telophase cells. Equal accumulation of actin, myosin, anillin, and RhoA was scored when the fluorescence intensity of the protein on both sides of the equatorial cortex were within 80% of each other. Normal localization of Ect2, Mklp1, Plk1, and Prc1 was defined as a strong band of protein across the entire equatorial width of telophase cells, defined by DIC imaging. All statistical tests were performed using the Student t test and P < 0.05 was used as the significance cutoff.
Nascent protein synthesis labeling.
Nascent Chlamydia protein synthesis was detected with Click-iT AHA protein synthesis labeling kit (Invitrogen). Azithromycin was added to Chlamydia-infected cells at 28 hpi and incubated for 7 h at a final concentration of 20 μg/ml. Methionine-free medium (Invitrogen) was supplemented with 1 mM sodium pyruvate, 4 mM l-glutamine, and 0.2 mM l-cysteine–2 HCl and was added to infected cells for 1 h to deplete any preexisting free methionine. Serum was not included in this medium to prevent serum contribution of free methionine, while azithromycin was always present at 20 μg/ml until the cells were fixed. Methionine-free media containing 50 μM l-azidohomoalanine (AHA) was added to the cells for 3 h before fixation with 4% PFA. After permeabilization with 0.25% Triton X-100 in PBS for 20 min, AHA incorporation was visualized using 50 μM Alexa Fluor 555 alkyne, triethylammonium (A20013; Invitrogen) in manufacturer-recommended reaction buffer (C10269; Invitrogen). The incubation time and washes were performed exactly as described in the manufacturer's protocol.
For experiments involving cycloheximide, following 1 h of methionine starvation, cycloheximide (Sigma-Aldrich) was added alone or together with azithromycin, with final concentrations of 100 μM and 20 μg/ml, respectively, to Chlamydia-infected cells at 30 hpi. AHA was added at the same time as the cycloheximide, and the cells were incubated for 2 h before fixation.
RESULTS
Chlamydia infection disrupts host telophase during mitosis.
To determine the precise cell division defect induced by Chlamydia infection, a modified aphidicolin and nocodazole synchronization protocol (44) was used to obtain mitotic cells after Chlamydia infection. Chlamydia infection was carried out at such a time point that it would be 24 hpi by the time of mitotic cell collection (see Fig. S1A in the supplemental material). Cells at each mitotic stage were quantified based on previously established phenotypes (see Fig. S1B in the supplemental material) (1, 4, 46, 51), and the mitotic stage composition was monitored for each time point (see Fig. S1C to I in the supplemental material). At 0.5 h postrelease (hpr), more than 96% of the collected cells were in prophase or metaphase, suggesting that the tap-off procedure produced a relatively pure population of mitotic cells (see Fig. S1C and D in the supplemental material). No significant change was detected in prophase, metaphase, and anaphase progression between uninfected and Chlamydia-infected cells when they were collected at 24 hpi (see Fig. S1C to E in the supplemental material). Interestingly, there was a significant increase of Chlamydia-infected telophase cells at 1.5 hpr compared to control cells (see Fig. S1F in the supplemental material). There was also a significant decrease in Chlamydia-infected cells in cytokinesis at 1.5 hpr, compared to control cells (see Fig. S1G in the supplemental material). Together, these results suggested that telophase was prolonged after Chlamydia infection.
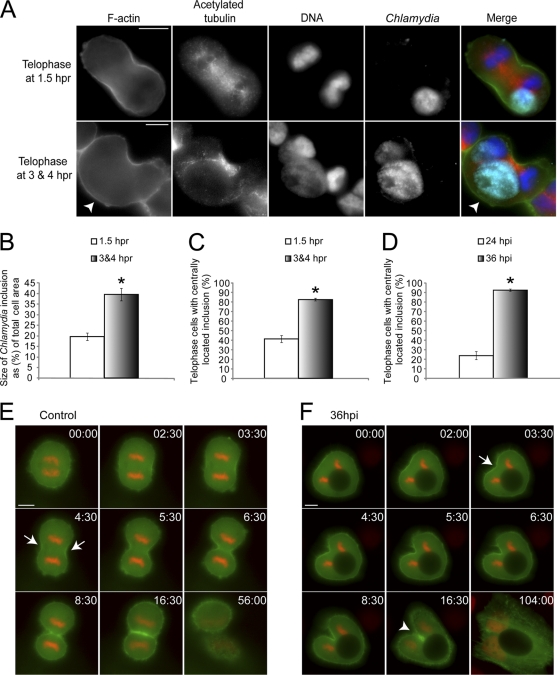
Next, we examined telophase cells more closely at different time points after release to identify any possible defects that could be attributed to Chlamydia infection. Interestingly, many infected telophase cells at 3 and 4 hpr, which had been held in telophase for an extend period of time, lacked an ingressing furrow on one side (Fig. 1A). Accumulation of actin occurred at both ingressing furrows in most telophase cells at 1.5 hpr, while actin only accumulated at a single cleavage furrow in infected cells at 3 and 4 hpr (Fig. 1A). Inclusion size analysis revealed that the average size of Chlamydia inclusions was 20% of the total cell area for telophase cells at 1.5 hpr (Fig. 1B). The inclusion size was 40% in telophase cells at 3 and 4 hpr (Fig. 1B), indicating a strong correlation between large inclusions and severely prolonged telophase. This significant increase in inclusion size is likely due to difference in the number of bacteria that initially infected the cells. We also observed that 44% of telophase cells contained centrally located Chlamydia inclusions at 1.5 hpr, and this number increased to 88% in telophase cells at 3 and 4 hpr (Fig. 1C). In order to consistently obtain large Chlamydia inclusions, we increased the infection time to 36 h. Significantly more Chlamydia inclusions localized to the host cell center during telophase at 36 hpi than at 24 hpi (Fig. 1D).
Fig. 1.
Chlamydia infection causes unilateral cleavage furrow formation and leads to host cell multinucleation. (A) Chlamydia-infected telophase cells were obtained using aphidicolin and nocodazole. Mitotic apparatus (red), Chlamydia (cyan), F-actin (green), and DNA (blue) were visualized as described in Materials and Methods. A representative telophase cells at 1.5 hpr showed normal cleavage furrow formation (top), while telophase cells at 3 and 4 hpr often exhibited a single unilateral cleavage furrow (arrowhead). (B) Telophase cells at 3 and 4 hpr contained significantly larger Chlamydia inclusions than those at 1.5 hpr (P < 0.01). (C) Telophase cells at 3 and 4 hpr had significantly more centrally located Chlamydia inclusions than those at 1.5 hpr (P < 0.005). (D) Infected telophase cells at 36 hpi had significantly more centrally located Chlamydia inclusions than cells at 24 hpi (P < 0.005). (E and F) Cerulean-Lifeact (green) was transiently transfected into HeLa cells stably expressing mCherry-H2B (red). Numbers indicate minutes:seconds. (E) Uninfected cells always showed bilateral actin accumulation and furrow ingression at the cell equator during telophase as indicated by arrows (n = 29 of 29). (F) Chlamydia-infected cells containing centrally located inclusions often exhibited a unilateral cleavage furrow as marked by arrow (n = 13 out of 14). The unilaterally ingressing furrow eventually became blocked by the inclusion and could not ingress further (arrowhead). Error bars represent the standard error of the mean (SEM) from three independent experiments. Scale bars, 10 μm.
We monitored mitotic events with live cell imaging to confirm the cleavage furrow defect after Chlamydia infection. Chlamydia infection was carried out for 36 h to obtain large Chlamydia inclusions that produced a prominent telophase defect (Fig. 1A). Cerulean-Lifeact (32, 41) was transiently transfected into HeLa cells stably expressing mCherry-H2B to dual label actin and chromosomes. Uninfected cells formed normal cleavage furrows on both sides of the cell during telophase (n = 29 out of 29; Fig. 1E and see Movie S1 in the supplemental material). In contrast, infected cells with equatorial inclusions often formed a single, F-actin-rich, cleavage furrow on the equatorial cortex away from the Chlamydia inclusion (n = 13 out of 14; Fig. 1F and see Movie S2 in the supplemental material). The ingressing furrow was blocked by Chlamydia when it reached and pressed against the inclusion (Fig. 1F). Our fluorescent and DIC analysis showed that host cells with unilateral cleavage furrows typically exited mitosis without dividing into two daughter cells, forming multinucleated interphase cells (n = 27 out of 33; Fig. 1F and see Movie S2 in the supplemental material). However, when the Chlamydia inclusion was located in the polar region of the host cell, F-actin-rich bilateral cleavage furrows formed, and the host cell successfully divided into two daughter cells (see Movie S3 in the supplemental material).
Chlamydia inclusion disrupts RhoA and downstream signaling events necessary for cleavage furrow formation.
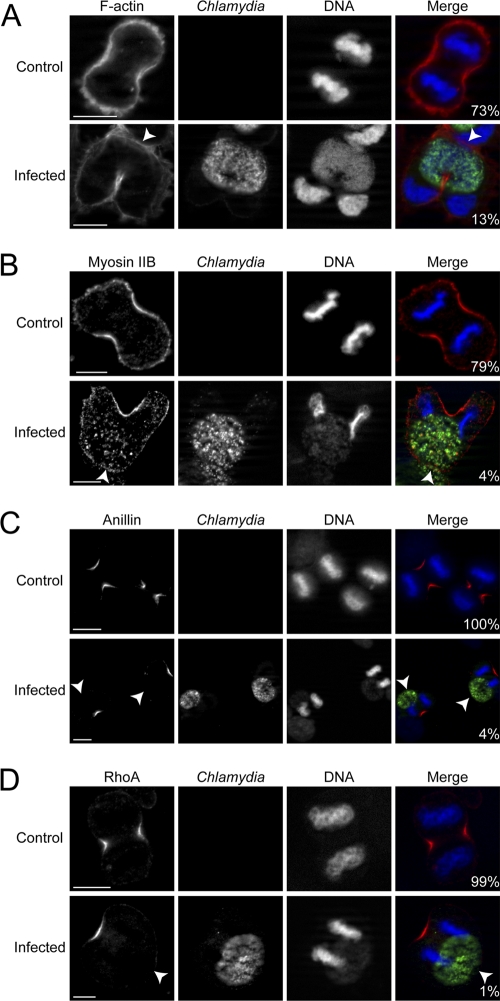
Since unilateral cleavage furrows were routinely induced in Chlamydia-infected telophase cells, we next monitored the recruitment of relevant cleavage furrow cytoskeletal and signaling proteins. Monastrol was used to synchronize HeLa cells to avoid potential disruption of signaling events caused by MT-depolymerizing agents (35). Chlamydia infection was carried out for 36 h to obtain large inclusions, and phalloidin staining revealed that 73% of uninfected versus 13% of infected telophase cells had equal accumulation of F-actin at the equatorial cortices (Fig. 2A). We next examined the localization of the F-actin motor, myosin II, and observed that 79% of uninfected but only 4% of infected telophase cells had equal myosin II accumulation at the equatorial cortices (Fig. 2B). These results indicated a strong contractile defect at the cell cortex near Chlamydia inclusion during furrow ingression.
Fig. 2.
Chlamydia infection disrupts the accumulation of actin, myosin II, anillin, and RhoA during telophase. (A to D) Chlamydia (green) and DNA (blue) were stained as described in Materials and Methods. Arrowheads indicated prospective furrowing plasma membrane lacking normal cytoskeletal or signaling protein accumulation. Numbers indicate the percentages of cells with normal bilateral accumulation for each protein (red). Scale bars, 10 μm.
Due to the lack of actomyosin filament assembly at one side of the cell, we looked at upstream signaling proteins responsible for the stabilization and activation of the contractile machinery. We first examined anillin, a scaffold protein responsible for stabilizing actomyosin filaments (20), and observed equal accumulation at the cell equator in 100% of uninfected but only 4% of infected telophase cells (Fig. 2C). We next examined the accumulation of RhoA, which is the activator of actomyosin filament assembly and recruiter of anillin (8). RhoA was equally accumulated on both equatorial cortices in 99% of uninfected but just 1% of Chlamydia-infected telophase cells (Fig. 2D). Attenuated F-actin, myosin II, anillin, and RhoA accumulation always occurred on the side of telophase cells closer to the Chlamydia inclusion (Fig. 2A to D). Notably, the displacement of myosin II, anillin, and RhoA in Chlamydia-infected cells was already prominent during anaphase (see Fig. S2B to D in the supplemental material). It is also worth noting that the localization of actin, myosin II, anillin, and RhoA appeared to be normal when the Chlamydia inclusions were located in the polar region of the host cells (see Fig. S3A to D in the supplemental material).
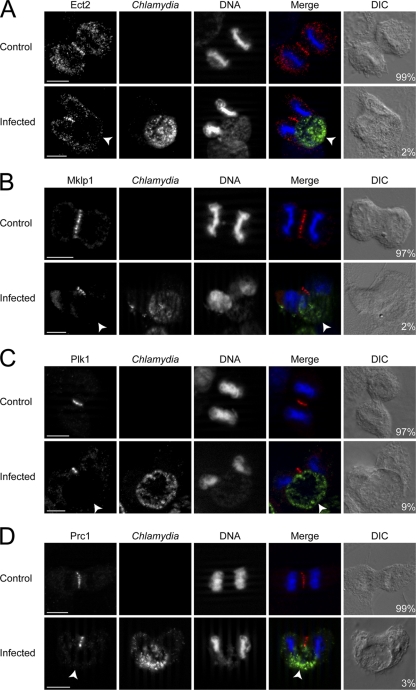
Ect2 and Plk1 localization becomes disrupted in the presence of Chlamydia inclusions.
To understand how RhoA became mislocalized on only one side of the cell, we investigated the localization of Ect2, a RhoGEF known to activate and promote RhoA localization (35, 45, 56). The typical Ect2 staining, showing a linear distribution across the equatorial plane (35, 37, 56), was only observed in 2% of Chlamydia-infected telophase cells versus 99% of uninfected cells (Fig. 3A). Instead, infected cells showed a linear, truncated staining pattern that rarely extended to both cell cortices (Fig. 3A). Similarly, the distribution of Mklp1, which is part of the centralspindlin complex that recruits Ect2 to the spindle midzone (35, 56, 57), was normal in 97% of control telophase cells, while only 2% of infected telophase cells showed Mklp1 staining that spanned the equatorial plane (Fig. 3B).
Fig. 3.
Chlamydia infection disrupts the equatorial localization of Ect2, Mklp1, Plk1, and Prc1 during telophase. (A to D) Chlamydia (green) and DNA (blue) were stained as described in Materials and Methods. Arrowheads mark the equatorial cell cortex lacking normal signaling protein targeting. Numbers indicate percentages of cells with normal equatorial accumulation for each protein, which spanned the equatorial width (red). Scale bars, 10 μm.
Plk1 is known to play very important roles in recruiting Ect2 to the centralspindlin complex and to the cell equator (37, 53). Upon studying the localization of Plk1, we observed that 97% of uninfected versus 9% of Chlamydia-infected telophase cells had normal Plk1 distribution, which spanned the equatorial width (Fig. 3C). We subsequently examined the distribution of Prc1, a protein that recruits Plk1 to the cell equator (5, 9, 33). While Prc1 showed normal distribution in 99% of uninfected cells, only 3% infected telophase cells showed typical Prc1 staining that traversed the equatorial plane (Fig. 3D). Analysis of anaphase cells showed similar patterns of disruption for Ect2, Mklp1, Plk1, and Prc1 in Chlamydia-infected cells (see Fig. S4A to D in the supplemental material). Notably, when Chlamydia inclusions were in the polar region of the dividing host cells, Ect2, Mklp1, Plk1, and Prc1 all exhibited the normal linear distribution across the equatorial width of the host cells (see Fig. S3E to H in the supplemental material).
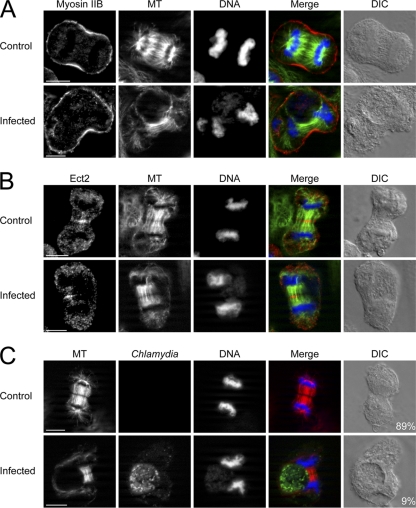
Mislocalization of signaling proteins occurs in concert with displaced astral and central spindle MTs.
A consistent defect was observed in Ect2, Mklp1, Plk1, Prc1, (Fig. 3A to D) and Aurora B (see Fig. S5 in the supplemental material) localization after Chlamydia infection in mitotic cells. We hypothesized that delivery of these molecules was impaired due to MT organization disruption in Chlamydia-infected cells. Control cells showed normal MT targeting to the furrowing cell cortices that were enriched for myosin II (Fig. 4A). In contrast, only the cell cortex further away from Chlamydia inclusions in infected cells showed robust MT targeting and myosin II accumulation (Fig. 4A). The inclusion side had negligible or very sparse MT penetration with corresponding attenuated myosin II signal (Fig. 4A). F-actin, anillin, and RhoA showed identical localization as myosin II in relation to MT density (data not shown). We next investigated the distribution of central spindle MTs and Ect2 in Chlamydia-infected cells. Ect2 was recruited as a perpendicular line across the mitotic midzone in uninfected telophase cells as expected (Fig. 4B). Ect2 in Chlamydia-infected cells similarly decorated the central spindle MTs, but both Ect2 and central spindle MTs were severely displaced away from the cell cortex adjacent to the inclusion (Fig. 4B). Mklp1, Plk1, and Prc1 showed localization patterns identical to that of Ect2 in relation to MT distribution (data not shown).
Fig. 4.
The mislocalization of myosin II and Ect2 coincides with MT displacement. (A to C) MT, Chlamydia and DNA were stained as described in Materials and Methods. (A) Similar density of MTs (green) reached both equatorial cortices and myosin II (red) accumulated equally on both sides in uninfected cells. In Chlamydia-infected cells, the lack of myosin II accumulation always coincided with the lack of MTs. (B) Ect2 (red) decorated the midzone MTs (green) across the equatorial width in uninfected telophase cells. After Chlamydia infection, the midzone and astral MTs were severely shifted to one side of the telophase cell and Ect2 only accumulated where the MTs were present. (C) Both equatorial cortices received equal amounts of MTs (red) in 89% of uninfected but only 9% of infected telophase cells. Scale bars, 10 μm.
We quantified the fluorescence intensity of MTs at each cleavage furrow and found that 89% of uninfected and only 9% of infected telophase cells had equal density of MTs at both equatorial cortices (Fig. 4C). Fine step Z-stacks taken using spinning disk confocal revealed that MTs were equally distributed on both sides of uninfected telophase cells, while both central spindle and astral MTs were severely displaced to one side of the cell and rarely extended to the cell cortex adjacent to the Chlamydia inclusion in infected cells (see Fig. S6 in the supplemental material).
Ingested latex beads mimic the Chlamydia inclusion as a physical barrier and can induce unilateral cleavage furrow defect.
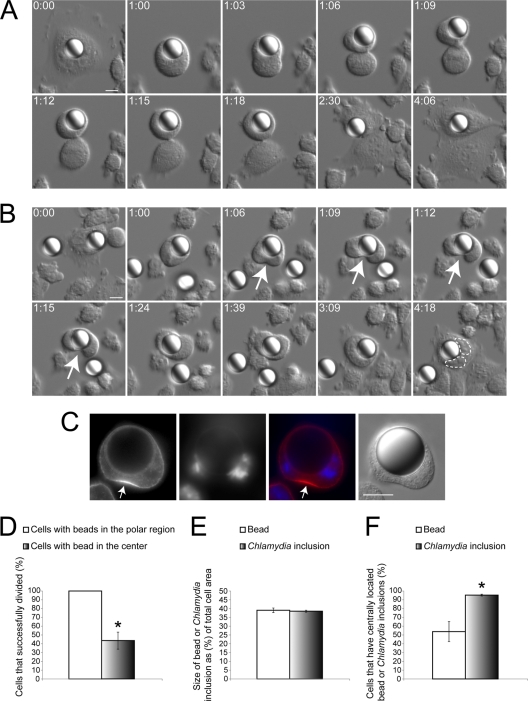
We sought to determine whether the Chlamydia inclusion caused the displacement of cytoskeletal and signaling proteins by simply acting as a physical barrier. To mimic such an intracellular physical obstacle, Chinese hamster ovary (CHO) cells stably expressing Fcγ receptors (CHO-IIA cells) were used to internalize IgG-opsonized 15-μm polystyrene beads. Although the phagocytic uptake of these large beads was extremely low (data not shown), we were able to obtain sufficient numbers of cells with internalized beads to compare the progress through cell division with the more reliably generated Chlamydia inclusion-containing cells. Using DIC live cell imaging, we observed two phenotypes in dividing cells. CHO-IIA cells that contained a bead in the polar region successfully divided into two daughter cells (Fig. 5A and see Movie S4 in the supplemental material). When the beads were centrally located in CHO-IIA cells, a unilateral cleavage furrow formed (Fig. 5B), similar to Chlamydia-infected cells. These cells were often prevented from dividing into two daughter cells (Fig. 5B and see Movie S5 in the supplemental material). Cells with internalized beads that failed to divide eventually formed binucleated interphase cells (n = 18 out of 18, Fig. 5B). We confirmed that signaling proteins such as RhoA were unable to accumulate at the equatorial cell cortex near the latex bead, which closely resembled the defect caused by Chlamydia inclusion (Fig. 5C). The success rate of division for cells with centrally located beads was significantly lower than that of cells with beads in the polar region (Fig. 5D). This result suggested that the mechanical hindrance due to the presence of a large vacuole at the cell equator is sufficient to disrupt cytokinesis.
Fig. 5.
Centrally located large polystyrene beads can recapitulate the unilateral cleavage furrow defect. (A and B) CHO-IIA cells that internalized a bead were subject to DIC imaging. Numbers indicate hours:minutes. Scale bars, 10 μm. (A) Cells with beads in the polar region were able to form bilateral cleavage furrows and successfully divide into two discrete daughter cells (n = 16 out of 16). (B) When the bead was in the cell center, a unilateral cleavage furrow (arrows) could be induced. Cytoplasmic division frequently failed in cells with centrally positioned beads (n = 18 out of 27). Two nuclei were outlined with dashed lines. (C) RhoA (red) only accumulated at the equatorial cell cortex away from the centrally located latex bead in CHO IIA cells. DNA was stained with DAPI (blue) (n = 10 out of 10). (D) Cells containing centrally located beads had a significantly lower success rate of division than cells containing beads in the polar region (P < 0.005). (E) The relative size of 15-μm beads compared to host cells was indistinguishable from that of the Chlamydia inclusions at 36 hpi (P > 0.74). (F) Chlamydia inclusions localized to the cell equator significantly more often than similarly sized beads during telophase (P < 0.005). (D to F) Error bars represent the SEM from at least three independent experiments.
The polystyrene beads were chosen to have a size comparable to that of Chlamydia inclusions at 36 hpi (Fig. 5E). Despite having identical relative sizes, more than 95% of the Chlamydia inclusions at 36 hpi localized to the cell equator, whereas less than 54% of the beads were present in the cell equator during telophase (Fig. 5F). These results showed that Chlamydia inclusions localized to the cell equator much more efficiently than large inert particles, suggesting that Chlamydia may be actively localizing to the cell equator.
Active bacterial protein synthesis contributes to central localization of Chlamydia inclusions.
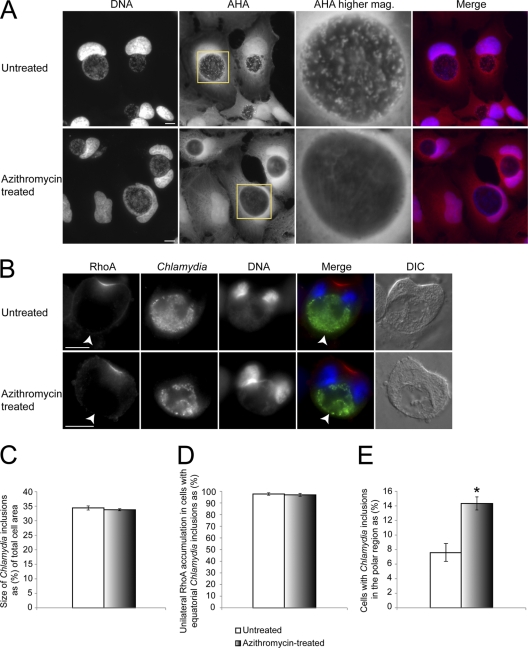
Since genetic manipulation methods do not currently exist for Chlamydia (18, 48), to address whether Chlamydia was actively localizing to the center of dividing cells, we created metabolically inactive Chlamydia inclusions using azithromycin, one of the most commonly prescribed antibiotics for treating Chlamydia infection (54). To block Chlamydia protein synthesis, azithromycin was added to cells at 28 hpi to allow initial growth of the inclusion to an intrusive size. We assessed bacterial protein synthesis with a nonradioactive nascent protein synthesis labeling technique using AHA. Chlamydia particles in untreated cells were brightly labeled with AHA and appeared as distinct puncta within the inclusion (Fig. 6A and see Fig. S7A in the supplemental material), while no bacteria showed visible incorporation of AHA 7 h after azithromycin treatment (Fig. 6A). AHA incorporation of Chlamydia inclusion was unaffected by cycloheximide, a host protein synthesis inhibitor (Fig. S7B). In addition, simultaneous cycloheximide and azithromycin treatment knocked out both host and bacterial AHA incorporation (see Fig. S7C in the supplemental material). Together, these results validated the use of AHA for detecting bacterial protein synthesis.
Fig. 6.
Bacterial protein synthesis plays a role in localizing the inclusion to the cell center during telophase. (A and B) Chlamydia-infected cells were treated with azithromycin for 7 h. AHA (red) and DNA (blue) were stained as described in Materials and Methods. Scale bars, 10 μm. (A) In untreated cells, Chlamydia showed distinct punctate staining within the inclusion, while inclusions in azithromycin-treated cells lacked detectable accumulation of AHA. (B) Similar to untreated cells, after azithromycin treatment, centrally located Chlamydia inclusions could still cause RhoA mislocalization during telophase (arrowheads). (C) Telophase cells with similarly sized Chlamydia inclusions (P > 0.56) were chosen, with or without azithromycin treatment, for subsequent RhoA distribution and inclusion positioning analysis. (D) Azithromycin treatment did not rescue the RhoA mislocalization defect when the Chlamydia inclusion was centrally located (P > 0.72). (E) A significant increase of cells containing Chlamydia inclusions in the polar region was detected after azithromycin treatment (P < 0.05). Error bars represent the SEM from three independent experiments.
To monitor cleavage furrow formation, we examined the localization of RhoA in these cells. Following antibiotic treatment, we observed that RhoA was still severely mislocalized in the presence of centrally positioned inclusions (Fig. 6B and D). Cells with identical inclusion sizes were chosen for comparison to ensure that any difference we detected was not solely due to the difference in inclusion size (Fig. 6C). Interestingly, alteration of Chlamydia inclusion localization was observed. Analysis of antibiotic-treated cells revealed a significant increase of Chlamydia inclusions in the polar region of cells compared to untreated cells (Fig. 6E). After azithromycin treatment, cells with inclusions in the polar region were able to form normal bilateral cleavage furrows (see Fig. S7D in the supplemental material), similar to CHO-IIA cells with beads in the polar region (Fig. 5A). We confirmed the success rate of cell division in cells containing polar Chlamydia inclusions with DIC live cell imaging. More than 98% of cells with inclusions in the polar region successfully divided into two daughter cells, while less than 20% of cells with equatorial inclusions were able to complete furrow ingression (see Fig. S7E in the supplemental material). After antibiotic treatment, there was a significant increase in the percentage of cells that successfully completed furrow ingression (see Fig. S7F in the supplemental material). Thus, metabolic activities of Chlamydia influenced the spatial distribution of this bacterial compartment in dividing cells, which largely determined the efficiency of cleavage furrow disruption.
DISCUSSION
This study shows that Chlamydia inclusions at later stages of infection can disrupt cleavage furrow formation on one side of the host cell, thus leading to a unique unilateral furrow ingression that often failed to complete. Numerous epidemiological studies have identified C. trachomatis as both an independent risk factor and a cofactor with human papillomavirus to cause cervical cancer (25, 43). In addition, Chlamydia pneumonia, whose life cycle is similar to C. trachomatis, has also been shown to increase the risks of lung cancer (23, 28). Although the tumorigenic effects of C. pneumonia may be caused by chronic inflammation (7), it has been demonstrated that C. trachomatis infection can lead to supernumerary centrosome formation (17, 24, 27). Our study provides a mechanism by which Chlamydia can cause cytokinesis failure and multinucleation; both the centrosome and cytokinesis defects are linked to tumorigenesis (14, 15, 34, 36, 47).
We demonstrated the mechanism by which Chlamydia inclusions efficiently disrupted host cleavage furrow formation. Starting as far upstream as Plk1 and Prc1, we observed a consistent and sharp reduction of all of the major downstream signaling elements, including the critical actomyosin machinery, at the inclusion side. Our live cell imaging showed no indication of partially formed or regressed furrows on the inclusion side, suggesting that cleavage furrow initiation was impaired.
Chlamydia inclusions reproducibly mimicked the elegant oil droplet studies performed by Rappaport in echinoderm eggs (40), where unilateral cleavage formation was also induced. We were able to recapitulate the unilateral cleavage furrow using centrally located, large latex beads in CHO-IIA cells, suggesting the asymmetric cleavage formation was caused by the physical presence of a large vacuole. Green urchin and Hydractinia embryo cells spontaneously form unilateral cleavage furrows, which have been attributed to naturally occurring eccentric mitotic spindles (3, 39). After Chlamydia infection, the mitotic spindle also became eccentrically positioned, due to the presence of a large inclusion.
Our study is the first to systematically assess the localization of key signaling elements during unilateral cleavage formation. Previous studies that observed unilateral cleavage furrows lacked large-scale immunofluorescence quantifications due to the transiency of furrow ingression (6). In our study, we consistently obtained large numbers of telophase cells exhibiting unilateral furrows with monastrol synchronization and Chlamydia infection. Also unique about this system was that endogenous proteins were not experimentally knocked down and were functional based on their recruitment and ability to promote at least one cleavage furrow. It is important to note that normal bilateral cleavage furrows formed, when the Chlamydia inclusion was located in the polar region. Together, these observations suggested that Chlamydia-induced cleavage furrow defect was the result of physical displacement instead of degradation of host signaling proteins essential for furrow initiation. Ect2 is recruited to the midzone MTs to promote RhoA activation at the overlying cell cortex (37, 56). In the presence of Chlamydia inclusions, Ect2 was physically blocked from reaching one of the prospective furrowing regions, and the absence of RhoA at this site suggested that Ect2 could only activate RhoA locally. This provided direct evidence that Ect2 was responsible for the spatial control of RhoA activation during mitosis, without using global protein knock down techniques such as siRNA (38). Chlamydia inclusion-containing cells therefore serve as a good model to study cleavage furrow protein recruitment, as the signaling protein of interest is still present and functional on one side of the furrow, thus acting as an internal positive control. Similarly, activated RhoA could only exert its function locally in the cell, as we only observed recruitment of anillin and actomyosin filament assembly on the same side where RhoA accumulated.
We showed that Chlamydia inclusions could localize to the cell equator more efficiently than similarly sized vacuoles generated by internalized beads, indicating the bacteria are actively localizing to the cell center. The different nature of the vacuole membrane surrounding latex beads and inclusions may account for the spatial differences between the two (13, 26, 42). The Chlamydia inclusion is known to be devoid of endosomal markers but can reroute Golgi body-derived vesicles from the exocytic pathways (13). Rab6 and 11 are both recruited to the C. trachomatis inclusion membrane (42); moreover, Rab6- or Rab11-positive vesicles localize to the equatorial cell cortex during anaphase (21, 52), implicating a possible role for these GTPases in trafficking the inclusion during cell division. A recent study on Theileria showed that the parasite can recruit host Plk1 and localize to the host central spindle (50). Unlike Theileria, none of the host signaling proteins that we examined were noticeably recruited to the Chlamydia inclusion membrane. Due to the lack of Chlamydia genetic manipulation tools, we had to generate metabolically inactive Chlamydia inclusions to address whether Chlamydia proteins contributed to the central localization of the inclusions during telophase. The significant increase of inclusions in the host polar region during telophase after antibiotic treatment suggested that Chlamydia protein synthesis played a role in positioning this bacterial compartment. It is possible that the antibiotic treatment reduced the expression of some Chlamydia proteins on the inclusion surface that are important for its positioning. Interestingly, the majority of Chlamydia inclusions devoid of detectable levels of protein synthesis still localized to the cell center, suggesting factors essential for Chlamydia positioning may have been expressed or recruited before antibiotic treatment at 28 hpi.
Chlamydia is known to alter host processes by directly or indirectly degrading host proteins (10–12, 19). We observed that cells with large inclusions in the polar region were able to form normal cleavage furrows and have normal signaling protein localization; moreover, almost 100% of Chlamydia-infected cells that contained inclusions in the polar region successfully divided into two daughter cells. Therefore, it was unlikely that the cleavage furrow formation defect was caused by generic host protein degradation. Our results demonstrated that conventional antibiotic treatment such as azithromycin was effective in blocking Chlamydia protein synthesis; however, it could not fully rescue the unilateral cleavage furrow defect that frequently resulted in the formation of multinucleated host cells. It has been previously shown that Chlamydia could exit by extrusion in more than 50% of infected host cells, which could remain viable afterward (2, 22). Therefore, it is reasonable to speculate that Chlamydia may contribute to tumorigenesis by causing host genome duplication and leaving behind viable tetraploid host cells following extrusion exit. Future studies should investigate the tumorigenic potential of these multinucleated progeny and aim to understand the mechanisms by which Chlamydia is able to efficiently localize itself to the cell equator even in the absence of detectable protein synthesis.
Supplementary Material
ACKNOWLEDGMENTS
This project is funded by a MOP-68992 grant from Canadian Institutes of Health Research (CIHR) to R.E.H. R.E.H. is a recipient of a CIHR New Investigator Award and an Ontario Early Researcher Award.
We are grateful to William Trimble (The Hospital for Sick Children, Toronto, Canada), Michael Glotzer (University of Chicago), and David Hackstadt (Rocky Mountain Laboratories NIAID/NIH) for their generous gifts of antibodies. We thank Daniel Gerlich (Swiss Institute of Technology Zurich), Lise Munsie (McMaster University), and Ray Truant (McMaster University) for their kind donation of fluorescent constructs. We thank Sergio Grinstein (The Hospital for Sick Children) for kindly providing the CHO-IIA stable cell line. We are grateful to Ari Chow (University of Toronto at Scarborough) and Guang Yu Hu for their advice on generating stable HeLa cell lines. We appreciate the assistance of Mark Jen, Dorothy Zhao, and Sam Khalouei (University of Toronto at Scarborough) for their help with live and fixed cell imaging and the Mauricio Terebiznik lab (University of Toronto at Scarborough) for stimulating discussions.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 3 October 2011.
REFERENCES
- 1. Barr F. A., Gruneberg U. 2007. Cytokinesis: placing and making the final cut. Cell 131:847–860 [DOI] [PubMed] [Google Scholar]
- 2. Beatty W. L. 2007. Lysosome repair enables host cell survival and bacterial persistence following Chlamydia trachomatis infection. Cell Microbiol. 9:2141–2152 [DOI] [PubMed] [Google Scholar]
- 3. Bement W. M., Benink H. A., von Dassow G. 2005. A microtubule-dependent zone of active RhoA during cleavage plane specification. J. Cell Biol. 170:91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Birkenfeld J., et al. 2007. GEF-H1 modulates localized RhoA activation during cytokinesis under the control of mitotic kinases. Dev. Cell 12:699–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burkard M. E., et al. 2009. Plk1 self-organization and priming phosphorylation of HsCYK-4 at the spindle midzone regulate the onset of division in human cells. PLoS Biol. 7:e1000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Campbell S., Richmond S. J., Yates P. 1989. The development of Chlamydia trachomatis inclusions within the host eukaryotic cell during interphase and mitosis. J. Gen. Microbiol. 135:1153–1165 [DOI] [PubMed] [Google Scholar]
- 7. Chaturvedi A. K., et al. 2010. Chlamydia pneumoniae infection and risk for lung cancer. Cancer Epidemiol. Biomarkers Prev. 19:1498–1505 [DOI] [PubMed] [Google Scholar]
- 8. D'Avino P. P. 2009. How to scaffold the contractile ring for a safe cytokinesis: lessons from Anillin-related proteins. J. Cell Sci. 122:1071–1079 [DOI] [PubMed] [Google Scholar]
- 9. D'Avino P. P., et al. 2007. Recruitment of Polo kinase to the spindle midzone during cytokinesis requires the Feo/Klp3A complex. PLoS One 2:e572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dong F., Su H., Huang Y., Zhong Y., Zhong G. 2004. Cleavage of host keratin 8 by a Chlamydia-secreted protease. Infect. Immun. 72:3863–3868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dong F., Zhong Y., Arulanandam B., Zhong G. 2005. Production of a proteolytically active protein, chlamydial protease/proteasome-like activity factor, by five different Chlamydia species. Infect. Immun. 73:1868–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fan P., Dong F., Huang Y., Zhong G. 2002. Chlamydia pneumoniae secretion of a protease-like activity factor for degrading host cell transcription factors required for major histocompatibility complex antigen expression. Infect. Immun. 70:345–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fields K. A., Hackstadt T. 2002. The chlamydial inclusion: escape from the endocytic pathway. Annu. Rev. Cell Dev. Biol. 18:221–245 [DOI] [PubMed] [Google Scholar]
- 14. Fujiwara T., et al. 2005. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 437:1043–1047 [DOI] [PubMed] [Google Scholar]
- 15. Ganem N. J., Storchova Z., Pellman D. 2007. Tetraploidy, aneuploidy, and cancer. Curr. Opin. Genet. Dev. 17:157–162 [DOI] [PubMed] [Google Scholar]
- 16. Greene W., Zhong G. 2003. Inhibition of host cell cytokinesis by Chlamydia trachomatis infection. J. Infect. 47:45–51 [DOI] [PubMed] [Google Scholar]
- 17. Grieshaber S. S., Grieshaber N. A., Miller N., Hackstadt T. 2006. Chlamydia trachomatis causes centrosomal defects resulting in chromosomal segregation abnormalities. Traffic 7:940–949 [DOI] [PubMed] [Google Scholar]
- 18. Heuer D., Kneip C., Maurer A. P., Meyer T. F. 2007. Tackling the intractable: approaching the genetics of Chlamydiales. Int. J. Med. Microbiol. 297:569–576 [DOI] [PubMed] [Google Scholar]
- 19. Heuer D., et al. 2009. Chlamydia causes fragmentation of the Golgi compartment to ensure reproduction. Nature 457:731–735 [DOI] [PubMed] [Google Scholar]
- 20. Hickson G. R., O'Farrell P. H. 2008. Anillin: a pivotal organizer of the cytokinetic machinery. Biochem. Soc. Trans. 36:439–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hill E., Clarke M., Barr F. A. 2000. The Rab6-binding kinesin, Rab6-KIFL, is required for cytokinesis. EMBO J. 19:5711–5719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hybiske K., Stephens R. S. 2007. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc. Natl. Acad. Sci. U. S. A. 104:11430–11435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jackson L. A., Wang S. P., Nazar-Stewart V., Grayston J. T., Vaughan T. L. 2000. Association of Chlamydia pneumoniae immunoglobulin A seropositivity and risk of lung cancer. Cancer Epidemiol. Biomarkers Prev. 9:1263–1266 [PubMed] [Google Scholar]
- 24. Johnson K. A., Tan M., Sutterlin C. 2009. Centrosome abnormalities during a Chlamydia trachomatis infection are caused by dysregulation of the normal duplication pathway. Cell Microbiol. 11:1064–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kalliala I., Anttila A., Pukkala E., Nieminen P. 2005. Risk of cervical and other cancers after treatment of cervical intraepithelial neoplasia: retrospective cohort study. BMJ 331:1183–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kinchen J. M., Ravichandran K. S. 2008. Phagosome maturation: going through the acid test. Nat. Rev. Mol. Cell. Biol. 9:781–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Knowlton A. E., et al. 2011. Chlamydia trachomatis infection causes mitotic spindle pole defects independently from its effects on centrosome amplification. Traffic 12:854–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Littman A. J., et al. 2004. Chlamydia pneumoniae infection and risk of lung cancer. Cancer Epidemiol. Biomarkers Prev. 13:1624–1630 [PubMed] [Google Scholar]
- 29. Madeleine M. M., et al. 2007. Risk of cervical cancer associated with Chlamydia trachomatis antibodies by histology, HPV type, and HPV cofactors. Int. J. Cancer 120:650–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matsumura F. 2005. Regulation of myosin II during cytokinesis in higher eukaryotes. Trends Cell Biol. 15:371–377 [DOI] [PubMed] [Google Scholar]
- 31. McClarty G., Tipples G. 1991. In situ studies on incorporation of nucleic acid precursors into Chlamydia trachomatis DNA. J. Bacteriol. 173:4922–4931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Munsie L. N., Caron N., Desmond C. R., Truant R. 2009. Lifeact cannot visualize some forms of stress-induced twisted F-actin. Nat. Methods 6:317. [DOI] [PubMed] [Google Scholar]
- 33. Neef R., et al. 2007. Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat. Cell Biol. 9:436–444 [DOI] [PubMed] [Google Scholar]
- 34. Nigg E. A. 2002. Centrosome aberrations: cause or consequence of cancer progression? Nat. Rev. Cancer 2:815–825 [DOI] [PubMed] [Google Scholar]
- 35. Nishimura Y., Yonemura S. 2006. Centralspindlin regulates ECT2 and RhoA accumulation at the equatorial cortex during cytokinesis. J. Cell Sci. 119:104–114 [DOI] [PubMed] [Google Scholar]
- 36. Olaharski A. J., et al. 2006. Tetraploidy and chromosomal instability are early events during cervical carcinogenesis. Carcinogenesis 27:337–343 [DOI] [PubMed] [Google Scholar]
- 37. Petronczki M., Glotzer M., Kraut N., Peters J. M. 2007. Polo-like kinase 1 triggers the initiation of cytokinesis in human cells by promoting recruitment of the RhoGEF Ect2 to the central spindle. Dev. Cell 12:713–725 [DOI] [PubMed] [Google Scholar]
- 38. Piekny A., Werner M., Glotzer M. 2005. Cytokinesis: welcome to the Rho zone. Trends Cell Biol. 15:651–658 [DOI] [PubMed] [Google Scholar]
- 39. Rappaport R., Conrad G. W. 1963. An experimental analysis of unilateral cleavage in invertebrate eggs. J. Exp. Zool. 153:99–112 [DOI] [PubMed] [Google Scholar]
- 40. Rappaport R., Rappaport B. 1983. Cytokinesis: effects of blocks between the mitotic apparatus and the surface on furrow establishment in flattened echinoderm eggs. J. Exp. Zool. 227:213–227 [Google Scholar]
- 41. Riedl J., et al. 2008. Lifeact: a versatile marker to visualize F-actin. Nat. Methods 5:605–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rzomp K. A., Scholtes L. D., Briggs B. J., Whittaker G. R., Scidmore M. A. 2003. Rab GTPases are recruited to chlamydial inclusions in both a species-dependent and species-independent manner. Infect. Immun. 71:5855–5870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Samoff E., et al. 2005. Association of Chlamydia trachomatis with persistence of high-risk types of human papillomavirus in a cohort of female adolescents. Am. J. Epidemiol. 162:668–675 [DOI] [PubMed] [Google Scholar]
- 44. Sauer G., et al. 2005. Proteome analysis of the human mitotic spindle. Mol. Cell Proteomics 4:35–43 [DOI] [PubMed] [Google Scholar]
- 45. Somers W. G., Saint R. 2003. A RhoGEF and Rho family GTPase-activating protein complex links the contractile ring to cortical microtubules at the onset of cytokinesis. Dev. Cell 4:29–39 [DOI] [PubMed] [Google Scholar]
- 46. Steigemann P., et al. 2009. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell 136:473–484 [DOI] [PubMed] [Google Scholar]
- 47. Storchova Z., Pellman D. 2004. From polyploidy to aneuploidy, genome instability and cancer. Nat. Rev. Mol. Cell. Biol. 5:45–54 [DOI] [PubMed] [Google Scholar]
- 48. Valdivia R. H. 2008. Chlamydia effector proteins and new insights into chlamydial cellular microbiology. Curr. Opin. Microbiol. 11:53–59 [DOI] [PubMed] [Google Scholar]
- 49. Vieira O. V., et al. 2004. Acquisition of Hrs, an essential component of phagosomal maturation, is impaired by mycobacteria. Mol. Cell. Biol. 24:4593–4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. von Schubert C., et al. 2010. The transforming parasite Theileria co-opts host cell mitotic and central spindles to persist in continuously dividing cells. PLoS Biol. 8:e1000499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Walczak C. E., Cai S., Khodjakov A. 2010. Mechanisms of chromosome behaviour during mitosis. Nat. Rev. Mol. Cell. Biol. 11:91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wilson G. M., et al. 2005. The FIP3-Rab11 protein complex regulates recycling endosome targeting to the cleavage furrow during late cytokinesis. Mol. Biol. Cell 16:849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wolfe B. A., Takaki T., Petronczki M., Glotzer M. 2009. Polo-like kinase 1 directs assembly of the HsCyk-4 RhoGAP/Ect2 RhoGEF complex to initiate cleavage furrow formation. PLoS Biol. 7:e1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Workowski K. A., Berman S. M. 2006. Sexually transmitted diseases treatment guidelines, 2006. MMWR Morb. Mortal. Wkly. Rep. 55(RR11):1–94 [PubMed] [Google Scholar]
- 55. World Health Organization 2001. Global prevalence and incidence of selected curable sexually transmitted diseases: overview and estimates. World Health Organization, Geneva, Switzerland [Google Scholar]
- 56. Yuce O., Piekny A., Glotzer M. 2005. An ECT2-centralspindlin complex regulates the localization and function of RhoA. J. Cell Biol. 170:571–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhao W. M., Fang G. 2005. MgcRacGAP controls the assembly of the contractile ring and the initiation of cytokinesis. Proc. Natl. Acad. Sci. U. S. A. 102:13158–13163 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.