Abstract
Subtilase cytotoxin (SubAB) from verotoxin (VT)-producing Escherichia coli (VTEC) strains was first described in the 98NK2 strain and has been associated with human disease. However, SubAB has recently been found in two VT-negative E. coli strains (ED 591 and ED 32). SubAB is encoded by two closely linked, cotranscribed genes (subA and subB). In this study, we investigated the presence of subAB genes in 52 VTEC strains isolated from cattle and 209 strains from small ruminants, using PCR. Most (91.9%) VTEC strains from sheep and goats and 25% of the strains from healthy cattle possessed subAB genes. The presence of subAB in a high percentage of the VTEC strains from small ruminants might increase the pathogenicity of these strains for human beings. Some differences in the results of PCRs and in the association with some virulence genes suggested the existence of different variants of subAB. We therefore sequenced the subA gene in 12 strains and showed that the subA gene in most of the subAB-positive VTEC strains from cattle was almost identical (about 99%) to that in the 98NK2 strain, while the subA gene in most of the subAB-positive VTEC strains from small ruminants was almost identical to that in the ED 591 strain. We propose the terms subAB1 to describe the SubAB-coding genes resembling that in the 98NK2 strain and subAB2 to describe those resembling that in the ED 591 strain.
INTRODUCTION
Verotoxin (VT)-producing Escherichia coli (VTEC), also referred to as Shiga toxin-producing E. coli, can cause diarrhea, hemorrhagic colitis, and hemolytic-uremic syndrome (HUS) in humans (9). VTEC produces two major toxin types, VT1 and VT2, as well as different variants (5, 6). Ruminants represent an important reservoir for VTEC and a source of infection for humans (3, 10–12). VT production alone is not sufficient to cause VTEC-related disease (1); the eae gene affects the virulence of VTEC and is required for the production of attaching and effacing lesions in the intestinal mucosa (1). In addition to the eae gene, a novel cytotoxin called subtilase cytotoxin (SubAB) might also contribute to the virulence of eae-negative VTEC strains in humans (17). It has been suggested previously that this toxin may be able to augment the effects of VT or to cause pathology in its own right (14, 15). SubAB was first described in the VTEC O113:H21 strain 98NK2, associated with an outbreak of HUS in Australia (17). SubAB in the 98NK2 strain is encoded by two closely linked, cotranscribed genes (subA and subB), located on a large, conjugative virulence plasmid designated pO113 (17). This plasmid also contains other putative virulence genes such as saa and ehxA, which encode an autoagglutinating adhesin and a hemolysin called EHEC (enterohemorrhagic E. coli) enterohemolysin, respectively (16–18). subAB genes have subsequently been identified only in VTEC strains (2, 14). However, SubAB has recently been identified in two VT-negative E. coli strains (ED 591 and ED 32) isolated from unrelated cases of childhood diarrhea (21). The ED 591 and ED 32 strains were positive for the tia gene, a genetic determinant of invasion previously described in enterotoxigenic E. coli (4), but were negative for the saa gene (21). In both strains, subAB genes were located close to the tia gene (21).
A limited number of studies have investigated the presence of subAB in VTEC from healthy cattle (2, 7, 8), but to the best of our knowledge, the presence of these genes in VTEC from diarrheic calves and small ruminants has not been studied. In the present study, we therefore examined the distribution of subAB in a large collection of VTEC strains isolated from healthy and diarrheic cattle, sheep, and goats to determine their association with ruminant VTEC strains.
MATERIALS AND METHODS
Bacterial strains.
A total of 261 VTEC strains were used in this study. All strains were isolated between 1993 and 2005 in Spain from healthy cattle (36 strains), sheep (60 strains), and goats (145 strains) and from calves (16 strains), lambs (3 strains), and goat kids (1 strain) with diarrhea. Most of these strains have previously been serotyped and tested for vt types and for the presence of the eae, ehxA, and saa genes (3, 6, 10–13). Only one strain per animal was included in the study.
Detection of virulence genes by PCR.
One colony from each VTEC strain was harvested after overnight culture, suspended in sterile water, incubated at 100°C for 10 min, and centrifuged. The supernatant was used for PCR. Primer pairs RTsubABF/RTsubABR, SubHCDF/SubSCDR, and tia_lo/tia_sense were used to detect subAB, subA, and tia genes, respectively. The 16S rRNA housekeeping gene was used as an internal amplification control. Primer sequences and PCR conditions are given in Table 1.
Table 1.
PCR primers and conditions used for detection of subAB and tia genes and for amplification of the subA gene
| Target gene | Primer | Nucleotide sequence (5′-3′) | Annealing conditions | Reference |
|---|---|---|---|---|
| subAB | RTsubABF | GCAGATAAATACCCTTCACTTG | 55°C, 60 s | 17 |
| subAB | RTsubABR | ATCACCAGTCCACTCAGCC | 55°C, 60 s | 17 |
| subA | SubHCDF | TATGGCTTCCCTCATTGCC | 50°C, 60 s | 14 |
| subA | SubSCDR | TATAGCTGTTGCTTCTGACG | 50°C, 60 s | 14 |
| tia | tia_lo | TCCATGCGAAGTTGTTATCA | 56°C, 60 s | 21 |
| tia | tia_sense | TTCTCTTTTTACCCTGCTTTTTGC | 56°C, 60 s | 21 |
| subA | Sub-F1 | CAATACGGCGCTCTGTTGACG | 60°C, 60 s | This study |
| subA | Sub-F2 | AACATATTGACCAGCAATAC | 60°C, 30 s; 50°C, 30 s | This study |
| subA | Sub-R | AAACATGCCATCCCGGGCATC | 60°C, 60 s, with Sub-F1; 60°C, 30 s, and 50°C, 30 s, with Sub-F2 | This study |
| 16S rRNA | 16SF | CATTGACGTTACCCGCAGAA | 56°C, 60 s | 19 |
| 16S rRNA | 16SR | CGCTTTACGCCCAGTAATTCC | 56°C, 60 s | 19 |
Sequencing of the subA gene.
One bovine saa-positive O113 VTEC strain, one bovine O8 VTEC strain, one caprine saa-positive O91:H14 VTEC strain, and nine VTEC strains that were considered representative of the most frequent serotypes of VTEC from sheep and goats (O5:HNM [NM indicates nonmotile], O76:H19, O81:H21, O91:HNM, O128:HNM, O146:H21, O166:H28, ONT [NT indicates nontypeable]:HNM, and ONT:H21) were used to sequence subA. The sequence of the whole subA gene was obtained by PCR amplification using two different sets of primer pairs (Sub-F1 and Sub-R for the saa-positive strains and Sub-F2 and Sub-R for the saa-negative strains) (Table 1). The PCR products were purified using the SpinPrep PCR cleanup kit (Novagen, Darmstadt, Germany), and DNA was sequenced at the Sequencing Unit of CIB-CSIC (Madrid, Spain) and Secugen S.L. (Madrid, Spain). The VECTOR NTI program was used to align the sequences of these strains, which were compared with those of the prototype 98NK2 and ED 591 strains.
Production and detection of SubAB in CHO cells.
Bacteria were grown for 20 h in Luria-Bertani medium at 37°C. Cultures were then disrupted by sonication at 4°C, and the lysates were centrifuged (15,700 × g) for 5 min at 4°C. In addition, culture supernatants were obtained by centrifuging at 15,700 × g for 5 min. The sonicated extracts and culture supernatants were tested for SubAB production by cytotoxicity assays in Chinese hamster ovary (CHO) cells, as previously described (17), with minor modifications. Briefly, CHO culture assays were performed on confluent cell monolayers in 96-well plates. A total of 50 μl per well of each undiluted culture supernatant or sonicated extract was added, and cells were incubated at 37°C for 60 min. After incubation, 150 μl of F-12K medium supplemented with 2% fetal calf serum was added per well. The cytopathic effect (characterized by rounding cells and detachment from the substratum) was observed after 3 days of incubation.
Nucleotide sequence accession numbers.
The sequences of the subA gene from the subAB2 VTEC strains B218, C20, C36, C91, C103, C137, K93, K95, K300, and K394 have been deposited into GenBank under the accession numbers JN638552 to JN638561, respectively.
RESULTS AND DISCUSSION
A total of 261 VTEC strains from ruminants were examined for the presence of subAB genes by PCR using the primer pairs RTsubABF/RTsubABR and SubHCDF/SubSCDR. Primers RTsubABF and RTsubABR are able to amplify a region bridging the end of subA and the beginning of subB (17, 21), while primers SubHCDF and SubSCDR are specific for two of the three critical functional domains in the SubA-coding sequence (14). The only strain from a diarrheic goat kid, 135 of the 145 (93.1%) strains from healthy goats, 55 of the 60 (91.7%) strains from healthy sheep, one of the three (33.3%) strains from diarrheic lambs, and 9 of the 36 (25%) strains from healthy cattle gave positive reactions with the two primer pairs used to detect the subAB genes. In contrast, the subAB genes were not detected in any of the 16 strains from diarrheic calves (Tables 2 to 4 ). In agreement with previous studies (2, 7, 8, 14, 21), subAB genes were found only in eae-negative VTEC strains. The percentage of subAB-positive VTEC strains from healthy cattle in this study (25%) was identical to that found by Khaitan et al. (8) but lower than those reported by Cergole-Novella et al. (2) and Karama et al. (7) (41.2 and 54.2%, respectively). VTEC strains from sheep and goats have been considered a minor human health hazard (1, 3, 12); however, the presence of subAB genes in a high percentage (91.9%) of VTEC strains from small ruminants might increase the pathogenicity of these strains for humans.
Table 2.
Serotypes and virulence genes in VTEC strains isolated from cattlea
| Source and serotype | No. of strains | vt genotype (no. of strains) | No. of strains positive for: |
|||||
|---|---|---|---|---|---|---|---|---|
| subAB1 | subAB2 | saa | tia | ehxA | eae | |||
| Healthy cattle | ||||||||
| O2:HNDb | 1 | vt2 (1) | 1 | 0 | 1 | 0 | 0 | 0 |
| O4:HND | 1 | vt2 (1) | 0 | 0 | 0 | 1 | 0 | 0 |
| O5:HNMc | 1 | vt1 (1) | 0 | 0 | 0 | 0 | 1 | 1 |
| O8:HND | 4 | vt2 (4) | 0 | 2 | 0 | 2 | 0 | 0 |
| O21:HND | 1 | vt2 (1) | 0 | 0 | 1 | 0 | 1 | 0 |
| O22:HND | 3 | vt2 (1), vt1 + vt2 (2) | 0 | 0 | 2 | 1 | 3 | 0 |
| O23:HND | 1 | vt1 + vt2 (1) | 0 | 0 | 1 | 0 | 1 | 0 |
| O74:HND | 1 | vt2 (1) | 0 | 0 | 1 | 0 | 1 | 0 |
| O84:H2 | 1 | vt1 (1) | 0 | 0 | 0 | 0 | 1 | 1 |
| O87:HND | 1 | vt1 (1) | 0 | 0 | 0 | 0 | 0 | 0 |
| O91:HND | 1 | vt2 (1) | 0 | 0 | 1 | 1 | 1 | 0 |
| O98:HNM | 1 | vt1 (1) | 0 | 0 | 0 | 0 | 1 | 1 |
| O103:H8 | 1 | vt1 (1) | 0 | 0 | 0 | 0 | 0 | 1 |
| O110:HND | 1 | vt1 (1) | 0 | 0 | 0 | 0 | 0 | 0 |
| O111:HNM | 2 | vt1 + vt2 (2) | 0 | 0 | 0 | 0 | 2 | 2 |
| O113:HND | 4 | vt2 (4) | 3 | 0 | 3 | 1 | 3 | 0 |
| O116:HND | 1 | vt2 (1) | 1 | 0 | 1 | 0 | 1 | 0 |
| O136:HND | 1 | vt1 (1) | 0 | 0 | 0 | 1 | 0 | 0 |
| O172:HNM | 1 | vt2 (1) | 0 | 0 | 0 | 0 | 0 | 1 |
| O174:HND | 2 | vt2 (2) | 0 | 0 | 0 | 2 | 1 | 0 |
| O175:HND | 1 | vt2 (1) | 0 | 0 | 0 | 1 | 0 | 0 |
| ONTd:HND | 5 | vt1 (2), vt2 (3) | 2 | 0 | 4 | 0 | 4 | 0 |
| Diarrheic calves | ||||||||
| O4:HND | 1 | vt2 (1) | 0 | 0 | 0 | 1 | 0 | 0 |
| O5:HNM | 1 | vt1 (1) | 0 | 0 | 0 | 0 | 1 | 1 |
| O23:HND | 1 | vt1 + vt2 (1) | 0 | 0 | 1 | 0 | 1 | 0 |
| O26:HNM | 2 | vt1 (2) | 0 | 0 | 0 | 0 | 2 | 2 |
| O26:H11 | 3 | vt1 (3) | 0 | 0 | 0 | 1 | 2 | 3 |
| O39:HND | 1 | vt1 (1) | 0 | 0 | 0 | 0 | 0 | 0 |
| O91:HND | 2 | vt2 (2) | 0 | 0 | 0 | 0 | 1 | 0 |
| O113:HND | 1 | vt2 (1) | 0 | 0 | 0 | 1 | 0 | 0 |
| O128:H16 | 1 | vt1 (1) | 0 | 0 | 0 | 0 | 1 | 1 |
| O128:HND | 1 | vt1 (1) | 0 | 0 | 1 | 0 | 1 | 0 |
| O171:HND | 1 | vt2 (1) | 0 | 0 | 0 | 0 | 0 | 0 |
| ONT:HND | 1 | vt1 (1) | 0 | 0 | 0 | 1 | 0 | 0 |
Table 4.
Serotypes and virulence genes in VTEC strains isolated from goatsa
| Source and serotype | No. of strains | vt genotype (no. of strains) | No. of strains positive for: |
|||||
|---|---|---|---|---|---|---|---|---|
| subAB1 | subAB2 | saa | tia | ehxA | eae | |||
| Healthy goats | ||||||||
| O5:HNMb | 9 | vt1c (2), vt1c + vt2 (7) | 0 | 9 | 1 | 2 | 7 | 0 |
| O5:H21 | 1 | vt1c + vt2 (1) | 0 | 1 | 0 | 0 | 1 | 0 |
| O7:H21 | 1 | vt1c (1) | 0 | 0 | 0 | 0 | 0 | 0 |
| O18:H28 | 1 | vt1c (1) | 0 | 1 | 0 | 1 | 0 | 0 |
| O58:H21 | 1 | vt1c + vt2 (1) | 0 | 1 | 0 | 1 | 1 | 0 |
| O64:H21 | 1 | vt1c (1) | 0 | 0 | 0 | 0 | 0 | 0 |
| O76:HNTc | 1 | vt1c (1) | 0 | 1 | 0 | 1 | 0 | 0 |
| O76:H19 | 30 | vt1c (30) | 0 | 30 | 0 | 30 | 30 | 0 |
| O81:HNM | 2 | vt1c + vt2 (2) | 0 | 2 | 0 | 1 | 0 | 0 |
| O81:H21 | 9 | vt1c (9) | 0 | 9 | 0 | 8 | 8 | 0 |
| O87:H38 | 1 | vt1 + vt2 (1) | 1 | 0 | 1 | 1 | 1 | 0 |
| O91:H14 | 3 | vt1 (3) | 3 | 0 | 3 | 3 | 2 | 0 |
| O119:H19 | 1 | vt1c (1) | 0 | 0 | 0 | 0 | 1 | 0 |
| O126:H8 | 6 | vt1c (6) | 0 | 6 | 0 | 3 | 6 | 0 |
| O128:HNM | 2 | vt1c (2) | 0 | 2 | 0 | 2 | 1 | 0 |
| O128:H2 | 4 | vt1c (2), vt1c + vt2 (2) | 0 | 4 | 0 | 4 | 3 | 0 |
| O128:H19 | 1 | vt1c (1) | 0 | 1 | 0 | 1 | 1 | 0 |
| O145:H21 | 1 | vt1c (1) | 0 | 0 | 0 | 0 | 0 | 0 |
| O146:HNM | 2 | vt1c + vt2 (2) | 0 | 2 | 0 | 2 | 2 | 0 |
| O146:H21 | 10 | vt1c (2), vt1c + vt2 (8) | 0 | 10 | 0 | 10 | 10 | 0 |
| O156:H25 | 1 | vt1c (1) | 0 | 0 | 0 | 0 | 1 | 1 |
| O157:H7 | 3 | vt2 (3) | 0 | 0 | 0 | 0 | 3 | 3 |
| O166:H28 | 11 | vt1c (9), vt1c + vt2 (2) | 0 | 11 | 0 | 4 | 5 | 0 |
| O173:H8 | 1 | vt1c (1) | 0 | 1 | 0 | 1 | 0 | 0 |
| O174:H8 | 1 | vt1c (1) | 0 | 1 | 0 | 1 | 0 | 0 |
| O175:H38 | 1 | vt2 (1) | 0 | 1 | 0 | 1 | 1 | 0 |
| ONT:HNM | 8 | vt1c (3), vt1c + vt2 (5) | 0 | 8 | 0 | 7 | 3 | 0 |
| ONT:H2 | 2 | vt1c + vt2 (2) | 0 | 2 | 0 | 2 | 2 | 0 |
| ONT:H4 | 5 | vt1c (5) | 0 | 5 | 0 | 0 | 5 | 0 |
| ONT:H19 | 2 | vt1c (1), vt1c + vt2 (1) | 0 | 2 | 0 | 1 | 2 | 0 |
| ONT:H21 | 20 | vt1c (17), vt1c + vt2 (3) | 0 | 18 | 0 | 10 | 18 | 0 |
| ONT:H28 | 1 | vt1c + vt2 (1) | 0 | 1 | 0 | 1 | 1 | 0 |
| ONT:HNT | 2 | vt1c (2) | 0 | 2 | 0 | 1 | 2 | 0 |
| Diarrheic goat kid | ||||||||
| O166:H28 | 1 | vt1c (1) | 0 | 1 | 0 | 0 | 0 | 0 |
Seven of the nine subAB-positive VTEC strains from healthy cattle were positive for vt2 and saa, and all but one were also positive for ehxA (Table 2). Other studies (2, 7, 8) have also found an association between subAB and vt2, ehxA, and saa genes in VTEC strains from cattle. The other two subAB-positive VTEC strains from healthy cattle possessed the vt2 and tia genes (Table 2). Although all subAB-positive VTEC strains from cattle possessed the saa or tia gene, 10 saa-positive VTEC strains and eight tia-positive VTEC strains from cattle were subAB negative. The association of subAB with saa or tia is probably due to the possibility that the subAB genes may be located on a plasmid which also contains saa (17) or on a putative pathogenicity island together with tia (21).
A total of 188 of the 192 subAB-positive VTEC strains from sheep and goats were saa negative. Of these, 133 possessed the tia gene and 142 possessed the ehxA gene (Tables 3 and 4). The other four subAB-positive VTEC strains from small ruminants possessed the saa and tia genes, and three of these also had the ehxA gene (Table 4). Most of the VTEC strains from sheep and goats produced VT1 (alone or in combination with VT2) and possessed the vt1c gene (6). Interestingly, the four subAB-positive VTEC strains from goats that were also positive for saa and tia were the only VT1-producing isolates from that animal species that were negative for the vt1c gene (6). Although some of the subAB-positive VTEC strains from sheep and goats were negative for tia and saa, all the strains from small ruminants positive for tia and/or saa were also positive for subAB. As previously mentioned, the presence of saa or tia in the subAB-positive VTEC strains is probably due to a genetic linkage among these genes (17, 21).
Table 3.
Serotypes and virulence genes in VTEC strains isolated from sheepa
| Source and serotype | No. of strains | vt genotype (no. of strains) | No. of strains positive forb: |
|||
|---|---|---|---|---|---|---|
| subAB2 | tia | ehxA | eae | |||
| Healthy sheep | ||||||
| O5:HNMc | 13 | vt1c (7), vt1c + vt2 (6) | 13 | 3 | 13 | 0 |
| O6:H10 | 3 | vt1c (3) | 0 | 0 | 0 | 0 |
| O21:H21 | 1 | vt1c + vt2 (1) | 1 | 1 | 0 | 0 |
| O26:H11 | 1 | vt1 + vt2 (1) | 0 | 0 | 1 | 1 |
| O71:HNM | 1 | vt1c + vt2 (1) | 1 | 1 | 0 | 0 |
| O75:H8 | 1 | vt1c + vt2 (1) | 1 | 1 | 1 | 0 |
| O91:HNM | 6 | vt1 + vt2 (5), vt1c + vt2 (1) | 6 | 6 | 0 | 0 |
| O128:HNM | 6 | vt1c (3), vt1c + vt2 (3) | 5 | 5 | 0 | 0 |
| O128:H16 | 1 | vt1c (1) | 1 | 1 | 0 | 0 |
| O141:HNM | 1 | vt1c (1) | 1 | 1 | 1 | 0 |
| O146:H21 | 8 | vt1c + vt2 (8) | 8 | 8 | 8 | 0 |
| O163:HNM | 1 | vt1 + vt2 (1) | 1 | 0 | 1 | 0 |
| O163:H11 | 1 | vt1 + vt2 (1) | 1 | 1 | 0 | 0 |
| O166:HNM | 1 | vt1c + vt2 (1) | 1 | 1 | 1 | 0 |
| O166:H28 | 4 | vt1c (2), vt1c + vt2 (2) | 4 | 0 | 4 | 0 |
| ONTd:HNM | 9 | vt1c (3), vt1c + vt2 (6) | 9 | 8 | 3 | 0 |
| ONT:H4 | 2 | vt1c (2) | 2 | 0 | 2 | 0 |
| Diarrheic lambs | ||||||
| O71:HNM | 1 | vt1c + vt2 (1) | 1 | 1 | 0 | 0 |
| O110:HNM | 1 | vt1c (1) | 0 | 0 | 0 | 0 |
| ONT:HNM | 1 | vt1c (1) | 0 | 0 | 0 | 0 |
Of the 261 VTEC strains included in this study, 21 were positive for saa and 151 were positive for tia, but only five were positive for both genes (four subAB-positive strains from goats and one subAB-negative strain from a healthy cow). Tozzoli et al. (21) also observed that the E. coli strains positive for the tia gene were negative for the saa gene and vice versa. These results thus demonstrate a negative association between the saa and tia genes.
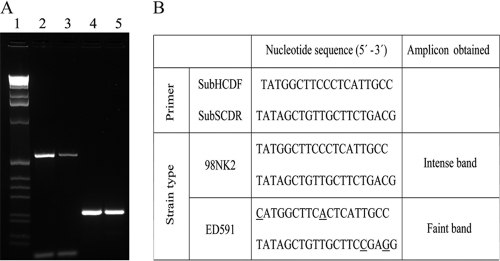
All VTEC strains shown to be positive for subAB using the primer pair RTsubABF/RTsubABR were also positive using the primer pair SubHCDF/SubSCDR and vice versa. All subAB-positive VTEC strains produced bands of similar intensities with the primer pair RTsubABF/RTsubABR, but the 11 subAB-positive, saa-positive VTEC strains gave a more intense band than that of the 190 subAB-positive, saa-negative strains with the primer pair SubHCDF/SubSCDR (Fig. 1). The differences in intensities of amplicons with the primer pair SubHCF/SubSCDR were due to two nucleotide mismatches and not to differences in the amounts of DNA template (Fig. 1). The differences in PCRs observed with the primer pair SubHCDF/SubSCDR and in the association with the saa and tia genes suggest that the subAB genes in most of the VTEC strains from cattle differed from most VTEC strains from sheep and goats. We therefore sequenced the subA gene in 12 strains. The three critical functional domains characteristic of members of the subtilase family (17) were conserved in the predicted SubA sequences of all the strains tested. In addition, the alignment of the subA sequences and the translated amino acid sequences from these strains with the corresponding sequences from the prototype 98NK2 and ED 591 strains showed that the two saa-positive strains that gave an intense band with the SubHCDF/SubSCDR primer pair demonstrated nucleotide and amino acid homologies of 99.9% and 100%, respectively, with the 98NK2 strain. The 10 saa-negative strains that gave a faint band with the SubHCDF/SubSCDR primer pair demonstrated homologies of 97.8 to 99.9% with the ED 591 strain at the nucleotide level and 97.1 to 100% at the amino acid level. These results therefore demonstrated that the subA gene in the strains producing an intense band with the SubHCDF/SubSCDR primer pair was almost identical to that of the 98NK2 strain, while that in the strains producing a faint band was almost identical to that of the ED 591 strain.
Fig. 1.
(A) PCR amplifications of subA in B173 (VTEC O113 strain isolated from cattle) and C137 (VTEC O146:H21 strain isolated from sheep) using the primer pair SubHCDF/SubSCDR (lanes 2 and 3, respectively) and of subAB in the same strains using the primer pair RTsubABF/RTsubABR (lanes 4 and 5, respectively). Equal amounts of bacterial DNA template were used in each PCR amplification as is shown by the internal 16S rRNA amplification controls in lanes 2 and 3 (100 bp). Lane 1, molecular size marker X (0.07 to 12.2 kbp; Roche, Mannheim, Germany). B173 showed 99.9% homology at the nucleotide level and 100% at the amino acid level with the prototype strain 98NK2. The subA gene in C137 showed 99.4% homology at the nucleotide level and 99.8% at the amino acid level with the prototype strain ED 591. (B) Alignment of primers used in PCR loaded in lanes 2 and 3 and counterpart sequence from subA of the two reference type strains. The nucleotide mismatches according to the PCR primers used for the amplifications are underlined.
SubAB production in the 12 VTEC strains with sequenced subA genes (2 98NK2-like and 10 ED 591-like) and in two subAB-negative strains was tested in CHO cells, which are susceptible to SubAB but refractory to VT (17). Sonicated extracts and culture supernatants of the subAB-positive but not the subAB-negative VTEC strains caused a cytopathic effect in CHO cells. SubAB is not secreted very efficiently from live VTEC (J. C. Paton, personal communication); however, we were able to detect SubAB production in undiluted culture supernatants of the bacterial strains.
The subAB genes in the 98NK2 and ED 591 strains show a nucleotide sequence homology of 90% (21). In addition, the results of the current study, as well as those of previous studies, showed that the subAB genes in E. coli strains with subA genes almost identical to that in the 98NK2 strain and the subAB genes in strains with subA genes resembling that in the ED 591 strain have different main hosts and different associated virulence genes. We therefore propose calling the 98NK2-like SubAB-coding genes subAB1 and the ED 591-like SubAB-coding genes subAB2.
SubAB has been detected almost exclusively in VTEC strains to date, but the plasmid carrying subAB is capable of conjugative transmission, at least in VTEC O113:H21 (20), and Paton and Paton (15) suggested the potential for wider dissemination among other E. coli pathotypes. In support of this hypothesis, Tozzoli et al. (21) found two VT-negative E. coli strains that produced SubAB. VTEC detection in our laboratory has been performed by testing for VT production using the Vero cell cytotoxicity assay in four isolates for each animal sampled. In addition to the 261 VTEC strains included in this study, the presence of the subAB genes was examined by PCR in 14 E. coli strains negative for the vt genes responsible for the cytopathic effect in Vero cells, but none of these was positive for subAB (data not shown). Because SubAB has a cytopathic effect in Vero cells (17), the results indicate that SubAB production was restricted to VTEC strains, at least in those strains used in the current study. The results of the present study showed that many of the VTEC strains from small ruminants and two strains from healthy cattle possessed a subA gene almost identical to that previously identified in two VT-negative E. coli strains producing SubAB (21). The two VTEC strains from healthy cattle belonged to the same serogroup (O8) as did one of the two VT-negative E. coli strains found by Tozzoli et al. (21), and it is thus possible that the two VT-negative strains represented strains that have lost their vt genes.
subAB genes in cattle were found only in VTEC strains from healthy animals, and the fact that VTEC is rarely found in diarrheic lambs and goat kids (12) suggests that SubAB is not associated with diarrhea in neonatal ruminants.
The PCR results using the primer pair SubHCDF/SubSCDR and the association with virulence genes suggest that all the human SubAB-producing VTEC strains in Australia (14, 17) possess subAB genes similar to that in the 98NK2 strain. However, three of the five subAB-positive VTEC strains found by Tozzoli et al. (21) in Italy probably possessed subAB genes resembling those found in the two VT-negative E. coli strains (ED 591 and ED 32) described in that same study. It would therefore be interesting to determine the distribution of the two subAB variants in other human VTEC collections and to establish if these SubAB variants display differences in potency and clinical outcome, as is the case for variants and types of VT (5).
In conclusion, the results of this study showed that subAB genes are widely distributed among VTEC strains from sheep and goats, suggesting that these strains might be of more pathogenic significance for humans than previously believed. Our data also showed that subAB-positive VTEC strains from ruminants possess two variants of the genes, associated with different hosts and virulence genes. We propose the terms subAB1 and subAB2 to distinguish between these variants.
ACKNOWLEDGMENTS
This study was supported by a grant from the Banco Santander Central Hispano-Universidad Complutense (INBAVET 920338).
We thank J. C. Paton for information about the detection of SubAB in CHO cells.
Footnotes
Published ahead of print on 30 September 2011.
REFERENCES
- 1. Beutin L., Geiger D., Zimmermann S., Karch H. 1995. Virulence markers of Shiga-like toxin-producing Escherichia coli strains originating from healthy domestic animals of different species. J. Clin. Microbiol. 33: 631–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cergole-Novella M. C., et al. 2007. Distribution of virulence profiles related to new toxins and putative adhesins in Shiga toxin-producing Escherichia coli isolated from diverse sources in Brazil. FEMS Microbiol. Lett. 274: 329–334 [DOI] [PubMed] [Google Scholar]
- 3. Cortés C., et al. 2005. Serotypes, virulence genes and intimin types of verotoxin-producing Escherichia coli and enteropathogenic E. coli isolated from healthy dairy goats in Spain. Vet. Microbiol. 110: 67–76 [DOI] [PubMed] [Google Scholar]
- 4. Fleckenstein J. M., Kopecko D. J., Warren R. I., Elsinghorst E. A. 1996. Molecular characterization of the tia invasion locus from enterotoxigenic Escherichia coli. Infect. Immun. 64: 2256–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fuller C., Pellino C. A., Flagler M. J., Strasser J. E., Weiss A. A. 2011. Shiga toxin subtypes display dramatic differences in potency. Infect. Immun. 79: 1329–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Horcajo P., Domínguez-Bernal G., De La Fuente R., Ruiz-Santa-Quiteria J. A., Orden J. A. 2010. Association of vt1c with verotoxin-producing Escherichia coli from goats and sheep. J. Vet. Diagn. Invest. 22: 332–334 [DOI] [PubMed] [Google Scholar]
- 7. Karama M., Johnson R. P., Holtslander R., McEwen S. A., Gyles C. L. 2008. Prevalence and characterization of verotoxin-producing Escherichia coli (VTEC) in cattle from an Ontario abattoir. Can. J. Vet. Res. 72: 297–302 [PMC free article] [PubMed] [Google Scholar]
- 8. Khaitan A., Jandhyala D. M., Thorpe C. M., Ritchie J. M., Paton A. W. 2007. The operon encoding SubAB, a novel cytotoxin, is present in Shiga toxin-producing Escherichia coli isolates from the United States. J. Clin. Microbiol. 45: 1374–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mainil J. 1999. Shiga/verocytotoxins and Shiga/verotoxigenic Escherichia coli in animals. Vet. Res. 30: 235–257 [PubMed] [Google Scholar]
- 10. Orden J. A., et al. 1998. Verotoxin-producing Escherichia coli (VTEC) and eae-positive non-VTEC in 1-30-days-old diarrhoeic dairy calves. Vet. Microbiol. 63: 239–248 [DOI] [PubMed] [Google Scholar]
- 11. Orden J. A., et al. 2002. Verotoxin-producing Escherichia coli (VTEC), enteropathogenic E. coli (EPEC) and necrotoxigenic E. coli (NTEC) isolated from healthy cattle in Spain. J. Appl. Microbiol. 93: 29–35 [DOI] [PubMed] [Google Scholar]
- 12. Orden J. A., et al. 2003. Prevalence and characterization of vero cytotoxin-producing Escherichia coli isolated from diarrhoeic and healthy sheep and goats. Epidemiol. Infect. 130: 313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Orden J. A., Cortés C., Ruiz-Santa-Quiteria J. A., Martínez S., De la Fuente R. 2005. Detection of the saa gene in verotoxin-producing Escherichia coli from ruminants. J. Vet. Diagn. Invest. 17: 65–67 [DOI] [PubMed] [Google Scholar]
- 14. Paton A. W., Paton J. C. 2005. Multiplex PCR for direct detection of Shiga toxigenic Escherichia coli strains producing the novel subtilase cytotoxin. J. Clin. Microbiol. 43: 2944–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Paton A. W., Paton J. C. 2010. Escherichia coli subtilase cytotoxin. Toxins 2: 215–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Paton A. W., Srimanote P., Woodrow M. C., Paton J. C. 2001. Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect. Immun. 69: 6999–7009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paton A. W., Srimanote P., Talbot U. M., Wang H., Paton J. C. 2004. A new family of potent AB5 cytotoxins produced by Shiga toxigenic Escherichia coli. J. Exp. Med. 200: 35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schmidt H., Beutin L., Karch H. 1995. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect. Immun. 63: 1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Spano G., Beneduce L., Terzi V., Stanca A. M., Massa S. 2005. Real-time PCR for the detection of Escherichia coli O157:H7 in dairy and cattle wastewater. Lett. Appl. Microbiol. 40: 164–171 [DOI] [PubMed] [Google Scholar]
- 20. Srimanote P., Paton A. W., Paton J. C. 2002. Characterization of a novel type IV pilus locus carried on the large plasmid of human-virulent strains of locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli. Infect. Immun. 70: 3094–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tozzoli R., et al. 2010. Production of the subtilase AB5 cytotoxin by Shiga toxin-negative Escherichia coli. J. Clin. Microbiol. 48: 178–183 [DOI] [PMC free article] [PubMed] [Google Scholar]