Abstract
The Warburg effect describes an increase in aerobic glycolysis and enhanced lactate production in cancer cells. Lactate dehydrogenase A (LDH-A) regulates the last step of glycolysis that generates lactate and permits the regeneration of NAD+. LDH-A gene expression is believed to be upregulated by both HIF and Myc in cancer cells to achieve increased lactate production. However, how oncogenic signals activate LDH-A to regulate cancer cell metabolism remains unclear. We found that the oncogenic receptor tyrosine kinase FGFR1 directly phosphorylates LDH-A. Phosphorylation at Y10 and Y83 enhances LDH-A activity by enhancing the formation of active, tetrameric LDH-A and the binding of LDH-A substrate NADH, respectively. Moreover, Y10 phosphorylation of LDH-A is common in diverse human cancer cells, which correlates with activation of multiple oncogenic tyrosine kinases. Interestingly, cancer cells with stable knockdown of endogenous LDH-A and rescue expression of a catalytic hypomorph LDH-A mutant, Y10F, demonstrate increased respiration through mitochondrial complex I to sustain glycolysis by providing NAD+. However, such a compensatory increase in mitochondrial respiration in Y10F cells is insufficient to fully sustain glycolysis. Y10 rescue cells show decreased cell proliferation and ATP levels under hypoxia and reduced tumor growth in xenograft nude mice. Our findings suggest that tyrosine phosphorylation enhances LDH-A enzyme activity to promote the Warburg effect and tumor growth by regulating the NADH/NAD+ redox homeostasis, representing an acute molecular mechanism underlying the enhanced lactate production in cancer cells.
INTRODUCTION
Cancer cells take up more glucose than normal tissue and favor aerobic glycolysis, generating lactate through a NADH-dependent enzyme, lactate dehydrogenase A (LDH-A), which catalyzes the conversion of pyruvate to lactate during glycolysis. This is the last step of glycolysis that permits the regeneration of NAD+, which is needed as an electron acceptor to maintain cytosolic glucose catabolism (2). Therefore, most tumor cells are reliant on lactate production for their survival.
LDH-A gene expression is believed to be upregulated by both HIF and Myc in cancer cells to achieve increased lactate production (1, 7, 16, 25-27). In addition, expression of LDH-A was previously implicated to be involved in tumor initiation and growth. Targeting LDH-A by short hairpin RNA (shRNA) in several tumor cell lines is sufficient to stimulate oxidative phosphorylation in these cells, which is accompanied by an increase in the rate of oxygen consumption and a decrease in mitochondrial membrane potential (5). This provides evidence of the direct link between glycolysis and oxidative phosphorylation that involves LDH-A. Moreover, RNA interference (RNAi)-mediated reduction of LDH-A expression compromises the ability of tumor cells to proliferate under hypoxia and induce tumorigenesis (5). Recently, it was reported that targeting LDH-A by a small-molecule inhibitor, FX11, induced significant oxidative stress and cell death, as well as attenuated tumor growth in xenograft nude mouse models of human lymphoma and pancreatic cancer (17).
However, how oncogenic signals activate LDH-A to regulate cancer cell metabolism remains unclear. The molecular mechanisms underlying the Warburg effect are complex. Cell surface growth factor receptors, which often carry tyrosine kinase activities in their cytoplasmic domains, are overexpressed in many human cancers and are believed to play a key role in determining cell metabolism (9, 10). We have previously explored the hypothesis that tyrosine kinase signaling, which is commonly increased in tumors, regulates the Warburg effect and contributes to tumorigenesis and maintenance of the tumor. Using a phosphoproteomics-based study, we found that tyrosine phosphorylation inhibits a metabolic enzyme, pyruvate kinase M2 isoform (PKM2) in cancer cells, which represents a common mechanism to promote the Warburg effect and provide a metabolic advantage to tumorigenesis and tumor growth (12). We report here that tyrosine phosphorylation activates LDH-A to promote cancer cell metabolism and tumor growth by regulating NADH/NAD+ redox homeostasis in cancer cells, which represents an acute molecular mechanism underlying the Warburg effect and lactate production, in addition to the chronic mechanism that is believed to be regulated by HIF and Myc.
MATERIALS AND METHODS
Phosphoproteomics studies.
Phosphoproteomics studies were performed as described previously (12) by using a PhosphoScan kit (Cell Signaling Technologies [CST]). Tandem mass spectra were collected in a data-dependent manner with an LTQ ion trap mass spectrometer (ThermoFinnigan).
Reagents.
Tyrosine kinase inhibitor (TKI258) was provided by Novartis Pharma. shRNA constructs for LDH-A knockdown were purchased from Open Biosystems. LDH-A variants were subcloned into pDEST27 (Invitrogen) and pET53 (Novagen) vectors for glutathione S-transferase (GST)-tagged LDH-A expression in mammalian cells and His-tagged LDH-A expression in bacterial cells, respectively. The mutations Y10F, Y83F, and Y172F were introduced into LDH-A by using a QuikChange-XL site-directed mutagenesis kit (Stratagene). Phospho-Tyr pY99, FGFR1, c-ABL, and FLT-3 antibodies were obtained from Santa Cruz Biotechnology; LDH-A, His, and JAK2 antibodies were from CST; and antibodies against GST, Flag, and β-actin were from Sigma. Specific antibody against phospho-LDH-A (Y10) was generated by CST. Polyclonal antibody was produced by immunizing rabbits with a synthetic phosphopeptide (KLH-coupled) corresponding to residues surrounding Y10 of human LDH-A. Antibodies were purified by protein A and peptide affinity chromatography at CST.
Cell culture.
H1299, KG-1a, K562, HEL, Molm14, EOL-1, and MDA-MB231 cells were cultured in RPMI 1640 medium with 10% fetal bovine serum (FBS). H157, H358, and H226 cells were cultured in RPMI 1640 with glutamine and 5% FBS. 293T and MCF-7 cells were cultured in Dulbecco modified Eagle medium (DMEM) with 10% FBS. The 212LN, 686LN, Tu212, and Tu686 cell lines were cultured in DMEM-Ham F-12 50/50 mix medium in the presence of 10% FBS. 22RV cells were cultured in RPMI 1640 medium with 10% FBS, 1 mM sodium pyruvate, and 10 mM HEPES. PC3 cells were cultured in F12 Kaighn's medium with 5% FBS. A cell proliferation assay was performed by seeding 5 × 104 cells in a six-well plate and culturing the cells at 37°C in normoxia (5% CO2 and 95% air). At 24 h after seeding, cells that were used for further culture under hypoxia were cultured at 37°C in a sealed hypoxia chamber filled with 1% O2, 5% CO2, and 94% N2. Cell proliferation was determined by cell numbers recorded at 48 and 96 h after being seeded and normalized to that of each of the cell lines at the starting time (T = 0 h). LDH-A rescue H1299 cell lines were generated as previously described (12) using Flag-tagged human LDH-A WT, Y10F, and Y172F in retroviral vector pLHCX (Clontech). Stable knockdown of endogenous hLDH-A was achieved using lentiviral vector harboring shRNA construct (Open Biosystems; 5′-CCGGGATCTGTGATTAAAGCAGTAACTCGAGTTACTGCTTTAATCACAGATCTTTTT-3′). The shRNA is designed to target the 3′ noncoding region of hLDH-A mRNA and shows no effect on the plasmid-directed expression of LDH-A cDNA in cells.
Purification of recombinant LDH-A proteins and in vitro kinase assays.
His6-tagged LDH-A proteins were purified by sonication of BL21(DE3)pLysS cells obtained from 250 ml of culture with IPTG (isopropyl-β-d-thiogalactopyranoside) induction at 37°C for 4 h. The cell lysates were resolved by centrifugation and loaded onto a Ni-NTA column in 20 mM imidazole. After two washes, the protein was eluted with 250 mM imidazole. Proteins were desalted on a PD-10 column, and the purification efficiency was examined by Coomassie staining and Western blotting. In vitro FGFR1, ABL, FLT3, and JAK2 kinase assays were previously described (12).
LDH enzyme activity and lactate production assays.
LDH activity was determined by measuring the decrease of fluorescence intensity at 340 nm from the oxidation of NADH in 20 mM HEPES-K+ (pH 7.2), 0.05% bovine serum albumin, 20 μM NADH, and 2 mM pyruvate (Sigma) using a spectrofluorimeter (excitation, 340 nm; emission, 460 nm) as previously described (24). Cellular lactate production was measured under normoxia with a fluorescence-based lactate assay kit (MBL). Phenol red-free RPMI medium without FBS was added to a six-well plate of subconfluent cells and incubated for 1 h at 37°C. After incubation, 1 μl of medium from each well was assessed by using a lactate assay kit. Cell numbers were counted by using a microscope (magnification, ×400).
Oxygen consumption rate, NADH/NAD+ ratio, intracellular ATP concentration, glucose utilization, and glycolytic rate assays.
Oxygen consumption rates were measured with a Clark type electronode equipped with a 782 oxygen meter (Strathkelvin Instruments). A total of 107 cells were resuspended in RPMI 1640 medium with 10% FBS and placed in a water-jacked chamber RC300 (Strathkelvin Instruments), and recording was started immediately. The NADH/NAD+ ratios were determined by measuring NADH/NAD+ concentrations according to the protocol (BioAssay Systems). In brief, NADH and NAD+ were extracted from 105 H1299 rescue cells separately and measured at 565 nm after an enzyme-catalyzed kinetic reaction, in which the intensity of the product color is proportionate to the NADH/NAD+ concentrations in the samples. Intracellular ATP concentration was measured by an ATP bioluminescent somatic cell assay kit (Sigma). A total of 106 cells were treated with trypsin and resuspended in ultrapure water. Luminescence was measured with spectrofluorometer (Spectra Max Gemini; Molecular Probe) immediately after the addition of ATP enzyme mix to cell suspension. Glucose utilization assay was performed as described previously (11), and 106 cells were plated onto a 6-cm dish 1 day prior to the assay. The media were replaced with phenol-red free RPMI with 1% FBS prior to continuous culture for 3 days. Medium samples were collected each day. Glucose concentrations in media were measured using a colorimetric glucose assay kit (Biovision) and normalized with cell numbers. A glycolytic rate assay was performed as described previously (29). In brief, 0.5 × 106 cells were washed once in phosphate-buffered saline (PBS) prior to incubation in 1 ml of Krebs buffer without glucose for 30 min at 37°C. The Krebs buffer was replaced with Krebs buffer containing 10 mM glucose spiked with 10 μCi of [3H]glucose. After incubation for 1 h at 37°C, triplicate 50-μl aliquots were transferred to uncapped PCR tubes containing 50 μl of 0.2 N HCl, and each tube was transferred into an Eppendorf tube containing 0.5 ml of H2O for diffusion. The tubes were sealed, and diffusion was allowed to occur for a minimum of 24 h at 34°C. The amounts of diffused 3H2O were determined by scintillation counting.
NADH binding ability assay.
The NADH binding ability of LDH-A was determined by measuring the affinity of LDH-A to agarose-immobilized Cibacron Blue 3GA, which mimics NADH. Then, 400 ng of purified recombinant LDH-A variants were incubated with recombinant active FGFR1 in the presence or absence of ATP, followed by incubation with 30 μl of Cibacron Blue agarose (Sigma) at 4°C for 2 h. After a washing step with 20 mM Tris-HCl (pH 8.6), bound LDH-A was purified and eluted in PBS and subjected to SDS-PAGE, followed by Western blotting. The same amount of protein was loaded as input to ensure equivalent protein amounts in each reaction. The relative NADH binding activities were determined from the ratio of the amounts of bound LDH-A to input protein.
Xenograft studies.
Nude mice (nu/nu, female 6 to 8 weeks old, Harlan Labs) were subcutaneously injected with 2 × 107 H1299 cells stably expressing LDH-A wild type (WT) and Y10F mutant in conjunction with stable knockdown of endogenous LDH-A on the left and right flanks, respectively. Tumor formation was assessed every 2 to 3 days. Tumor growth was recorded by measuring two perpendicular diameters of the tumors over a 4-week time course according to the formula 4π/3 × (width/2)2 × (length/2). The tumors were harvested and weighed at the experimental endpoint, and the masses of tumors (in grams) derived from cells expressing LDH-A WT or Y10F mutant in both flanks of each mouse were compared. Statistical analyses were performed in comparison to the control group by using a paired Student t test.
Statistical analysis.
Statistical analysis and graphical presentation was done using GraphPad Prism 4.0. The data shown are from one representative experiment of multiple independent experiments and are given as mean ± the standard deviation. Statistical analysis of significance (P values) was based on a two-tailed Student t test. The P value of the xenograft experiment was determined by a paired Student t test.
RESULTS
LDH-A is tyrosine phosphorylated and activated by FGFR1 in cancer cells.
To better understand how tyrosine kinase signaling, commonly upregulated in tumors, regulates the Warburg effect, we previously performed a mass spectrometry-based proteomics study (12) using murine hematopoietic Ba/F3 cells stably expressing ZNF198-FGFR1, a constitutively active fusion tyrosine kinase associated with the t(8;13)(p11;q12) stem cell myeloproliferative disorder (30). We identified a group of enzymes that regulate metabolism, including LDH-A, pyruvate kinase M2 isoform (PKM2), glucose-6-phosphate dehydrogenase, and malate dehydrogenase 2 as tyrosine phosphorylated in Ba/F3 cells containing ZNF198-FGFR1 but not in control cells grown in the absence of interleukin-3. We further demonstrated that tyrosine phosphorylation at Y105 inhibits PKM2 as a common mechanism to promote the Warburg effect in cancer cells and tumor growth (12).
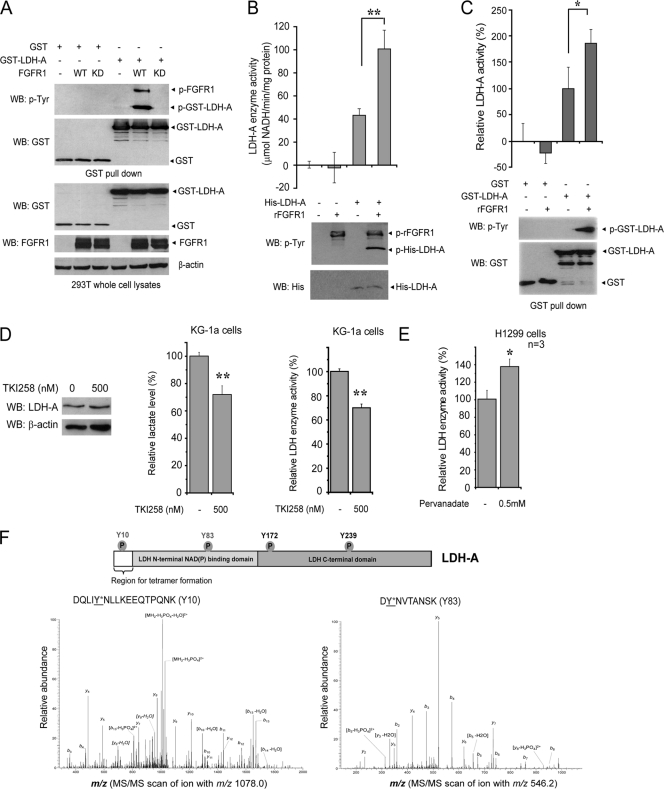
We next sought to explore the effect of tyrosine phosphorylation on LDH-A activity and cancer cell metabolism. GST-tagged LDH-A was tyrosine phosphorylated in 293T cells transiently cotransfected with plasmids encoding FGFR1 wild type (WT) but not in cells coexpressing GST-LDH-A and a kinase- dead (KD) form of FGFR1 (Fig. 1A). Moreover, in an in vitro kinase assay, incubation with recombinant FGFR1 (rFGFR1) results in significantly elevated LDH-A enzyme activity with increased phosphorylation levels at tyrosine residues of recombinant His-tagged LDH-A (Fig. 1B) or purified GST-LDH-A (Fig. 1C). Overexpression of FGFR1 or its mutational activation have been implicated in various human solid tumors, including breast cancer, pancreatic adenocarcinoma, and malignant astrocytoma (15, 18, 20, 21). We found that treatment with the FGFR1 inhibitor TKI258 (Fig. 1D, left) significantly decreased lactate production (Fig. 1D, middle) and reduced LDH enzymatic activity (Fig. 1D, right) in human myeloid leukemia KG-1a cells harboring the FOP2-FGFR1 fusion protein (8). Moreover, treatment with the protein tyrosine phosphatase inhibitor pervanadate resulted in increased LDH enzyme activity in human lung cancer NCI-H1299 cells overexpressing FGFR1 (19) (Fig. 1E).
Fig. 1.
Oncogenic FGFR1 phosphorylates and activates LDH-A. (A) Immunoblotting of 293T cell lysates for tyrosine phosphorylation of GST-LDH-A when coexpressed with FGFR1 wild type (WT) or a kinase-dead form (KD). (B) Active rFGFR1 directly phosphorylates purified, recombinant His-tagged LDH-A at tyrosine residues in an in vitro kinase assay (lower) and activates LDH-A enzyme activity (upper; **, P < 0.01). (C) GST-LDH-A was pulled down by beads from transfected 293T cell lysates and treated with active rFGFR1. Tyrosine phosphorylation of GST-LDH-A activates LDH-A enzyme activity (*, 0.01 < P < 0.05). (D) Inhibition of FGFR1 by tyrosine kinase inhibitor TKI258 does not alter LDH-A protein levels (left) but results in decreased lactate production (middle) and reduced LDH enzyme activity (right) in leukemia KG-1a cells (FOP2-FGFR1) (**, P < 0.01). Relative lactate production and LDH activity were normalized to those in control cells without TKI258 treatment. (E) Treatment with the tyrosine phosphatase inhibitor pervanadate (0.5 mM for 15 min) results in increased LDH enzyme activity in lung cancer H1299 cells expressing FGFR1 (*, 0.01 < P < 0.05). Relative LDH activity was normalized to that in control cells without pervanadate treatment. (F) In the top panel a schematic representation of LDH-A is shown. The four phosphorylated tyrosine residues are indicated, including FGFR1-direct phosphorylation sites (Y10 and Y83) and FGFR1-indirect phosphorylation sites (Y172 and Y239). In the bottom panel, Y10 and Y83 phosphorylation of LDH-A was determined by mass spectrometry. Recombinant LDH-A phosphorylated by active, recombinant FGFR1 was applied to SDS-PAGE, and the band containing LDH-A was excised, followed by in-gel digestion with trypsin. The resulting peptides were harvested and analyzed by reversed-phase liquid chromatography-tandem mass spectrometry (LC-MS/MS). MS/MS scan of the precursor ions m/z 1,078.0 (left) and m/z 546.2 (right), which were fragmented into multiple labeled product ions (b and y ions), led to identification of two peptides harboring Y10 and Y83, respectively, according to the mass shift (+80 Da) due to phosphorylation. The neutral loss of water also occurred during the fragmentation. Y10 was also identified as phosphorylated in another peptide (DQLIYNLLK) with m/z 600.3 (data not shown).
FGFR1 activates LDH-A through direct phosphorylation at Y10 and Y83 of LDH-A, which promotes the formation of active, tetrameric LDH-A and the binding of substrate NADH, respectively.
Figure 1F shows a schematic illustration of LDH-A and potential FGFR1-dependent tyrosine phosphorylation sites. Our previous phospho-proteomics studies showed that LDH-A is phosphorylated at Y172 and Y239 in Ba/F3 cells transformed by active FGFR1 fusion tyrosine kinases (12). However, further mass spectrometry-based studies revealed that rFGFR1 directly phosphorylates purified, recombinant LDH-A at Y10 and Y83 in an in vitro kinase assay, but not at Y172 and Y239 as predicted. FGFR1 may activate alternative tyrosine kinases in cells which subsequently phosphorylate LDH-A at Y172 and Y239; phosphorylation of LDH-A Y239 was previously implied to correlate with LDH-A nuclear localization in cancer cells (32). Mouse LDH-A harbors V10 instead of Y10, which explains why we could not detect Y10 phosphorylation in murine hematopoietic Ba/F3 cells expressing ZNF198-FGFR1 in the phosphoproteomics studies. However, our coworkers at CST have found in phosphoproteomics-based studies that Y10 of LDH-A is phosphorylated in both human cancer tissue samples and cancer cell lines established from different malignancies (http://www.phosphosite.org/siteAction.do?id=19619). We also proposed that the phosphorylation stoichiometry of Y83 in murine Ba/F3 cells might not be sufficient to be detected by mass spectrometry in the phosphoproteomics studies. Thus, we decided to focus on the two FGFR1 direct phosphorylation sites, Y10 and Y83.
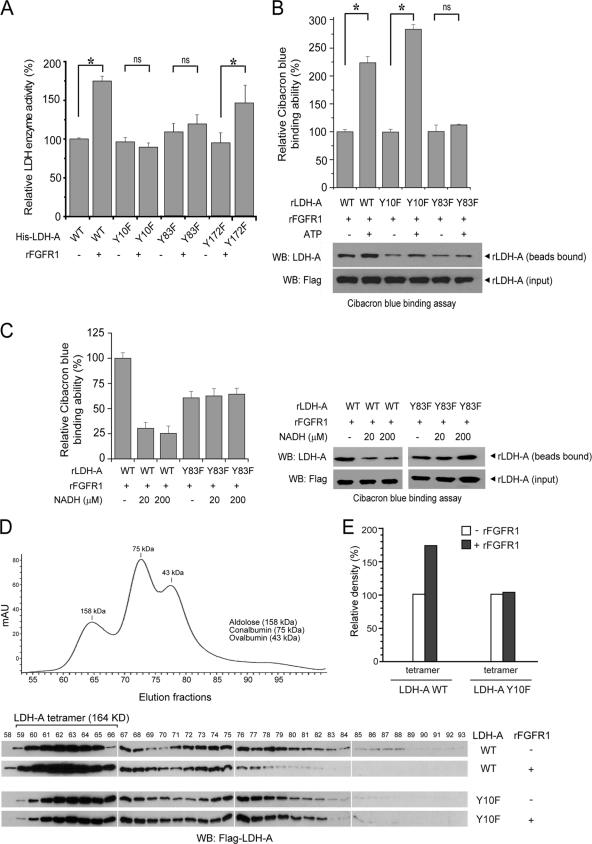
We performed in vitro kinase assays followed by an LDH-A enzyme activity assay, in which active rFGFR1 was incubated with purified His-tagged LDH-A WT or mutants including Y10F, Y83F, and control Y172F. We observed that phosphorylation by FGFR1 significantly increased the enzyme activity of LDH-A WT and Y172F. In contrast, substitution of Y10 or Y83 abolished the FGFR1-dependent increase in the LDH-A enzyme activity (Fig. 2A). Previous structural studies have shown that Y83 is directly proximal to the substrate NADH binding site in the human muscle l-lactate dehydrogenase when in complex with NADH and oxamate (22), suggesting that FGFR1 may phosphorylate LDH-A at Y83 to alter substrate (NADH) binding to LDH-A, respectively. To test this hypothesis, we incubated active rFGFR1 with purified, recombinant LDH-A WT, Y10F, or Y83F in an in vitro kinase assay, followed by incubation with Cibacron Blue 3GA agarose. Cibacron Blue 3GA mimics NADH and is a pseudo-affinity dye ligand of many dehydrogenases, including LDH-A (4), which use NADH (or NADPH) as a substrate. As shown in Fig. 2B (top and bottom), phosphorylation of LDH-A WT or Y10F mutant by FGFR1 resulted in a significant increase in the amount of LDH-A bound to the Cibacron Blue agarose beads, indicating increased binding between LDH-A and substrate NADH. In contrast, substitution of Y83 abolished the enhanced binding between LDH-A and NADH in the presence of rFGFR1 and ATP. Moreover, in a NADH competition experiment, incubation with NADH results in decreased LDH-A WT but not Y83F mutant amounts bound to the Cibacron Blue agarose beads upon phosphorylation by rFGFR1 (Fig. 2C, left and right).
Fig. 2.
FGFR1 activates LDH-A via phosphorylation at Y10 and Y83, which promotes formation of active, tetrameric LDH-A and cofactor NADH binding, respectively. (A) Mutational analysis revealed that substitution of Y10 or Y83, but not Y172, abolishes FGFR1-dependent increase of LDH-A enzyme activity (ns, not significant; *, 0.01 < P < 0.05). Relative enzyme activity was normalized to that of sample using His-LDH-A WT without rFGFR1 treatment. (B) His-LDH-A WT, Y10F, or Y83F were incubated with rFGFR1 in the presence or absence of ATP, followed by incubation with Cibacron Blue 3GA agarose. Relative Cibacron Blue-binding (NADH binding [top]) that was normalized to each His-LDH-A variant in the absence of ATP was determined based on the relative intensity of each bound His-LDH-A protein (bottom), which was normalized to the individual input amount of protein (*, 0.01 < P < 0.05; ns, not significant). All experiments were performed at least three times with similar results. (C) Incubation with NADH results in decreased LDH-A WT but not Y83F mutant amounts bound to the Cibacron Blue agarose beads upon phosphorylation by rFGFR1. Relative Cibacron Blue binding (left) that was normalized to His-LDH-A WT in the presence of rFGFR1 and ATP but absence of NADH was determined based on the relative intensity of each bound His-LDH-A protein (right), which was normalized to the individual input amount of protein. (D) Purified recombinant LDH-A WT and Y10F proteins were incubated with active, recombinant FGFR1 in the in vitro kinase assay, followed by gel filtration chromatography. The collected fractions were analyzed by Western blotting. (E) Summary results of combined densitometry of LDH-A bands in fractions 59 to 66, which represent tetrameric LDH-A proteins.
Although Y10 is distal from the binding sites of both substrates, pyruvate and NADH, in the human muscle l-lactate dehydrogenase when in complex with NADH and oxamate (22), Y10 is part of an N-terminal region of approximately 20 residues that was reported to contribute to the stabilization of tetrameric LDH (23, 31), which represents “a dimer of dimers” and is catalytically more active than the dimeric form of LDH-A (6, 13). Thus, we hypothesized that FGFR1 may phosphorylate LDH-A at Y10 to alter formation of active, tetrameric LDH-A. We performed a gel filtration chromatography experiment to determine the oligomeric state of LDH-A upon treatment with FGFR1. Purified recombinant rLDH-A WT and Y10F proteins were incubated with recombinant, active rFGFR1 in the in vitro kinase assay, followed by gel filtration chromatography. The collected fractions were analyzed by Western blotting. As shown in Fig. 2D, based on the results of Western blot and three markers of the gel filtration chromatography (ovalbumin, 43 kDa; conalbumin, 75 kDa; aldolose, 158 kDa), we determined that fractions 59 to 66 contain tetrameric LDH-A proteins (164 kDa). Figure 2E shows the summary results of densitometry, indicating that FGFR1-dependent phosphorylation enhances formation of tetrameric LDH-A WT, while substitution of Y10 abolishes the enhancement of tetramer formation.
LDH-A is specifically phosphorylated at Y10 in various cancer cell lines and by diverse oncogenic tyrosine kinases.
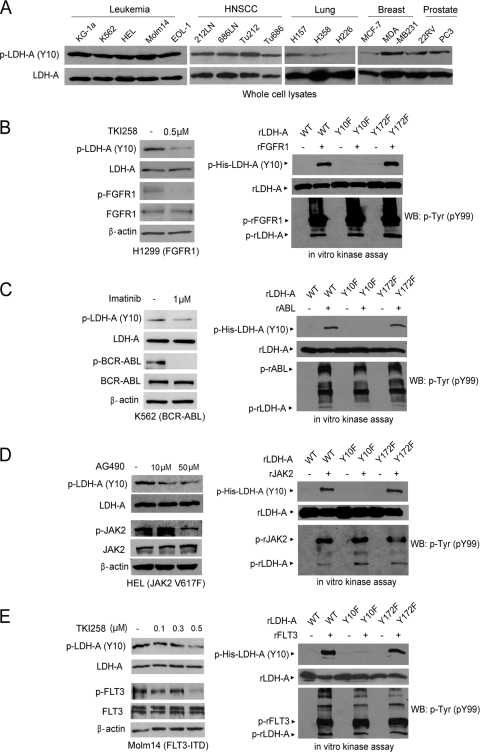
We also generated an antibody that specifically recognizes LDH-A phospho-Y10. We found that LDH-A was phosphorylated at Y10 in diverse hematopoietic cancer cell lines associated with various constitutively activated tyrosine kinase mutants. These included KG-1a (FOP2-FGFR1), K562 (BCR-ABL), HEL (JAK2 Val617Phe mutant), Molm14 (FLT3-internal tandem duplication [ITD] mutant), and EOL-1 (FIP1L1-PDGFRA). Moreover, we also found that Y10 phosphorylation of LDH-A was common in various human solid tumor cell lines, including 212LN, 686LN, Tu212, and Tu686 human head and neck squamous cell carcinoma cells, H157 and H358 lung cancer cells, MDA-MB231 breast cancer cells, and 22RV and PC3 prostate cancer cells. However, phosphorylation levels of Y10 of LDH-A were relatively low in H226 lung cancer and MCF-7 breast cancer cells (Fig. 3A).
Fig. 3.
LDH-A is specifically phosphorylated at Y10 in various cancer cell lines and by diverse oncogenic tyrosine kinases. (A) Immunoblotting detected Y10 phosphorylation of LDH-A in diverse leukemia (KG-1a, K562, HEL, Molm14, and EOL-1) cell lines and solid tumor cell lines, including head and neck cancer (212LN, 686LN, Tu212, and Tu686), lung cancer (H157 and H358), breast cancer (MDA-MB231), and prostate cancer (22RV and PC3), whereas LDH-A is relatively less phosphorylated in lung cancer H226 and breast cancer MCF-7 cells. (B to E) Immunoblotting shows that targeting FGFR1 by TKI258 in H1299 cells (B; left), BCR-ABL by imatinib in K562 cells (C; left), JAK2 by AG490 in HEL cells (D; left), or FLT3 in Molm14 cells by TKI258 (E; left) decreases phosphorylation of LDH-A Y10. Active, recombinant FGFR1 (B; right), ABL (C; right), JAK2 (D; right), and FLT3 (E; right) directly phosphorylate LDH-A at Y10 in corresponding in vitro kinase assays. Experiments were performed at least three times with similar results.
We also observed that inhibiting FGFR1 by TKI258 (Fig. 3B, left) resulted in decreased LDH-A Y10 phosphorylation in H1299 lung cancer cells and that rFGFR1 phosphorylated purified His-LDH-A WT and Y172F mutant, but not the Y10F mutant, at Y10 (Fig. 3B, right). Moreover, we found that ABL (Fig. 3C), JAK2 (Fig. 3D), and FLT3 (Fig. 3E) also directly phosphorylated LDH-A in the in vitro kinase assays using recombinant proteins, and inhibition of BCR-ABL by imatinib, JAK2 by AG490, and FLT3-ITD by TKI258 resulted in decreased Y10 phosphorylation of LDH-A in the pertinent human cancer cell lines. However, Y83 phosphorylation of LDH-A was not detected by the extensive phosphoproteomics-based studies performed by our coworkers at CST (http://www.phosphosite.org/proteinAction.do?id=4103&showAllSites=true) using diverse human tissue samples and cancer cell lines. This may suggest a low stoichiometry of Y83 phosphorylation in human cancer cells. Therefore, we continued focusing on LDH-A Y10 phosphorylation that physiologically occurs in human cancer cells.
Expression of the LDH-A Y10F mutant in cancer cells leads to decreased proliferation under hypoxia and increased respiration.
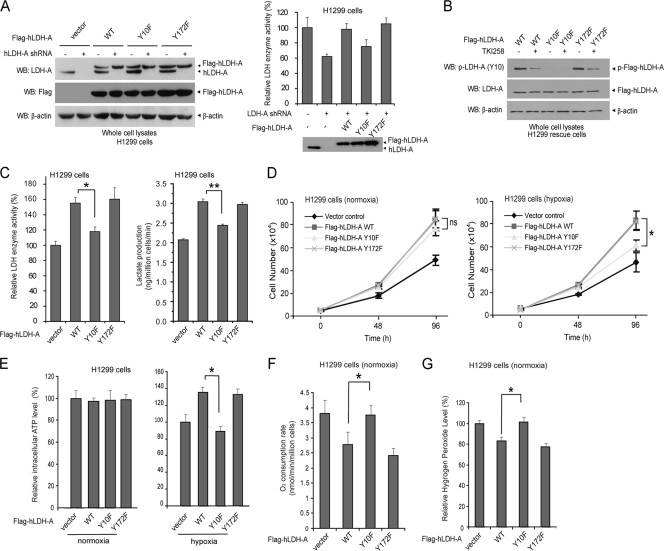
To elucidate the role of LDH-A Y10 phosphorylation in cancer cell metabolism and tumor growth, we used FGFR1-expressing human lung cancer H1299 cells to generate “rescue” cell lines as described previously (12) by RNAi-mediated stable knockdown of endogenous human LDH-A (hLDH-A) and rescue expression of Flag-tagged hLDH-A WT, Y10F, or Y172F mutants (Fig. 4A, left). The hLDH-A shRNA targets the 3′ noncoding region of hLDH-A mRNA and shows no effect on the plasmid directed expression of hLDH-A proteins in cells. Knockdown of endogenous LDH-A resulted in decreased LDH activity, while expression of LDH-A WT or Y172F mutant, but not Y10F mutant, rescued this phenotype in H1299 cells with LDH-A knockdown (Fig. 4A, right). Both Flag-hLDH-A WT and Y172F, but not Y10F, were phosphorylated at Y10 by FGFR1 in H1299 cells, and treatment with TKI258 decreased Y10 phosphorylation of WT and Y172F (Fig. 4B). In addition, the Y10F mutant showed decreased enzymatic activity (Fig. 4C, left) that led to a significant decrease in lactate production in Y10F rescue cells compared to cells with hLDH-A WT and Y172F (Fig. 4C, right). We also observed that, under normoxic conditions, cells rescued with any of the hLDH-A variants showed a comparable rate of proliferation that was greater than that of parental cells, in which endogenous hLDH-A was stably knocked down. However, Y10F rescue cells showed a significantly slower proliferation rate under hypoxic conditions than did cells rescued with WT or Y172F mutant (Fig. 4D). Moreover, although intracellular ATP concentrations in the parental and all of the rescue cells were comparable under normoxia, the parental cells with endogenous hLDH-A knocked down and Y10F rescue cells showed a significant decrease in the intracellular ATP concentrations under hypoxic conditions compared to hLDH-A WT or Y172F mutant rescued cells (Fig. 4E). These results are consistent with previous observations in murine mammary cancer cells with stable knockdown of mouse LDH-A (5). In addition, compared to cells rescued with hLDH-A WT and Y172F mutant, Y10F rescue cells and parental cells with stable knockdown of endogenous hLDH-A had a higher rate of oxygen consumption (Fig. 4F) and an increased production of hydrogen peroxide (Fig. 4G).
Fig. 4.
Expression of a catalytically less active LDH-A mutant, Y10F, in H1299 cells leads to decreased proliferation and ATP levels under hypoxic conditions with increased mitochondrial respiration. (A) Immunoblotting (left panel) shows shRNA-mediated stable knockdown of endogenous hLDH-A in H1299 cells using lentiviral transduction and rescue expression of Flag-tagged hLDH-A proteins, including WT, Y10F, and Y172F mutants. An LDH enzyme activity assay (right panel) using LDH-A knockdown and diverse rescue cell lines was performed. (B) Rescue expressed Flag-LDH-A WT and Y172F mutant, but not Y10F mutant, were phosphorylated at Y10 in H1299 cells expressing FGFR1. Treatment with TKI258 resulted in decreased Y10 phosphorylation of LDH-A in WT and Y172F rescue cells. (C) Y10F has significantly lower enzyme activity than Flag-hLDH-A WT or Y172F in rescue H1299 cells (left). Y10F rescue cells show significantly reduced lactate production under normoxia (right). (D to E) Rescue expression of hLDH-A Y10F in H1299 cells results in reduced cell proliferation rate (D) and intracellular ATP level (E) under hypoxic conditions (1%) but not at normal oxygen tension (normoxic; 17% oxygen) compared to cells expressing LDH-A WT or Y172F mutant. Cell proliferation was determined by the increase in cell number 96 h after seeding compared to that at seeding for each cell line (T = 0). The error bars represent mean values ± standard deviation from three independent experiments (*, 0.01 < P < 0.05). (F and G) Y10F rescue cells have a significantly elevated oxygen consumption rate (F) and an increased hydrogen peroxide production under normoxia (G) compared to cells expressing hLDH-A WT or Y172F mutant (*, 0.01 < P < 0.05). All of the experiments were performed at least three times with similar results.
Cancer cells expressing LDH-A Y10F mutant remain glycolytic and show increased mitochondrial respiration to sustain glycolysis by providing NAD+.
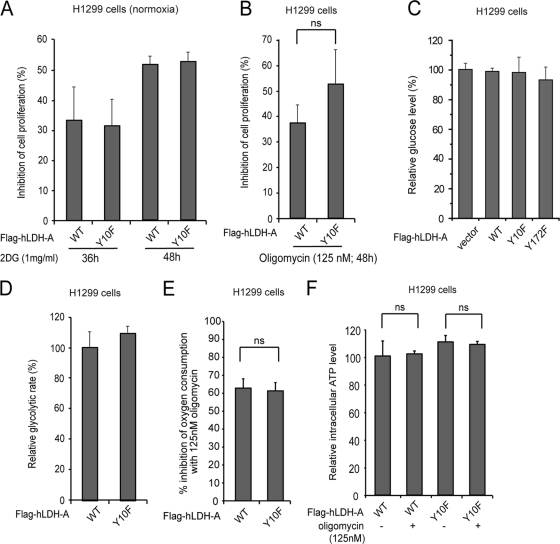
Tumor cells in general rely on cytosolic glycolysis and show reduced oxidative phosphorylation (OXPHOS) activity. We hypothesized that cells expressing LDH-A Y10F mutant might exhibit reduced glycolysis and rely more on OXPHOS that may correlate with the elevated oxygen consumption rate in these cells. However, treatment with the glycolytic inhibitor, 2-deoxy-d-glucose (2DG) (Fig. 5A) or oligomycin, a specific inhibitor of mitochondrial ATP synthase (Fig. 5B), resulted in reduced proliferation rates of Y10F cells that were comparable to those of cells with hLDH-A WT. Moreover, there was no significant difference in the glucose consumption rates and glycolytic rates between cells expressing hLDH-A WT and Y10F (Fig. 5C and D, respectively). Furthermore, although treatment with oligomycin resulted in comparable inhibition (∼60%) of oxygen consumption in both LDH-A WT and Y10F rescue cells (Fig. 5E), oligomycin treatment did not alter the ATP levels between rescue cells expressing hLDH-A WT or Y10F (Fig. 5F). These results together suggest that Y10F cells have increased O2 consumption but still rely on glycolysis instead of oxidative phosphorylation for ATP production, similar to cells with hLDH-A WT. Thus, the increased mitochondrial respiration in these cells contributes to ATP production in a manner that is independent of ATP synthase, probably by sustaining the cytosolic glycolysis.
Fig. 5.
Y10F rescue H1299 cells do not show increased oxidative phosphorylation. (A) Treatment with 2DG (1 mg/ml) resulted in a reduced proliferation rate of Y10F cells that was comparable to that of cells with hLDH-A WT. (B) Treatment with oligomycin (125 nM) for 48 h resulted in a reduced proliferation rate of Y10F cells that was comparable to that of cells with hLDH-A WT (ns = not significant). (C and D) Rescue cells expressing hLDH-A WT or Y10F have comparable glucose consumption rates (C) and glycolytic rates (D). (E and F) Oligomycin treatment resulted in comparable inhibition of oxygen consumption rates in both LDH-A WT and Y10F rescue cells (E) but did not affect the intracellular ATP levels in these cells (F).
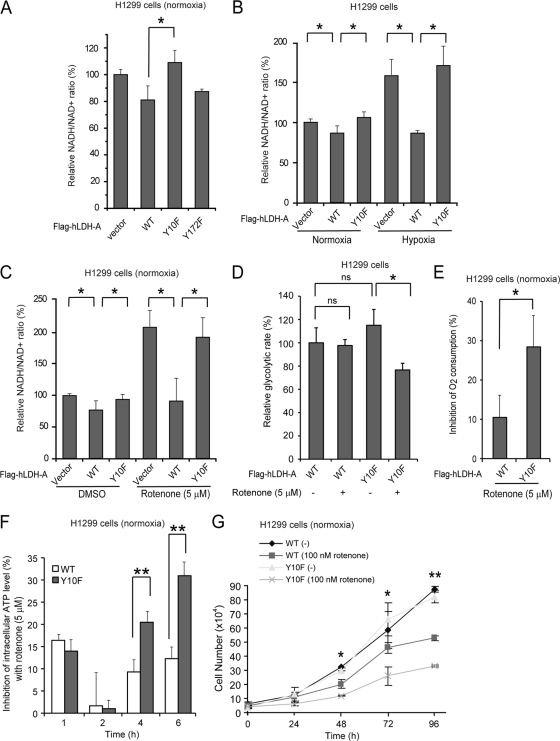
NADH could be shuttled by the malate/aspartate shuttle from the cytosol to the mitochondrial electron transport chain. Thus, one possibility is that Y10F cells may oxidize cytosolic NADH through the electron transport chain to sustain glycolysis by providing NAD+. To test this hypothesis, we examined the NADH/NAD+ ratio of these cells. Y10F rescue cells had a higher NADH/NAD+ ratio than did cells with hLDH-A WT or Y172F mutant under normoxia (Fig. 6A to C). In addition, switching to hypoxia condition or treatment with rotenone, a specific inhibitor of mitochondrial respiration chain complex I (NADH-coenzyme Q oxidoreductase, also known as NADH dehydrogenase), led to further increases in the NADH/NAD+ ratio (Fig. 6B and C, respectively), which corresponded to a significantly reduced glycolytic rate (Fig. 4D), increased inhibition of oxygen consumption (Fig. 6E) and ATP levels (Fig. 6F), and decreased proliferation rate (Fig. 6G) in Y10F rescue cells compared to those in cells with hLDH-A WT. Together, these data suggest that cells with a phosphorylation-deficient, catalytically less active form of hLDH-A (Y10F) rely more on mitochondrial respiration to supply NAD+ to sustain cytosolic glycolysis for ATP production and cell proliferation.
Fig. 6.
Y10F rescue H1299 cells rely more on mitochondrial complex I to sustain glycolysis by providing NAD+ compared to cells with LDH-A WT. (A) Y10F rescue cells show a significantly increased NADH/NAD+ ratio compared to cells with hLDH-A WT or Y172F mutant (*, 0.01 < P < 0.05). (B) Control LDH-A knockdown cells harboring an empty vector and Y10F rescue cells show a significantly increased NADH/NAD+ ratio compared to cells with hLDH-A WT under normoxia, and switching to hypoxic condition results in a significantly elevated NADH/NAD+ ratio compared to cells expressing LDH-A WT (*, 0.01 < P < 0.05). (C) Treatment of rescue cells harboring an empty vector or Y10F with rotenone leads to a significantly elevated NADH/NAD+ ratio compared to cells expressing LDH-A WT (*, 0.01 < P < 0.05). (D) Rotenone treatment resulted in a significantly decreased glycolytic rate in the Y10F rescue cells but not cells expressing LDH-A WT (*, 0.01 < P < 0.05). (E to G) Inhibition of mitochondrial complex I by rotenone results in a significantly increased inhibition of oxygen consumption rate (E) and ATP levels (F; after 4 h treatment), as well as a slower proliferation rate (G) in Y10F rescue cells, compared to those in cells with hLDH-A WT. At 48, 72, and 96 h, the decreased proliferation rates of the Y10F cells compared to cells with hLDH-A WT were statistically significant, as assessed by using the Student t test (*, 0.01 < P < 0.05; **, P < 0.01). All experiments were performed at least three times with similar results.
The presence of the LDH-A Y10F mutant in cancer cells leads to reduced tumor growth.
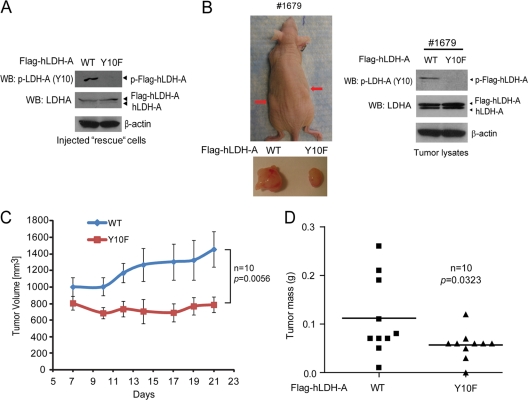
We next functionally validated these findings by performing xenograft experiments in which nude mice were injected with Flag-hLDH-A WT and Y10F rescue H1299 cells (Fig. 7A). Twenty million cells each were injected (Flag-hLDH-A WT rescue cells on the left flank and Y10F cells on the right flank; n = 10), and the mice were monitored for tumor growth over a 4-week time period. The growth rates and masses of tumors derived from Y10F rescue cells were significantly reduced compared to those of tumors formed by Flag-hLDH-A WT rescue cells (Fig. 7B to D). These results demonstrate that the presence of LDH-A Y10F in cancer cells results in attenuated tumor growth in vivo, suggesting that tyrosine phosphorylation of LDH-A confers a proliferative advantage.
Fig. 7.
Expression of LDH-A Y10F mutant in cancer cells leads to reduced tumor growth in xenograft nude mice. (A) Expression of Flag-tagged hLDH-A WT and Y10F detected by immunoblotting in injected rescue cells. Phosphorylation of LDH-A at Y10 was detected in LDH-A WT cells but not in cells expressing Y10F mutant. (B) The left panel shows dissected tumors (indicated by red arrows) in a representative nude mouse (animal 1679) injected with hLDH-A WT H1299 cells on the left flank and Y10F H1299 cells on the right flank are shown. In the right panel, the expression of hLDH-A WT and Y10F in tumor lysates was evaluated. (C and D) Y10F rescue cells show significantly reduced tumor growth rate (C; the P value was determined by a one-tailed Student t test) and masses (D; the P value was determined by a paired Student t test) in xenograft nude mice compared to cells with hLDH-A WT.
DISCUSSION
Our finding that tyrosine phosphorylation activates LDH-A may, at least in part, explain the enhanced lactate production in cancer cells. This could represent a common, short-term molecular mechanism underlying the Warburg effect in both leukemias and solid tumors, in addition to the chronic changes, including the upregulation of LDH-A gene expression, believed to be regulated by transcription factors, including HIF and Myc. Thus, tyrosine phosphorylation may provide a molecular switch upregulating LDH-A activity to provide a metabolic advantage facilitating tumor growth. Interestingly, Y10 is not evolutionarily conserved (Table 1). The occurrence of Y10 in the human LDH-A amino acid sequence is unique among mammals. This suggests that the Y10 phosphorylation-dependent regulation of LDH-A is specific for human cells.
Table 1.
Phylogenetic analysis of LDH-A in different species
| Species | LDH-A sequence (aa 1 to 10)a |
|---|---|
| Homo sapiens (human) | MATLKDQLIY |
| Mus musculus (mouse) | MATLKDQLIV |
| Cricetulus griseus (Chinese hamster) | MATLKDQLIV |
| Oryctolagus cuniculus (rabbit) | MAALKDQLIH |
| Pan troglodytes (chimpanzee) | MATLKDQLIH |
| Pongo abelii (Sumatran orangutan) | MATLKDQLIH |
| Chromis xanthochira (yellow-axil chromis) | MSTKDKLISH |
| Sus scrofa domesticus (domestic pig) | MATLKDQLIH |
| Sus scrofa (pig) | MATLKDQLIH |
| Capra hircus (goat) | MATLKNQLIQ |
| Equus caballus (horse) | MATVKDQLIQ |
| Monodelphis domestica (gray short-tailed opossum) | MGTVKDQLIL |
aa, amino acid.
Our findings demonstrate that tyrosine phosphorylation-dependent activation of LDH-A is important for redox homeostasis in cancer cells. The increased mitochondrial respiration in Y10F cells contributes to ATP production in a manner that appears to be independent of productive OXPHOS. These cells may still predominantly rely on cytosolic glycolysis, but they depend more on the increased mitochondrial respiration to generate NAD+ to sustain the levels of glycolysis. This explains the higher oxygen consumption rate in Y10F rescue cells compared to cells with hLDH-A WT. One concern about this model is that the slow rate of NADH shuttling from the cytosol to the mitochondrial electron transport chain, probably mediated by the malate/aspartate shuttle, may limit the supply of NADH to complex I. However, we observed that, in the stable “rescue” cells expressing LDH-A Y10F mutant, the total LDH activity is ca. 70% of that in cells expressing LDH-A WT (Fig. 4C). Therefore, the glycolysis in these cells may not entirely rely on NAD+ produced from the mitochondria. Thus, the slow rate of cytosolic NADH shuttling may still be sufficient to generate enough NAD+ from the mitochondria to essentially compensate the decreased supply of NAD+ in Y10F cells due to attenuated LDH-A activity.
However, such a compensatory increase in mitochondrial respiration in Y10F cells is unlikely to be sufficient to fully sustain glycolysis that is metabolically advantageous to the proliferative and tumorigenic potential of these cells, particularly under hypoxia. This may, in part, be due to the relatively slow rate of NADH shuttling from the cytosol to the mitochondrial electron transport chain (2). These findings are consistent with and would explain previous observations that targeting LDH-A by shRNA or small-molecule inhibitor attenuates cancer cell proliferation and tumor growth (5, 17). In addition, the finding that individuals with a complete genetic lack of LDH-A subunit production demonstrate only modest myoglobinuria after intense anaerobic exercise (14) identifies LDH-A as a promising therapeutic target to treat tumors that heavily rely on the Warburg effect for tumor cell survival and growth.
Our findings also suggest that oncogenic tyrosine kinase signaling may promote the Warburg effect by phosphorylating multiple metabolic enzymes, including LDH-A in the present report and previously reported PKM2 (12). Phosphorylation of Y105 inhibits PKM2 to promote a metabolic switch to aerobic glycolysis from oxidative phosphorylation in cancer cells, while phosphorylation at Y10 activates LDH-A to sustain the aerobic glycolysis by providing NAD+. It would be a bit difficult to reconcile the tyrosine phosphorylation-dependent enhanced lactate and NAD+ production with reduced PKM2 activity in cancer cells, since enhanced lactate production requires pyruvate produced by PKM2 but results in a net loss of carbon that could have been used for anabolic reactions. However, Vander Heiden et al. recently showed that the pyruvate kinase substrate, phosphoenolpyruvate (PEP) can transfer phosphate to the glycolytic enzyme phosphoglycerate mutase 1 (PGAM1) to phosphorylate the catalytic histidine 11 on PGAM1, producing pyruvate in the absence of PKM2 activity (28). In addition, it is possible that lactate production is fueled by glutamine rather than glucose carbons when PKM2 activity is suppressed. Glutamine can be converted to α-ketoglutarate (α-KG) by glutamate dehydrogenase. α-KG can be used by the tricarboxylic acid (TCA) cycle to produce ATP and other precursors for anabolic reactions for cell growth and proliferation, or it can exit the TCA cycle as malate to be converted into pyruvate and then lactate (3).
ACKNOWLEDGMENTS
We gratefully acknowledge the critical reading of the manuscript by Shannon E. Elf. We thank Anita Bellail and Chunhai Hao for their generous help with the gel filtration chromatography. The mass spectrometry based analysis of phospho-LDH-A was performed by Dongmei Cheng and Junmin Peng at Emory Proteomics Service Center.
This study was supported in part by National Institutes of Health grants CA120272 and CA140515 (J.C.). T.H. is a Fellow Scholar of the American Society of Hematology. G.Z.C., F.R.K., S.K., and J.C. are Georgia Cancer Coalition Distinguished Cancer Scholars. S.K. is an American Cancer Society Basic Research Scholar, a Special Fellow of the Leukemia and Lymphoma Society, and a Robbins Scholar. J.C. is an American Cancer Society Basic Research Scholar and a Scholar of the Leukemia and Lymphoma Society.
Footnotes
Published ahead of print on 3 October 2011.
REFERENCES
- 1. Brahimi-Horn M. C., Chiche J., Pouyssegur J. 2007. Hypoxia signalling controls metabolic demand. Curr. Opin. Cell Biol. 19:223–229 [DOI] [PubMed] [Google Scholar]
- 2. Bui T., Thompson C. B. 2006. Cancer's sweet tooth. Cancer Cell 9:419–420 [DOI] [PubMed] [Google Scholar]
- 3. Dang C. V. 2009. PKM2 tyrosine phosphorylation and glutamine metabolism signal a different view of the Warburg effect. Sci. Signal. 2:pe75. [DOI] [PubMed] [Google Scholar]
- 4. Durisova V., Vrbanova A., Ziegelhoffer A., Breier A. 1990. Interaction of Cibacron Blue 3GA and Remazol Brilliant Blue R with the nucleotide binding site of lactate dehydrogenase and (Na+ + K+)-ATPase. Gen. Physiol. Biophys. 9:519–528 [PubMed] [Google Scholar]
- 5. Fantin V. R., St-Pierre J., Leder P. 2006. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell 9:425–434 [DOI] [PubMed] [Google Scholar]
- 6. Girg R., Jaenicke R., Rudolph R. 1983. Dimers of porcine skeletal muscle lactate dehydrogenase produced by limited proteolysis during reassociation are enzymatically active in the presence of stabilizing salt. Biochem. Int. 7:433–441 [PubMed] [Google Scholar]
- 7. Gordan J. D., Thompson C. B., Simon M. C. 2007. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell 12:108–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gu T. L., et al. 2006. Phosphotyrosine profiling identifies the KG-1 cell line as a model for the study of FGFR1 fusions in acute myeloid leukemia. Blood 108:4202–4204 [DOI] [PubMed] [Google Scholar]
- 9. Hanahan D., Weinberg R. A. 2000. The hallmarks of cancer. Cell 100:57–70 [DOI] [PubMed] [Google Scholar]
- 10. Hanahan D., Weinberg R. A. 2011. Hallmarks of cancer: the next generation. Cell 144:646–674 [DOI] [PubMed] [Google Scholar]
- 11. Hatzivassiliou G., et al. 2005. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell 8:311–321 [DOI] [PubMed] [Google Scholar]
- 12. Hitosugi T., et al. 2009. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci. Signal. 2:ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Holbrook J. J., Liljas A., Steindel S. J., Rossmann M. G. 1975. Lactate dehydrogenase, p. 191–291 In Boyer P. D.(ed.), The enzymes, 3rd ed Academic Press, Inc., New York, NY [Google Scholar]
- 14. Kanno T., et al. 1988. Lactate dehydrogenase M-subunit deficiency: a new type of hereditary exertional myopathy. Clin. Chim. Acta Int. J. Clin. Chem. 173:89–98 [DOI] [PubMed] [Google Scholar]
- 15. Kobrin M. S., Yamanaka Y., Friess H., Lopez M. E., Korc M. 1993. Aberrant expression of type I fibroblast growth factor receptor in human pancreatic adenocarcinomas. Cancer Res. 53:4741–4744 [PubMed] [Google Scholar]
- 16. Kroemer G., Pouyssegur J. 2008. Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell 13:472–482 [DOI] [PubMed] [Google Scholar]
- 17. Le A., et al. 2010. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc. Natl. Acad. Sci. U. S. A. 107:2037–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Luqmani Y. A., Graham M., Coombes R. C. 1992. Expression of basic fibroblast growth factor, FGFR1 and FGFR2 in normal and malignant human breast, and comparison with other normal tissues. Br. J. Cancer 66:273–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marek L., et al. 2009. Fibroblast growth factor (FGF) and FGF receptor-mediated autocrine signaling in non-small-cell lung cancer cells. Mol. Pharmacol. 75:196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Morrison R. S., et al. 1994. Fibroblast growth factor receptor gene expression and immunoreactivity are elevated in human glioblastoma multiforme. Cancer Res. 54:2794–2799 [PubMed] [Google Scholar]
- 21. Penault-Llorca F., et al. 1995. Expression of FGF and FGF receptor genes in human breast cancer. Int. J. Cancer 61:170–176 [DOI] [PubMed] [Google Scholar]
- 22. Read J. A., Winter V. J., Eszes C. M., Sessions R. B., Brady R. L. 2001. Structural basis for altered activity of M- and H-isozyme forms of human lactate dehydrogenase. Proteins 43:175–185 [DOI] [PubMed] [Google Scholar]
- 23. Rossmann M. G., Liljas A., Brändén C.-I., Banaszak L. J. 1975. Dehydrogenase, p. 61–102 In Boyer P. D.(ed.), The enzymes, 3rd ed Academic Press, Inc., New York, NY [Google Scholar]
- 24. Sekine N., et al. 1994. Low lactate dehydrogenase and high mitochondrial glycerol phosphate dehydrogenase in pancreatic beta-cells. Potential role in nutrient sensing. J. Biol. Chem. 269:4895–4902 [PubMed] [Google Scholar]
- 25. Semenza G. L., Roth P. H., Fang H. M., Wang G. L. 1994. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J. Biol. Chem. 269:23757–23763 [PubMed] [Google Scholar]
- 26. Shim H., et al. 1997. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc. Natl. Acad. Sci. U. S. A. 94:6658–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tennant D. A., Duran R. V., Gottlieb E. 2010. Targeting metabolic transformation for cancer therapy. Nat. Rev. Cancer 10:267–277 [DOI] [PubMed] [Google Scholar]
- 28. Vander Heiden M. G., et al. 2010. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science 329:1492–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vander Heiden M. G., et al. 2001. Growth factors can influence cell growth and survival through effects on glucose metabolism. Mol. Cell. Biol. 21:5899–5912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xiao S., et al. 1998. FGFR1 is fused with a novel zinc-finger gene, ZNF198, in the t(8;13) leukaemia/lymphoma syndrome. Nat. Genet. 18:84–87 [DOI] [PubMed] [Google Scholar]
- 31. Zheng Y., Guo S., Guo Z., Wang X. 2004. Effects of N-terminal deletion mutation on rabbit muscle lactate dehydrogenase. Biochemistry (Mosc). 69:401–406 [DOI] [PubMed] [Google Scholar]
- 32. Zhong X. H., Howard B. D. 1990. Phosphotyrosine-containing lactate dehydrogenase is restricted to the nuclei of PC12 pheochromocytoma cells. Mol. Cell. Biol. 10:770–776 [DOI] [PMC free article] [PubMed] [Google Scholar]