Abstract
The goal of this study was to determine how enteric viruses persist within shellfish tissues. Several lines of novel evidence show that phagocytic blood cells (hemocytes) of Eastern oysters (Crassostrea virginica) play an important role in the retention of virus particles. Our results demonstrated an association of virus contamination with hemocytes but not with hemolymph. Live oysters contaminated overnight with hepatitis A virus (HAV) and murine norovirus (MNV) had 56% and 80% of extractable virus associated with hemocytes, respectively. Transfer of HAV-contaminated hemocytes to naïve (virus-free) oysters resulted in naïve oyster meat testing HAV positive for up to 3 weeks. Acid tolerance of HAV, MNV, poliovirus (PV), and feline calicivirus (FCV) correlated with the ability of each virus to persist within oysters. Using reverse transcription-PCR (RT-PCR) to evaluate persistence of these viruses in oysters, we showed that HAV persisted the longest (>21 days) and was most acid resistant, MNV and PV were less tolerant of acidic pH, persisting for up to 12 days and 1 day, respectively, and FCV did not persist (<1 day) within oysters and was not acid tolerant. This suggests that the ability of a virus to tolerate the acidic conditions typical of phagolysosomal vesicles within hemocytes plays a role in determining virus persistence in shellfish. Evaluating oyster and hemocyte homogenates and live contaminated oysters as a prelude to developing improved viral RNA extraction methods, we found that viruses were extracted more expediently from hemocytes than from whole shellfish tissues and gave similar RT-PCR detection sensitivities.
INTRODUCTION
Bivalve shellfish can filter as much as 40 liters of water/h (32), and as a consequence, they bioconcentrate many different enteric viruses, including hepatitis A virus (HAV) and human norovirus (HuNoV) strains, making shellfish important vectors for disease outbreaks. The ability to accumulate and concentrate waterborne viruses is well documented, and the processes by which virus pathogens are bioconcentrated have been characterized to some degree (13, 31, 35, 51, 52). The more critical issue from a seafood safety standpoint is in understanding the mechanism(s) of prolonged viral persistence within shellfish. Concentrated fecal bacteria can be purged substantially from shellfish by placing bivalves in bacterium-free water for 48 to 72 h, using a commercial process known as depuration (14). In contrast, human enteric nonenveloped viruses, despite their inability to replicate within shellfish tissues, may not be purged readily (19, 23). A review of the research literature suggests that viruses differ in the ability to persist within shellfish (6, 29, 39, 48, 53). For example, McLeod et al. (33) recently showed that HAV and HuNoV levels in oysters did not decrease significantly after a 23-h depuration period, while poliovirus (PV) levels declined substantially. Kingsley and Richards (29) demonstrated that HAV could be detected in oysters for as long as 6 weeks after exposure to virus-contaminated water, while Ueki et al. (53) showed that feline calicivirus (FCV), a HuNoV surrogate, does not persist in oysters.
The general scheme for bivalve feeding and digestive processes, as described by Newell and Langdon (36), is as follows. Waterborne particles are entrapped in the ciliated gill filaments and are swept toward the esophagus and into the stomach. The crystalline style then mechanically breaks up cells and particulates and mixes them with gastric enzymes. Subsequently, nutrient absorption and endocytosis of foreign materials by digestive epithelial cells lining the digestive diverticula occur. Intracellular digestion by hemocytes present in the gut lumen, whereby the hemocytes ingest and digest foreign materials and transport the digested materials to the gut lining or other tissues for assimilation, may also occur.
In addition to this digestive function, hemocytes can engulf and phagocytize foreign pathogens present on the mucosal surfaces of oysters or within the hemolymph as part of their innate immune functions. The hemolymph of bivalves normally contains moderate levels of indigenous microorganisms, which readily cross the epithelium of the gills (38). The importance of hemocytes in immune defense against microorganisms in the hemolymph was reviewed by Roch (46). Viruses and bacteria are commonly found on the mucosal surfaces and in the extrapallial fluid of bivalves. Hemocytes routinely traffic between the hemolymph and the outer surfaces of oysters (49). They also find their way into the extrapallial fluid between the oyster's body and shell, where they retain full immunological activity (1).
Unlike vertebrates, phylogenetically less advanced organisms such as bivalve mollusks do not have adaptive immune response mechanisms and lack major histocompatibility complex self/nonself-recognizing immune mechanisms. Therefore, hemocytes transferred from one oyster to another are unlikely to trigger allograft responses involving destruction of transplanted cells. In fact, Sullivan (50) demonstrated long-term survival of heterotropic molluscan allografts. Hemocytes transferred between tunicates retain normal function and can clear foreign particles posttransfer (42). Consequently, it is possible to transfer hemocytes from one oyster to another without resulting in self/nonself immune destruction or rejection.
The mechanism by which small virus particles (<50 nm in diameter) are bioconcentrated by bivalve shellfish is an open question, since ciliated gill filaments, which filter microorganisms from the water column, are spaced as far as 500 to 600 nm apart in shellfish. It has been suggested that rather than being trapped directly by gill filaments, virus particles become bound electrostatically to sulfate groups on the mucopolysaccharides of shellfish mucus (13). In more recent studies, Le Guyader et al. (31) and Tian et al. (51, 52) independently showed that HuNoV can interact with specific glycoproteins that may mimic human glycoproteins thought to function as cell surface receptors for initiation of norovirus infection within the human host. Le Guyader et al. (31) also reported detection of HuNoV in association with oyster hemocytes. Recent research also indicates that noroviruses can interact with zooplankton, providing a potential means of entrapment by bivalve gill cilia (17, 18). At present, it is not known if other viruses, such as HAV, hepatitis E virus, poliovirus, enteroviruses, adenoviruses, and astroviruses, are bioconcentrated via analogous mechanisms.
Postuptake localization of virus particles within shellfish tissues has been studied by several researchers (12, 22, 34, 35, 47). Metcalf and Styles (35) demonstrated that PV readily concentrates within the digestive organs of Pacific oysters (Crassostrea gigas) and Eastern oysters (Crassostrea virginica). DiGirolamo et al. (12) noted that approximately 15% of accumulated PV was disseminated throughout the body of Pacific oysters 48 h after contamination. Hay and Scotti (22) used a radiolabeled insect picornavirus to demonstrate that virus was concentrated intracellularly in regions adjacent to the digestive microtubules. For HuNoV, it was demonstrated that the stomach and digestive diverticula are the principal sites of HuNoV concentration 4 to 24 h after uptake (47). McLeod et al. (34) found that virus bioconcentration occurs within the digestive tract, and they identified PV in the gills and labial palps. Overall, the processes of uptake and dissemination of virus particles within bivalve tissues have been studied more extensively, while comparatively little research has been directed toward understanding the mechanism by which infectious viruses remain within bivalve shellfish and avoid being purged, digested, or otherwise destroyed by shellfish digestive processes.
Current diagnostic testing for viral contaminants in shellfish is cumbersome and time-consuming, thus preventing routine testing. Viral RNA extraction methods presently used by the U.S. Food and Drug Administration and U.S. Department of Agriculture laboratories utilize whole or dissected oyster tissues that require 1 to 2 days to process (10, 20). Studies using an RNeasy Mini kit (Qiagen, Valencia, CA) to extract viral RNA from oysters showed improved efficiency and reduced extraction time compared to previous methods (11, 18). The major limitation of kits is the small mass of starting material (1 g for the largest kit, the RNeasy Maxi kit [Qiagen]), which represents only a fraction of a whole oyster.
In this report, we evaluate the role of hemocytes in the persistence of enteric viruses in oysters and evaluate hemocytes as a starting material for improved viral RNA extraction and detection methods.
MATERIALS AND METHODS
Cells, viruses, and oysters.
FCV strain KCD was obtained from the American Type Culture Collection (ATCC), Manassas, VA, as culture VR-651 and was propagated in feline kidney cells (ATCC CCL-94). Working stocks of murine norovirus (MNV-1), obtained from Herbert W. Virgin (Washington University School of Medicine, St. Louis, MO), were prepared using confluent monolayers of RAW 264.7 cells (ATCC) cultured in high-glucose Dulbecco's modified Eagle medium (DMEM; Gibco-Invitrogen, Grand Island, NY) supplemented with 25 mM HEPES buffer, 10% fetal bovine serum (Gibco-Invitrogen), 2 mM Gluta-MAX-1 (Gibco-Invitrogen), 100 units/ml of penicillin, and 100 μg/ml of streptomycin sulfate (Gibco-Invitrogen), essentially as described by Wobus et al. (56). Hepatitis A virus was obtained from the ATCC as VR-1402, a cell culture-adapted cytopathic clone of strain HM-175/18f. Working HAV stocks were propagated on confluent FRhK-4 cells (ATCC CRL-1688) in DMEM supplemented with 10% fetal bovine serum. Poliovirus strain Chat was obtained as culture VR-192 from the ATCC and was propagated in buffalo green monkey kidney (BGM) cells provided by Daniel Dahling (Environmental Protection Agency, Cincinnati, OH). Medium market-sized oysters (Crassostrea virginica) and natural seawater were obtained from the University of Delaware Marine Laboratory (Lewes, DE). Oysters were acclimated for 1 to 3 days at 20 to 25°C in 40-liter aquaria containing 30 liters of natural seawater.
Plaque assays for infectious viruses.
Plaque assays were performed using serial dilutions of viruses in Earle's balance salt solution (EBSS; Gibco-Invitrogen), and the dilutions were assayed in triplicate, using 3 wells per dilution, for a total of 3 trials (N = 3; n = 9). Confluent six-well plates of RAW mouse monocytes or feline kidney cells were inoculated with 0.5 ml of diluted MNV or FCV, respectively. One-hundred-millimeter plates were inoculated with 2 ml of serially diluted PV or HAV. After 2 h of incubation at 37°C, the inocula were removed. Drained 6-well and 100-mm plates were overlaid with 0.75% low-melting-point agarose (Fisher Bioreagents, Fair Lawn, NJ) in minimal essential medium (MEM; Gibco-Invitrogen) for MNV and with 1.2% agarose (Sigma-Aldrich, St. Louis, MO) in DMEM for PV, FCV, and HAV. Murine norovirus, FCV, and PV plates were incubated for 3 days at 37°C with 5% CO2. Hepatitis A virus plates were incubated for 14 to 17 days at 37°C with 5% CO2. Enumeration of MNV and FCV plaques was performed after staining with 0.1% neutral red. Enumeration of PV and HAV plaques was performed on cultures after treatment with 10% formalin, removal of the agarose overlay, and crystal violet staining, as described previously (44).
Viral RNA extraction from oysters.
Viral RNA extraction from whole oysters was performed using the glycine, polyethylene glycol (PEG), Tri reagent, oligo(dT)25 (GPTT) protocol as described previously (27, 28). In brief, oyster homogenates were blended for 3 min in a laboratory blender with 250 ml of glycine buffer, pH 9.5. Fifty milliliters of homogenate (corresponding to approximately one-half of an oyster in glycine buffer) was incubated at 37°C for 30 min and clarified by centrifugation at 15,000 × g at 4°C. Supernatant was precipitated with an equal volume of 16% PEG solution for 1 h on ice, followed by centrifugation at 10,000 × g for 5 min at 4°C. The pellet was resuspended in 5 ml of Tri reagent (Sigma-Aldrich) and incubated for 5 min at room temperature. Chloroform (1.2 ml) was added and mixed by vortexing, and the mixture was incubated for 5 min at room temperature, followed by centrifugation at 12,000 × g for 20 min at 4°C. RNA in the top, aqueous layer was precipitated with 2.5 ml of isopropanol and centrifuged at 5,000 × g for 5 min. The pellet was washed with 1 ml of 75% ethanol to remove residual isopropanol and centrifuged at 5,000 × g for 5 min. The pellet was resuspended in 300 μl RNase-free water (Gibco-Invitrogen). Viral RNA was purified from total RNA by using Dynabeads following the procedure described below (see “RNA purification”), beginning with the binding step, and extracted RNA was stored on ice until assayed by real-time reverse transcription-PCR (RT-PCR).
Viral RNA extraction from hemocytes.
Hemocytes were harvested from oysters by two different methods.
(i) Method A.
For analysis of virus persistence, small V-shaped notches were ground into the oyster shell adjacent to the adductor muscle by use of a grinding wheel prior to contamination with virus to facilitate subsequent collection or transfer of hemocytes. After contamination of oysters by natural filtration processes, a 16-gauge needle was inserted into the oyster adductor muscle to collect approximately 3 ml of circulating hemocytes and hemolymph. For analysis of hemolymph, hemocytes were pelleted by centrifugation at 4,000 × g for 10 min, and the hemolymph was filtered (0.22 μm).
(ii) Method B.
To evaluate the utility of hemocytes for virus extraction, oysters were shucked, the top shell was removed, and the oyster tissues and hemolymph were reserved in the bottom shell. The hemolymph was separated from the tissue by direct draw with a pipette, and approximately 10 ml of hemolymph was collected from each oyster. A hemocytometer (Hausser Scientific, Horsham, PA) was used to quantify average hemocyte concentrations in pooled hemolymph from 20 oysters. Hemocytes were separated from the hemolymph by centrifugation at 500 × g for 5 min, followed by collection of the pellet.
RNA purification.
RNA extracts were prepared from samples containing hemocytes from one-half of an oyster (∼5 × 105 hemocytes) by using Dynabeads and RNeasy methods. The Dynabeads method was adapted from the manufacturer's instructions for preparation of lysate from cultured cells and cell suspensions (25). Hemocyte pellets were lysed in lysis/binding buffer (100 mM Tris-HCl, pH 7.5, 500 mM LiCl, 10 mM EDTA, 1% lithium dodecyl sulfate [LiDS], 5 mM dithiothreitol [DTT]) and homogenized by passage five times through a 21-gauge needle fitted to a 3-ml syringe. Fifty microliters of washed oligo(dT)25 Dynabeads (Invitrogen Dynal, Oslo, Norway), which bind to the poly(A) sequence of the HAV and MNV genomes, was added to the lysates. The lysates were placed on a LabQuake shaker rotisserie (Thermo Scientific, Waltham, MA) for 20 min at room temperature to facilitate binding. The Dynabeads were separated from the supernatant with a magnet and washed in 1-ml volumes, first in washing buffer A (10 mM Tris-HCl, pH 7.5, 0.15 M LiCl, 1 mM EDTA, 0.1% LiDS) and then in washing buffer B (10 mM Tris-HCl, pH 7.5, 0.15 M LiCl, 1 mM EDTA). RNA was eluted off the Dynabeads by heating at 80°C for 2 min in 40 μl RNase-free water. The samples were stored on ice until assayed by real-time RT-PCR.
RNeasy Mini kits were used according to the manufacturer's protocol for purification of total RNA from animal cells. The kit utilizes silica-binding membranes in a spin column format to extract total RNA (40). Hemocyte lysates were homogenized by passage five times through a 21-gauge needle fitted to a 3-ml syringe, mixed with 1 volume of 70% ethanol, and loaded on the spin column. RNA elution was performed using 40 μl of nuclease-free water. Samples were stored on ice pending assay by real-time RT-PCR.
For RT-PCR analysis of hemocytes, the four-step GPTT method (28) was modified by omitting the glycine and polyethylene glycol steps. Hemocytes were pelleted at 3,200 × g and treated with 5 ml Tri reagent and 1.2 ml chloroform. Aqueous and organic layers were separated by centrifugation at 12,000 × g for 5 min. The aqueous layer was removed and treated with 2.5 ml isopropanol to obtain a precipitate of virus RNA. One hundred microliters of Dynabeads was used to purify poly(A) RNA. Approximately 20 to 100 μl of sample was obtained. Reverse transcription-PCR was performed using 10 μl of the 20- to 100-μl extract. Reverse transcription-PCR analysis was performed using a Qiagen OneStep RT-PCR kit (Qiagen, Valencia, CA) according to the manufacturer's instructions.
Analysis of viral RNA.
RT-PCR was performed on shellfish extracts by using gene-specific primers and a OneStep RT-PCR kit from Qiagen in accordance with the manufacturer's instructions, with the addition of 10 U of RNase inhibitor (Gibco-Invitrogen). For PV, primers P6 (5′-CCTCCGGCCCCTGAATG-3′) and P7 (5′-ACCGGATGGCCAATCCAA-3′) were used to generate a 197-bp amplicon, as originally described by Jaykus et al. (26). For feline calicivirus, primer 4 (5′-TTGCAACTGATTATATTGTTCCTGG-3′) and primer 5 (5′-GCAGTGTTGGATATTTTCTTGTCACC-3′) were used to generate a 243-bp amplicon, as originally described by Radford et al. (41). For HAV, primers HAV 2949 (5′-TATTTGTCTGTCACAGAACAATCAG-3′) and 3192 (5′-AGGAGGTGGAAGCACTTCATTTGA-3′) were used to generate a 267-bp amplicon, as previously described (37, 45). For strain MNV-1, sense primer 6622s (5′-CGCCTTTACCAATTGGCC-3′) and antisense primer 6875 (5′-TGAAAGAGTTGGTTTGGAGC-3′) were used to generate a 273-bp amplicon (27). Positive-control mixtures for RT-PCR were composed of 1 μl of virus, 1 μl of RNase inhibitor, and 8 μl of water and were denatured at 99°C for 5 min prior to RT-PCR. Reverse transcription-PCR was performed starting with a reverse transcription step at 50°C for 30 min and then a 15-min Taq activation step at 95°C. Forty cycles of 60°C for 15 s with a 1-min extension at 72°C and 30 s of denaturation at 95°C were run. After RT-PCR, results were visualized by electrophoresis using 4 to 20% gradient polyacrylamide gels. RT-PCR50 titers for FCV, MNV, HAV, and PV, where one RT-PCR50 unit is the concentration of virus which theoretically yields a positive RT-PCR result 50% of the time, were calculated using serial 10-fold dilutions of virus stocks according to the method of Reed and Muench (43).
Real-time RT-PCR detection.
The primer and minor groove binder (MGB)-TaqMan probe set for HAV was originally described by Costafreda et al. (9) and consisted of Fw-HAV68 (5′-TCACCGCCGTTTGCCTAG-3′), Rv-HAV240 (5′-GGAGAGCCCTGGAAGAAAG-3′), and HAV150(−) (5′-FAM-CCTGAACCTGCAGGAATTAA-MGBNFQ-3′; FAM is 6-carboxyfluorescein). The primer and MGB-TaqMan probe set for MNV, originally described by Baert et al. (4), was modified with TET at the 5′ end for use in duplex assays and consisted of Fw-ORF1/ORF2 (5′-CACGCCACCGATCTGTTCTG-3′), Rv-ORF1/ORF2 (5′-GCGCTGCGCCATCACTC-3′), and MGB-ORF1/ORF2 (5′-TET-CGCTTTGGAACAATG-MGBNFQ-3′). Primers were purchased from Integrated DNA Technologies (Coralville, IA), and probes were purchased from Applied Biosystems (Carlsbad, CA). Reactions were performed using a OneStep RT-PCR kit (Qiagen) in accordance with the manufacturer's recommended procedures, using 25-μl reaction mixtures containing a 200 nM concentration of each primer and probe, 5 U of RNase inhibitor (Gibco-Invitrogen), and 5 μl of RNA extract. Positive-control mixtures containing standardized HAV and MNV RNAs and a negative-control mixture containing all of the reagents except for viral RNA were included with each set of reaction mixtures. The real-time RT-PCR assays were performed in a Smart Cycler (Cepheid, Sunnyvale, CA). The amplification profile consisted of 10 min at 95°C and 40 cycles of 15 s at 95°C, 1 min at 60°C, and 1 min at 70°C. The fluorescence was measured during the annealing step of each cycle. The cycle threshold (CT) was defined as the number of cycles at which the fluorescence of each sample crossed the threshold value of 30. Standards were produced by extraction of viral RNA from HAV and MNV stocks, using a ViralAmp RNA extraction kit (Qiagen). Standard curves were generated by plotting the regression of 10-fold serial dilutions of HAV and MNV standards from 104 to 10−4 PFU, using real-time RT-PCR. Standard curves were produced three times, with similar results. CT values obtained from the duplex reactions were similar to those obtained from reactions run individually. The CT values of the extracted RNAs were applied to the standard curves for a semiquantitative readout, reported as the number of recovered PFU equivalents of RNA.
Uptake and persistence of viruses in oysters.
Individual oysters were placed in 500 ml of natural seawater (salinity of 28 to 30 ppt) with various concentrations of FCV, MNV, HAV, and PV for approximately 16 h. Virus-contaminated oysters were depurated for various periods in 40-liter aquaria equipped with a recirculating UV sterilization unit (Aquafine Inc., Valencia, CA) until use.
Localization of viruses in hemocytes and whole oysters.
The distribution of viruses was determined in oyster hemocytes and whole oysters after removal of the hemocytes. In each experiment, three live oysters were placed in 30 liters of natural seawater in 40-liter aquaria, with one aquarium remaining uncontaminated, while the other was contaminated simultaneously with 106 PFU of both HAV and MNV overnight. After viral contamination, the external surfaces of the oysters were decontaminated at room temperature in a solution of 1% bleach in seawater for 5 min and then wiped dry. Oysters were shucked and hemocytes were separated as described in the RNA extraction section. The oyster tissues were homogenized for 1 min at maximum speed by use of a tissue homogenizer (Omni International, Kennesaw, GA), and approximately 300-mg samples were collected. RNAs were extracted from the tissue homogenates by using the RNeasy method as described previously. Extracts were analyzed by real-time RT-PCR assay. The CT values of the extracted RNAs were applied to the standard curves to calculate the virus PFU, from which the percentages of viruses in the hemocytes versus remaining tissue fractions were determined. Data are reported as the average percentage of total recovered PFU from the whole oyster (hemocyte fraction plus tissue fraction) for four independent experiments.
Comparison of viral RNA extraction methods for hemocytes.
Three live oysters were either left uncontaminated or contaminated individually with 103, 104, or 105 PFU of both HAV and MNV overnight in 1-liter beakers with 500 ml of natural seawater. After virus contamination, oyster shells were decontaminated at room temperature in a solution of 1% bleach in seawater for 5 min and then wiped dry. Oysters were shucked and hemocytes were separated as described in the viral RNA extraction section (method B). Hemocytes and whole oysters were pooled separately, and viral RNA was extracted from the hemocytes by using the Dynabeads and RNeasy methods or from whole-oyster homogenate by using the GPTT procedure (28). Extracts were analyzed by real-time RT-PCR assay. Laboratory contamination of oysters and subsequent extraction were performed in triplicate.
Virus sensitivity to low pH.
Poliovirus, FCV, MNV, and HAV were exposed to a pH range of 1 to 5 at 37°C for 30 min as described by Cannon et al. (7), using 100 mM phosphate buffer (pH 2), citrate buffer (pH 3 to 5), and HCl (pH 1). After neutralization with 2 N NaOH, plaque assays were performed to assess viability.
Transfer of hemocytes to naïve oysters.
V-shaped notches were ground into the oyster shell, and hemolymph was removed as described previously (method A). After initial removal of 0.5 to 1 ml of hemolymph from naïve oysters, 1 ml of pooled hemocytes from three virus-exposed oysters was introduced via injection.
Isolation of viable virus.
Isolation of viable PV from hemocytes was performed after exposure of two individual oysters to 1.1 × 108 PFU of PV each in 500 ml of seawater for 16 h, followed by depuration for 24 h. Approximately 1 ml of hemolymph was removed from the adductor muscle of each oyster and pooled. Hemocytes were pelleted at 200 × g. The pellet was washed with 5 ml of 0.15 M phosphate buffer, pH 9.5, and pelleted again at 200 × g, followed by the addition of 1 ml phosphate buffer, freezing at −80°C, thawing, vortexing, and pelleting at 2,800 × g. The supernatant was then neutralized with 2 N HCl, followed by serial dilution in Earle's balanced salt solution (EBSS), and was used for plaque assay.
Viral RNA recovery efficiency.
Experiments were conducted to determine the virus recovery from virus-seeded hemocytes and from whole oyster meats. Hemocytes were collected from oysters and separated into samples containing hemocytes from approximately one-half of an oyster per tube. Whole oysters were homogenized in a laboratory blender and separated into samples containing half of an oyster per tube. Samples were seeded with 10 μl of EBSS or with 10-fold serial dilutions of both HAV and MNV, from 104 to 10−2 PFU, prepared in EBSS. Viral RNA was extracted from hemocytes by using the Dynabeads and RNeasy methods. RNA extracts were prepared from whole-oyster homogenates by using the GPTT method as described above. Extracts were analyzed by real-time RT-PCR assay. Virus seeding and subsequent extraction were performed in triplicate. The limit of detection (LOD) of each method was determined by the minimum virus input required for positive detection in three consecutive experiments.
Data and statistical analyses.
Recovered PFU for hemocyte-based extraction methods were compared to those for the GPTT method at each dose by analysis of variance (ANOVA) after transforming the PFU values with a logarithmic (base 10) transformation, with 0 values being replaced by a random value between 0 and 1 to retain the “low-count” nature on the log scale. Mean separations at each level of HAV or MNV input were performed using the Bonferroni least significant difference method of the Mixed procedure (SAS software system v. 9.22; SAS Institute, Cary, NC).
RESULTS
Isolation of viruses from hemocytes.
One of the aims of this study was to determine whether viruses are associated with phagocytic hemocytes. Using RNA extraction and RT-PCR analysis, MNV, PV, and HAV were identified within oyster hemocytes and hemolymph drawn from the adductor muscle (method A) after overnight exposure of oysters to the viruses (data not shown). All attempts to identify FCV within hemocytes and whole oysters beyond the initial contamination period were unsuccessful. This result was expected, since limited FCV bioaccumulation by shellfish was reported previously (53).
In order to determine whether viruses were associated with hemocytes or were within the fluid portion of the hemolymph (plasma), whole hemolymph was withdrawn from the adductor muscle (method A) after overnight exposure of two oysters to HAV. Hemocytes were pooled and pelleted by centrifugation (4,000 × g for 10 min), and the supernatant was split into two samples. One sample was filtered with a 0.22-μm syringe filter which would retain cells and most oyster debris but would not retain small virus particles that might be free in the hemolymph. The other sample was not filtered. In three independent trials, HAV was detected by RT-PCR in association with pelleted hemocytes but was not within filtered hemolymph supernatant, indicating that the viruses were associated with hemocytes and not with the hemolymph (Fig. 1). Evaluation of hemocytes from oysters exposed to MNV and PV overnight in three independent trials also demonstrated that PV and MNV were associated with hemocytes but not with filtered (0.22 μm) hemolymph.
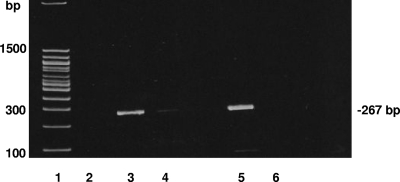
Fig. 1.
Separation of hemocytes from hemolymph. Oysters were contaminated with 4.3 × 107 PFU of HAV for 16 h, and then hemolymph was extracted. Hemocytes were separated from hemolymph by centrifugation, and the supernatant was tested or filtered (0.22 μm) and subsequently tested after extraction of the viral RNA. Lanes: 1, 100-bp DNA ladder; 2, filtered hemolymph supernatant; 3, hemocyte pellet fraction; 4, unfiltered supernatant; 5, positive-control mix for RT-PCR, containing 1 μl (4 × 104 PFU) of HAV, 8 μl of water, and 1 μl of RNase inhibitor; 6, negative-control mix for RT-PCR, containing 10 μl of water.
Assays for infectious viruses were performed on extracted hemocytes after exposure to 1 × 108 PFU of PV for 16 h, followed by depuration for 24 h, to confirm that viruses within hemocytes are viable. Assaying 100 μl of the 1 ml of unseparated hemolymph (containing hemocytes) identified 20 PFU of PV, demonstrating that viable virus was associated with circulating hemocytes. Murine norovirus, HAV, and FCV were not evaluated for viability.
Detection of viruses associated with hemocytes was consistent with detection of viruses within whole tissues.
Using RT-PCR and PAGE analysis, the ability of MNV, HAV, and PV to persist within oysters and in association with hemocytes was evaluated by testing one whole oyster and pooled hemocytes from two oysters, extracted by method A, at each time point (N = 3; n = 6) (Table 1). After exposing oysters to 6 × 105 PFU of MNV in 500 ml of seawater overnight, MNV RNA was detected within whole oysters and hemocytes for up to 8 days postcontamination, with a mean ± standard deviation (SD) of 6 ± 4 days, and was absent from all shellfish after day 11. After exposing oysters to 500 ml of seawater containing 1.1 × 107 PFU of PV, detectable virus RNA persisted within both whole oysters and hemocytes for a mean ± SD of 12 ± 6 days, with all oysters testing negative at day 20 postcontamination. Overnight exposure of oysters to 4.3 × 107 PFU of HAV and subsequent evaluation of viral RNA persistence within oyster meat and hemocytes were performed. A mean (±SD) persistence of 27 ± 4 days was observed, with two trials testing positive at 29 days postcontamination and one trial testing positive at day 22 postcontamination.
Table 1.
RT-PCR detection of viruses in whole oysters and hemocytes after waterborne exposure to single virus stocks at various PFUa
| Test day | RT-PCR detection in three independent trials |
|||||
|---|---|---|---|---|---|---|
| HAV |
MNV |
PV |
||||
| Oyster | Hemocyte | Oyster | Hemocyte | Oyster | Hemocyte | |
| 1 | +/+/+ | +/+/+ | +/+/+ | +/+/+ | +/+/+ | +/+/+ |
| 2 | −/−/− | −/−/+ | ||||
| 3 | −/−/− | +/+/+ | ||||
| 5 | +/+/− | +/+/− | +/−/+ | +/−/+ | ||
| 8 | +/−/+ | +/+/+ | ||||
| 9 | +/+/− | +/+/+ | +/+/− | +/+/− | ||
| 11 | −/−/− | −/−/− | ||||
| 13 | +/+/+ | −/−/+ | ||||
| 15 | +/+/− | +/+/− | ||||
| 16 | −/−/− | +/+/− | ||||
| 20 | −/−/+ | +/+/+ | −/−/− | −/−/− | ||
| 22 | +/+/− | +/−/− | ||||
| 24 | −/−/− | −/+/+ | ||||
| 27 | −/−/− | −/−/− | ||||
| 29 | −/+/+ | −/+/+ | ||||
Oysters were exposed to 6 × 105 PFU of MNV, 1.1 × 107 PFU of PV, or 4.3 × 107 PFU of HAV.
Viral RNA distribution in hemocyte and tissue fractions.
Enteric viruses have previously been shown to be distributed throughout oyster tissues, such as in the gills and the digestive tract (33, 54, 55). To determine the proportion of viruses distributed within hemocytes compared to tissues, hemolymph was removed from whole oysters by direct draw with a pipette after shucking. RNA extracts from hemocyte and tissue fractions were analyzed separately for HAV- and MNV-contaminated oysters to determine the percentage of virus contained within hemocytes versus the remaining tissues. Quantification of recovered viral RNA in each hemocyte fraction (extracted by method B) was expressed as a percentage of the total viral RNA recovered from the whole oyster, calculated as follows: % recovered viral RNA = [hemocyte fraction/(hemocyte fraction + tissue fraction)] × 100. The mean percentages ± SD of total HAV (N = 4; n = 12) and MNV (N = 6; n = 18) PFU recovered from the hemocyte fractions were 56% ± 34% and 79% ± 25%, respectively. Therefore, viral RNA can be detected in both hemocyte and tissue fractions from HAV- and MNV-contaminated oysters, with a substantial amount of the virus being associated with the hemocytes.
Variable virus persistence.
Direct comparison of oyster persistence times for different viruses by RT-PCR analysis is problematic due to a variable virus particle-to-PFU ratio, as well as normal physiological differences between individual oysters. To control for individual oyster variability as well as different virus particle-to-PFU ratios among the different virus stocks, individual oysters were exposed to 106.5 RT-PCR units of HAV, MNV, FCV, and PV simultaneously. In duplicate independent trials, individual oysters were tested at 0, 1, 3, 5, 7, 9, 12, 14, 17, 19, and 21 days postcontamination, using the GPTT viral RNA extraction protocol and RT-PCR (Table 2). After an overnight (16 h) contamination, the oysters tested positive for all four viruses. After 24 h in a laboratory-scale depuration tank, individual oysters tested positive for HAV, MNV, and PV in both trials but were negative for FCV. Poliovirus was no longer detected at 3 days postcontamination. MNV was detected in one trial at 3 days postcontamination, and in a second trial, it was detected for up to 12 days postcontamination. In both trials, HAV was present for at least 21 days postcontamination, when the study was terminated. Thus, HAV persisted the longest, followed by MNV, then PV, and finally FCV, which did not persist.
Table 2.
Viral RNA persistence within oysters after simultaneous exposure to 106.5 RT-PCR units of HAV, MNV, PV, and FCV
| Test day | RNA detection in two independent trials |
|||
|---|---|---|---|---|
| HAV | MNV | PV | FCV | |
| 0 (16 h) | +/+ | +/+ | +/+ | +/+ |
| 1 | +/+ | +/+ | +/+ | −/− |
| 3 | +/+ | +/+ | −/− | −/− |
| 5 | +/+ | +/− | −/− | −/− |
| 7 | +/+ | +/− | −/− | −/− |
| 9 | +/+ | +/− | −/− | −/− |
| 12 | +/+ | +/− | −/− | −/− |
| 14 | +/+ | −/− | −/− | −/− |
| 17 | +/+ | −/− | −/− | −/− |
| 19 | +/+ | −/− | −/− | −/− |
| 21 | +/+ | −/− | −/− | −/− |
Low-pH tolerance of viruses.
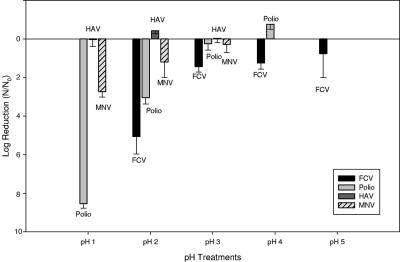
If viruses persist within phagocytic hemocytes, then viruses may be sequestered within low-pH endosomal/lysosomal intracellular compartments as a consequence of phagocytosis and may be subject to subsequent intracellular digestive processes. Thus, one would predict that more acid-tolerant viruses will survive longer within hemocytes. To address the relationship of acid tolerance and virus persistence within oysters, the low-pH resistance of different viruses was investigated. Viability was evaluated in buffers ranging from pH 1 to pH 5 for 30 min at 37°C. Results for acid pH tolerance of HAV, MNV, PV, and FCV are shown in Fig. 2. Overall, HAV was most tolerant to low pH, showing limited plaque reduction. Murine norovirus was the second most tolerant, with limited inactivation at pH 2. Feline calicivirus was completely inactivated (no plaques detected), and PV was substantially inactivated after exposure to pH 2. These results are essentially in agreement with those of Cannon et al. (7) and Hollinger and Emerson (24).
Fig. 2.
Virus pH tolerance. The bar graph depicts the log reduction of each virus, with standard deviation bars, after immersion in acidic buffers ranging from pH 1 to 5 for 30 min. Only FCV was evaluated in pH 5 buffer, and MNV and HAV stocks were not evaluated at pH 4. After immersion in pH 2 buffer, no FCV plaques were detected. After immersion in pH 1 buffer, no PV plaques were observed. Initial titers were 4.8 × 107 PFU/ml for HAV, 1.1 × 109 PFU/ml for PV, 7.5 × 105 PFU/ml for MNV, and 5.8 × 107 PFU/ml for FCV (N = 3; n = 9).
Persistence of transferred HAV-contaminated hemocytes.
Due to its superior ability to persist within shellfish compared to that of PV, MNV, and FCV, HAV was chosen for hemocyte transfer experiments. After overnight exposure to HAV-contaminated seawater, hemocytes and hemolymph were withdrawn from the adductor muscle sinus (method A) and transferred to the adductor muscle sinus of a naïve oyster after removal of an equivalent volume of hemolymph (0.5 to 1 ml). Three separate weekly trials were performed to test for the presence of HAV within individual whole oysters. We demonstrated that HAV-contaminated hemocytes transferred to a naïve unexposed oyster resulted in the oyster testing positive for virus at 14 days posttransfer in two trials (Fig. 3) and at 21 days posttransfer in one trial (not shown).
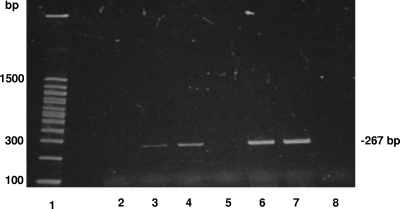
Fig. 3.
HAV persistence after transfer of contaminated hemocytes to naïve oysters. The presence of HAV RNA within whole oyster meat was determined by GPTT RNA extraction and RT-PCR. Lanes: 1, 100-bp DNA ladder; 2, uncontaminated oyster prior to hemocyte transfer; 3, whole-oyster sample 1 week after transfer of hemocytes; 4, whole-oyster sample at 2 weeks posttransfer; 5, whole-oyster sample at 3 weeks posttransfer; 6, a subsample of contaminated hemocytes before injection; 7, positive-control mix for RT-PCR, containing 1 μl of HAV, 1 μl of RNase inhibitor, and 8 μl of water; 8, negative-control mix for RT-PCR, containing 10 μl of water.
Viral RNA detection assay using hemocytes.
A laboratory contamination study was performed to address whether oyster hemocytes could be used instead of whole tissues for rapid diagnostic testing of MNV- and HAV-contaminated oysters. RNA extracts were prepared from hemocytes (method B) and from whole oysters after bioconcentration of 103 to 105 PFU of HAV or MNV by live oysters and extraction by the RNeasy, Dynabeads, or GPTT method, and these were analyzed by real-time RT-PCR. Results are shown in Table 3, which gives CT values and numbers of positive samples for triplicate experiments. After exposure of oysters to 105 PFU of HAV and MNV, all extraction methods detected HAV and MNV RNAs in all 3 experiments. After exposure to 104 PFU of HAV and MNV, each extraction method detected the presence of MNV, but HAV detection varied, with 1 of 3, 1 of 3, and 2 of 3 samples with positive CT values for the Dynabeads, RNeasy, and GPTT methods, respectively. After exposure of oysters to 103 PFU of HAV and MNV, there was no HAV detection with any extraction method. Murine norovirus was detected in 2 of 3, 3 of 3, and 2 of 3 extracts for the Dynabeads, RNeasy, and GPTT methods, respectively. Virus RNA PFU equivalents were determined by real-time RT-PCR assay. Each method was compared to GPTT at each dose by using ANOVA. Differences observed between extraction methods were not significant (P > 0.05). Overall, these findings indicate that HAV and MNV detection is similar in hemocyte and laboratory-contaminated whole-oyster samples.
Table 3.
Real-time RT-PCR detection of viruses in hemocyte and whole-tissue fractions from live oysters exposed to 103, 104, or 105 PFU of HAV or MNV overnight
| Virus and input PFU | Hemocytes |
Whole oysters and GPTT method |
||||
|---|---|---|---|---|---|---|
| Dynabeads |
RNeasy |
|||||
| CT (mean ± SD)a | No. of trials with positive CT/no. of trials | CT (mean ± SD)a | No. of trials with positive CT/no. of trials | CT (mean ± SD)a | No. of trials with positive CT/no. of trials | |
| HAV | ||||||
| 103 | NA | 0/3 | NA | 0/3 | NA | 0/3 |
| 104 | 39.74 | 1/3 | 37.71 | 1/3 | 36.95 | 2/3 |
| 105 | 36.03 ± 0.80 | 3/3 | 36.86 ± 1.73 | 3/3 | 36.73 ± 0.42 | 3/3 |
| MNV | ||||||
| 103 | 37.75 | 2/3 | 36.47 ± 0.98 | 3/3 | 39.01 | 2/3 |
| 104 | 34.65 ± 2.08 | 3/3 | 33.13 ± 2.80 | 3/3 | 35.45 ± 2.92 | 3/3 |
| 105 | 30.33 ± 1.23 | 3/3 | 30.12 ± 1.29 | 3/3 | 33.80 ± 1.39 | 3/3 |
Data are means of positive results or individual results if at least 1 of 3 trials was positive. NA, not applicable.
Viral RNA extracted from homogenates of seeded oyster hemocytes.
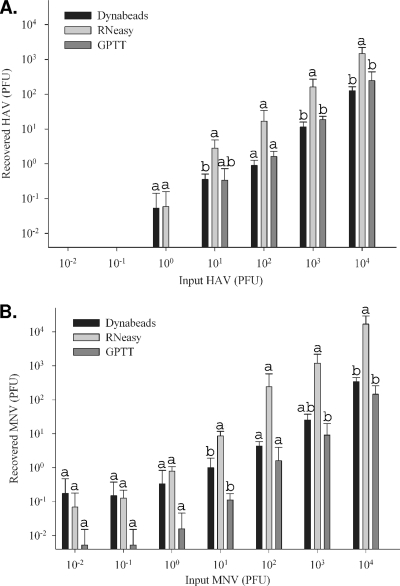
To compare the sensitivities of the RNA extraction methods, hemocyte (extracted by method B) and whole-oyster homogenates were seeded directly with virus. CT values and the numbers of samples with a positive CT value from triplicate experiments are detailed in Table 4. The LOD for HAV was 10 PFU for each of the hemocyte-based extraction methods tested and 100 PFU for the GPTT extraction method (Table 4). The LOD for MNV were 1 PFU, 0.1 PFU, and 10 PFU for the Dynabeads, RNeasy, and GPTT extraction methods, respectively (Table 4). Recovered PFU equivalents are shown in Fig. 4 A and B. Statistical differences in recovered PFU for hemocyte-based extraction methods were compared to the GPTT method at each dose by using ANOVA. Mean recovered PFU equivalents of HAV RNA extracted using the RNeasy method were higher than those obtained with the GPTT method. For HAV inputs of 101, 103, and 104 PFU, the amounts of RNA extracted by the RNeasy method were significantly higher (P < 0.05) (Fig. 4A). Similarly, mean recovered RNA PFU equivalents of MNV RNA extracted using the RNeasy method were higher than those obtained with the GPTT method (Fig. 4B). Mean recovered RNA PFU equivalents from MNV inputs of 101, 102, 103, and 104 PFU were higher than those obtained with the GPTT method. For MNV inputs of 101, 102, and 104 PFU, the amounts of viral RNA extracted using the RNeasy method were significantly higher (P < 0.05) than those obtained with the GPTT method (Fig. 4B). These studies show that detection of HAV and MNV is more sensitive with both of the hemocyte-based extraction methods than with the GPTT method using whole oysters.
Table 4.
Real-time RT-PCR detection of viruses in hemocyte and whole-tissue fractions from oysters seeded with 104 to 10−4 PFU of HAV and MNV
| Virus and input PFU | Hemocytes |
Whole oysters and GPTT method |
||||
|---|---|---|---|---|---|---|
| Dynabeads |
RNeasy |
|||||
| CT (mean ± SD)a | No. of trials with positive CT/no. of trials | CT (mean ± SD)a | No. of trials with positive CT/no. of trials | CT (mean ± SD)a | No. of trials with positive CT/no. of trials | |
| HAV | ||||||
| 10−2 | NA | 0/3 | NA | 0/3 | NA | 0/3 |
| 10−1 | NA | 0/3 | NA | 0/3 | NA | 0/3 |
| 100 | 39.87 | 1/3 | 39.61 | 1/3 | NA | 0/3 |
| 101 | 38.69 ± 0.72 | 3/3b | 35.72 ± 1.17 | 3/3b | 38.30 | 2/3 |
| 102 | 37.28 ± 0.65 | 3/3 | 33.31 ± 1.66 | 3/3 | 36.40 ± 0.72 | 3/3b |
| 103 | 33.39 ± 0.55 | 3/3 | 29.52 ± 1.01 | 3/3 | 32.62 ± 0.37 | 3/3 |
| 104 | 29.71 ± 0.44 | 3/3 | 26.07 ± 0.82 | 3/3 | 28.96 ± 1.15 | 3/3 |
| MNV | ||||||
| 10−2 | 33.56 | 1/3 | 37.31 | 2/3 | 39.66 | 2/3 |
| 10−1 | 35.79 | 2/3 | 35.93 ± 1.18 | 3/3b | 38.71 | 1/3 |
| 100 | 35.75 ± 2.78 | 3/3b | 33.02 ± 0.50 | 3/3 | 37.32 | 1/3 |
| 101 | 33.00 ± 1.30 | 3/3 | 29.71 ± 0.86 | 3/3 | 36.01 ± 0.93 | 3/3b |
| 102 | 30.53 ± 0.58 | 3/3 | 25.67 ± 2.22 | 3/3 | 33.46 ± 2.80 | 3/3 |
| 103 | 27.99 ± 0.80 | 3/3 | 22.59 ± 1.25 | 3/3 | 30.15 ± 1.84 | 3/3 |
| 104 | 24.11 ± 0.44 | 3/3 | 18.59 ± 1.06 | 3/3 | 25.93 ± 2.00 | 3/3 |
Data are means of positive results or individual results if at least 1 of 3 trials was positive. NA, not applicable.
Sample from which LOD was derived.
Fig. 4.
Comparison of extraction methods by use of seeded oyster extracts. Extraction methods were compared using real-time RT-PCR analysis of RNA extracts from hemocyte and whole-tissue fractions of oysters seeded with 10−2 to 104 PFU of hepatitis A virus (A) and murine norovirus (B). Data are reported as recovered PFU equivalents of viral RNA. Statistical differences in recovered PFU for hemocyte-based extraction methods (RNeasy and Dynabeads) relative to the GPTT method were compared using ANOVA. Like letters indicate no significant differences (P > 0.05) between extraction methods.
DISCUSSION
Numerous research papers have focused on the process of enteric virus bioaccumulation by bivalve shellfish, but to our knowledge, this study is the first to offer a putative explanation for why enteric viruses are not readily destroyed by digestive or innate immune processes. The hypothesis that hemocytes are a site of virus persistence is supported by several independent lines of evidence. First, there is an apparent relationship between RT-PCR detection of the virus within the whole oyster and its detection within hemocytes (Table 1). However, detection of the virus within hemocytes has been noted or suggested by other researchers (31) and, by itself, is not particularly surprising, given that the hemocytes interact with food vacuoles as part of the terminal phase of the bivalve digestion process.
Transfer of virus (HAV)-contaminated hemocytes to naïve oysters (Fig. 3) offers a second line of evidence showing that viruses can persist in association with these blood cells. Importantly, the transfer of hemocytes to previously uncontaminated oysters ensures that there is no virus “upstream” in the digestive tract of the oyster and establishes the time frame in which the hemocyte was contaminated. The demonstration that naïve oysters test positive for HAV by RT-PCR 2 weeks or more after hemocyte transfer clearly indicates that viruses persist in association with hemocytes and are not quickly destroyed or rapidly digested by hemocytes. While it cannot be said that hemocytes are the sole site of persistence within oyster tissues, persistence of virus within hemocytes shows that these phagocytic cells can provide an adequate vehicle for intraoyster virus persistence. Isolation of limited numbers of viable PV from hemocytes offers evidence that virus in association with hemocytes can be infectious. Future work will be needed to develop efficient extraction protocols for viable virus from hemocytes to evaluate and quantitate the persistence of viability as judged by in vitro infection instead of RNA detection. While RT-PCR is much more sensitive than tissue culture detection methods, detection of viral RNA does not necessarily offer any insights into the infectious state of virions from which the extracted viral RNA was originally derived.
A third line of evidence is that more acid-tolerant viruses are better able to persist within shellfish. Although direct studies comparing the abilities of different viruses to persist within hemocytes have been limited, a review of the literature indicates that certain viruses remain infectious within oyster tissues for extended periods, while other viruses decline more rapidly. For example, Enriquez et al. (15) demonstrated that HAV persisted longer than PV in mussels. Bosch et al. (6) demonstrated that within contaminated mussels, PV and adenovirus 40 had 2.7- and 3.0-log10 reductions after depuration for 48 h, while a 96-h depuration period reduced bacteriophage B40-8, HAV, and rotavirus serotype 3 less than 2 log10. McLeod et al. (34) reported that human norovirus and HAV persisted longer than PV within Pacific oysters. Ueki et al. (53) demonstrated that HuNoV readily persists within shellfish, but FCV does not. Results reported here are in general agreement with the findings of others regarding the relative persistence of HAV, PV, and FCV, and we confirmed that FCV does not persist well within shellfish, since FCV was undetectable 1 day after contamination of the oysters.
Oyster hemocytes contain acidic phagolysosomal vesicles. Beaven and Paynter (5) used fluorescent dyes and confocal microscopy to determine the internal pH of lysosomes within granular and agranular hemocytes of the Eastern oyster. They found that granular hemocytes and agranular phagolysosomes contained phagolysosomes that averaged pH 4.9 (ranging from 3.8 to 5.9) and 3.1 (ranging from 2.4 to 4.0), respectively. Given that smaller food particles are ingested by phagocytosis, it is plausible that viruses bioconcentrated from the water as potential food particles would eventually end up sequestered within these intracellular vesicles. Thus, we postulate that to persist within shellfish hemocytes, and consequently shellfish meat, viruses must be able to resist acidic digestion within these vesicles. We showed that the acid tolerance of four different viruses (HAV, MNV, PV, and FCV) to acidic buffers is closely linked with the ability of the viruses to persist within oysters. These findings provide evidence that the viruses are localized within phagolysosomes.
Sequestration of human and oyster pathogens within bivalve hemocytes is not unprecedented. Oyster parasites such as Perkinsus marinus (30) and Bonamia ostreae (8), as well as Vibrio vulnificus (21), a pathogenic human bacterium, have been identified in hemocytes. While the experiments in this study clearly implicate hemocytes as a site of persistence, it is not known if hemocytes are the sole site of virus persistence within bivalves. It has been suggested that HuNoV binding to histo-blood group-like glycoproteins within the main ducts which lead to the oyster digestive tract may prevent entry of HuNoV into the digestive system and its subsequent degradation (31).
Shellfish digestive diverticula are known to be sources of concentrated viruses within shellfish (47), inferring that virus uptake involves a digestive process. However, an alternate route of virus uptake by hemocytes that bypasses the digestive tract of bivalves is conceivable. In this scenario, hemocytes might take up human enteric viruses directly from water via association with the mucus-coated epithelial surfaces of the oyster gills, mantle, and other tissues. Hemocytes do routinely pass through oyster epidermis from the hemolymph, where they actively phagocytize and pinocytize foreign particles (2, 16). Due to the relative porosity of the epidermis of bivalves, it is conceivable that human viruses in the water column could diffuse through epidermal tissues to reach circulating hemocytes within hemolymph, which has been observed for bacteria (3). Thus, the relative degree to which digestive and innate immune uptake mechanisms contribute to virus uptake by hemocytes remains to be determined.
In this study, RT-PCR analysis was used as the primary means of assessing virus persistence. This method tests only for the presence of viral RNA and does not indicate the presence of viable viruses, although isolation of infectious PV from hemocytes was demonstrated in this study. While differences in persistence of viruses are noted when RNAs are extracted by the GPTT protocol, it may not be a valid assumption that these different viruses are extracted equally from oysters by this method, since variable extraction efficiencies have been noted (27). In evaluating the initial persistence of MNV, HAV, and PV, a high particle-to-PFU ratio for PV was noted, as judged by a comparison of the PFU titer of 1.1 × 107 and the RT-PCR50 value of 5.6 × 1010 per ml. Poliovirus initially appeared to persist well within shellfish during hemocyte and whole-shellfish comparison experiments. However, the relatively long initial persistence observed for PV was skewed by the high virus particle input, since subsequent direct comparison of identical RT-PCR units of the different viruses resulted in a shorter persistence of PV than of HAV and MNV.
The fact that hemocytes are a site of virus persistence has important implications for virus detection within bivalves. Hemocytes are relatively easy to extract from shellfish and should streamline viral RNA extraction methods, since current procedures often involve the extraction and assay of digestive tissues dissected from shellfish. We have shown that viral RNA extraction from hemocytes by using the Dynabeads and RNeasy methods can be completed in 30 min and is more efficient than our previously developed 8-h GPTT method for whole shellfish or dissected digestive tissues. The results of this study revealed that the RNeasy method had the best performance, recovering greater amounts of viral RNA in a short extraction time.
In summary, these studies (i) found an association between the abilities of different viruses to persist within shellfish and their resistance to low pH; (ii) identified the presence of HAV within hemocytes and noted that the length of virus persistence in hemocytes, as determined by RT-PCR analysis, corresponds with the persistence of viruses in whole oysters; (iii) determined that the transfer of HAV-contaminated hemocytes to unexposed oysters resulted in recipient oysters testing positive for virus for an extended period; and (iv) showed that hemocytes represent an expedient starting material for the development of rapid shellfish testing methods. These studies suggest an explanation for why shellfish depuration as a potential commercial intervention for enteric virus contamination is generally ineffective for purging viruses from shellfish tissues.
ACKNOWLEDGMENTS
We thank Gloria Meade for technical assistance. We thank Brendan Niemira, John Phillips, and Joshua Gurtler for critical reviews of the manuscript.
This work was funded by USDA NRI seed grant 2008-35201-04694 and by intramural funds from the USDA Agricultural Research Service.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
Published ahead of print on 23 September 2011.
REFERENCES
- 1. Allam B., Paillard C. 1998. Defense factors in clam extrapallial fluids. Dis. Aquat. Organ. 33: 123–128 [Google Scholar]
- 2. Allam B., Paillard C., Auffret M. 2000. Alterations in hemolymph and extrapallial fluid parameters in the Manila clam, Ruditapes philippinarum, challenged with the pathogen Vibrio tapetis. J. Invertebr. Pathol. 76: 63–69 [DOI] [PubMed] [Google Scholar]
- 3. Allam B., Paillard C., Ford S. E. 2002. Pathogenicity of Vibrio tapetis, the etiological agent of brown ring disease in clams. Dis. Aquat. Organ. 48: 221–231 [DOI] [PubMed] [Google Scholar]
- 4. Baert L., et al. 2008. Detection of murine norovirus 1 by using plaque assay, transfection assay, and real-time reverse transcription-PCR before and after heat exposure. Appl. Environ. Microbiol. 74: 543–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beaven A. E., Paynter K. T. 1999. Acidification of the phagosomes in Crassostrea virginica hemocytes following engulfment of zymosan. Biol. Bull. 196: 26–33 [DOI] [PubMed] [Google Scholar]
- 6. Bosch A., Pinto R. M., Abad F. X. 1995. Differential accumulation and depuration of human enteric viruses by mussels. Water Sci. Technol. 31: 447–451 [Google Scholar]
- 7. Cannon J. L., et al. 2006. Surrogates for the study of norovirus stability and inactivation in the environment: a comparison of murine norovirus and feline calicivirus. J. Food Prot. 69: 2761–2765 [DOI] [PubMed] [Google Scholar]
- 8. Cochennec-Laureau N., Auffret M., Renault T., Langlade A. 2003. Changes in circulation and tissue-infiltrating hemocytes parameters of European flat oysters, Ostrea edulis, naturally infected with Bonamia ostreae. J. Invertebr. Pathol. 83: 23–30 [DOI] [PubMed] [Google Scholar]
- 9. Costafreda M. I., Bosch A., Pinto R. M. 2006. Development, evaluation, and standardization of a real-time TaqMan reverse transcription-PCR assay for quantification of hepatitis A virus in clinical and shellfish samples. Appl. Environ. Microbiol. 72: 3846–3855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. DePaola A., et al. 2010. Bacterial and viral pathogens in live oysters: 2007 United States market survey. Appl. Environ. Microbiol. 76: 2754–2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Roda Husman A. M., et al. 2007. Rapid virus detection procedure for molecular tracing of shellfish associated with disease outbreaks. J. Food Prot. 70: 967–974 [DOI] [PubMed] [Google Scholar]
- 12. DiGirolamo R., Liston J., Matches J. 1975. Uptake and elimination of poliovirus by West Coast oysters. Appl. Microbiol. 29: 260–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. DiGirolamo R., Liston J., Matches J. 1977. Ionic bonding, the mechanism of virus uptake by shellfish mucus. Appl. Environ. Microbiol. 33: 19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dore W. J., Lees D. N. 1995. Behavior of Escherichia coli and male-specific bacteriophage in environmentally contaminated bivalve mollusks before and after depuration. Appl. Environ. Microbiol. 61: 2830–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Enriquez R., Frösner G. G., Hochstein-Mintzel V., Riedemann S., Rienhardt G. 1992. Accumulation and persistence of hepatitis A virus in mussels. J. Med. Virol. 37: 174–179 [DOI] [PubMed] [Google Scholar]
- 16. Feng S. Y., Feng J. S., Yamasu T. 1977. Roles of Mytilus coruscus and Crassostrea gigas blood cells in defense and nutrition. Comp. Pathobiol. 3: 31–67 [Google Scholar]
- 17. Gentry J., Vinjé J., Guandagnoli D., Lipp E. K. 2009. Norovirus distribution within the estuarine environment. Appl. Environ. Microbiol. 75: 5474–5480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gentry J., Vinjé J., Lipp E. K. 2009. A rapid and efficient method for quantitation of genogroups I and II norovirus from oysters and application in other complex environmental samples. J. Virol. Methods 16: 59–65 [DOI] [PubMed] [Google Scholar]
- 19. Grohmann G. S., Murphy A. M., Christopher P. J., Auty E., Greenberg H. B. 1981. Norwalk virus gastroenteritis in volunteers consuming depurated oysters. Aust. J. Exp. Biol. Med. Sci. 59: 219–228 [DOI] [PubMed] [Google Scholar]
- 20. Guillois-Becel Y., et al. 2009. An oyster-associated hepatitis A outbreak in France in 2007. Euro Surveill. 14: 1–6 [PubMed] [Google Scholar]
- 21. Harris-Young L., Tamplin M. L., Mason J. W., Aldrich H. C., Jackson J. K. 1995. Viability of Vibrio vulnificus in association with hemocytes of the American oyster (Crassostrea virginica). Appl. Environ. Microbiol. 61: 52–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hay B., Scotti P. 1986. Evidence for intracellular absorption of virus by the Pacific oyster, Crassostrea gigas. N. Z. J. Mar. Freshw. Res. 20: 655–659 [Google Scholar]
- 23. Heller D., et al. 1986. An outbreak of gastrointestinal illness associated with consumption of raw depurated oysters. Br. Med. J. 292: 1726–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hollinger F. B., Emerson S. U. 2001. Hepatitis A virus, p. 802 In Knipe D. M., et al. (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 25. Invitrogen 2008. Dynabeads oligo(dT)25: instruction manual. Invitrogen Dynal AS, Oslo, Norway [Google Scholar]
- 26. Jaykus L. A., De Leon R., Sobsey M. D. 1996. A virion concentration method for detection of human enteric viruses in oysters by PCR and oligoprobe hybridization. Appl. Environ. Microbiol. 62: 2074–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kingsley D. H. 2007. An RNA extraction protocol for shellfish-borne viruses. J. Virol. Methods 141: 58–62 [DOI] [PubMed] [Google Scholar]
- 28. Kingsley D. H., Richards G. P. 2001. Rapid and efficient extraction method for reverse transcription-PCR detection of hepatitis A and Norwalk-like viruses in shellfish. Appl. Environ. Microbiol. 67: 4152–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kingsley D. H., Richards G. P. 2003. Persistence of hepatitis A virus within oysters. J. Food Prot. 66: 331–334 [DOI] [PubMed] [Google Scholar]
- 30. La Peyre J. F., Chu F. L., Vogelbein W. K. 1995. In vitro interaction of Perkinsus marinus merozoites with Eastern and Pacific oyster hemocytes. Dev. Comp. Immunol. 19: 291–304 [DOI] [PubMed] [Google Scholar]
- 31. Le Guyader F. S., et al. 2006. Norwalk virus specific binding to oyster digestive tissues. Emerg. Infect. Dis. 12: 931–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Loosanoff V. L. 1958. Some aspects of behaviour of oysters at different temperatures. Biol. Bull. 114: 57–70 [Google Scholar]
- 33. McLeod C., Hay B., Grant C., Greening G., Day D. 2009. Inactivation and elimination of human enteric viruses by Pacific oysters. J. Appl. Microbiol. 107: 1809–1818 [DOI] [PubMed] [Google Scholar]
- 34. McLeod C., Hay B., Grant C., Greening G., Day D. 2009. Localization of norovirus and poliovirus in Pacific oysters. J. Appl. Microbiol. 106: 1220–1230 [DOI] [PubMed] [Google Scholar]
- 35. Metcalf T. G., Stiles W. C. 1965. The accumulation of enteric viruses by the oyster Crassostrea virginica. J. Infect. Dis. 115: 68–76 [DOI] [PubMed] [Google Scholar]
- 36. Newell R. I. E., Langdon C. J. 1996. Mechanisms and physiology of larval and adult feeding, p. 185–229 In Kennedy V. S., Newell R. I. E., Eble A. E. (ed.), The eastern oyster, Crassostrea virginica. Maryland Sea Grant, College Park, MD [Google Scholar]
- 37. Normann A., Graff J., Flehmig B. 1994. Detection of hepatitis A virus in a factor VIII preparation by antigen capture/PCR. Vox Sang. 67: S57–S61 [PubMed] [Google Scholar]
- 38. Olafsen J. A., Mikkelsen H. V., Giaever H. M., Hovik Hansen G. 1993. Indigenous bacteria in hemolymph and tissues of marine bivalves at low temperatures. Appl. Environ. Microbiol. 59: 1848–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Power U. F., Collins J. K. 1989. Differential depuration of poliovirus, Escherichia coli, and coliphage by the common mussel, Mytilus edulis. Appl. Environ. Microbiol. 55: 1386–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Qiagen 2006. RNeasy Mini handbook. Qiagen, Valencia, CA [Google Scholar]
- 41. Radford A. D., Dawson S., Wharmby C., Ryvar R., Gaskell R. M. 2000. Comparison of serological and sequence-based methods for typing feline calicivirus isolates from vaccine failures. Vet. Rec. 146: 117–123 [DOI] [PubMed] [Google Scholar]
- 42. Raftos D. 1998. Adoptive transfer of alloimmune memory in the solitary tunicate, Styela plicata. J. Exp. Zool. A Comp. Exp. Biol. 274: 310–316 [Google Scholar]
- 43. Reed L. J., Muench H. A. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. (London) 27: 493–497 [Google Scholar]
- 44. Richards G. P., Watson M. A. 2001. Immunochemiluminescent focus assays for the quantitation of hepatitis A virus and rotavirus in cell cultures. J. Virol. Methods 94: 69–80 [DOI] [PubMed] [Google Scholar]
- 45. Robertson B. H., et al. 1992. Genetic relatedness of hepatitis A virus strains recovered from different geographical regions. J. Gen. Virol. 73: 1365–1377 [DOI] [PubMed] [Google Scholar]
- 46. Roch P. 1999. Defense mechanisms and disease prevention in farmed marine invertebrates. Aquaculture 172: 125–145 [Google Scholar]
- 47. Schwab K. J., Neill F. H., Estes M. K., Metcalf T. G., Atmar R. L. 1998. Distribution of Norwalk virus within shellfish following bioaccumulation and subsequent depuration by detection using RT-PCR. J. Food Prot. 61: 1674–1680 [DOI] [PubMed] [Google Scholar]
- 48. Sobsey M. D., Davis A. L., Rullman V. A. 1987. Persistence of hepatitis A virus and other viruses in depurated Eastern oysters, p. 1740–1745 In Oceans '87 proceedings, vol. 5. Coastal and estuarine pollution. Marine Technology Society, Washington, DC [Google Scholar]
- 49. Stauber L. A. 1950. The fate of India ink injected intracardially into the oyster, Ostrea virginica Gmelin. Biol. Bull. 98: 227–241 [DOI] [PubMed] [Google Scholar]
- 50. Sullivan J. T. 1990. Long-term survival of heterotropic allografts of the amoebocyte-producing organ in Biomphalaria glabarata (Mollusca: Pulmonata). Trans. Am. Microsc. Soc. 109: 52–60 [Google Scholar]
- 51. Tian P., Bates A. H., Jensen H. M., Mandrell R. E. 2006. Norovirus binds to blood group A-like antigens in oyster gastrointestinal cells. Lett. Appl. Microbiol. 43: 645–651 [DOI] [PubMed] [Google Scholar]
- 52. Tian P., Engelbrektson A. L., Jiang X., Zhong W., Mandrell R. E. 2007. Norovirus recognizes histo-blood group antigen on gastrointestinal cells of clams, mussels, and oysters: a possible mechanism of bioaccumulation. J. Food Prot. 70: 2140–2147 [DOI] [PubMed] [Google Scholar]
- 53. Ueki Y., et al. 2007. Persistence of caliciviruses in artificially contaminated oysters during depuration. Appl. Environ. Microbiol. 73: 5698–5701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang D., Wu Q., Kou X., Yao L., Zhang J. 2008. Distribution of norovirus in oyster tissues. J. Appl. Microbiol. 105: 1966–1972 [DOI] [PubMed] [Google Scholar]
- 55. Wang D., et al. 2008. New target tissue for food-borne virus detection in oysters. Lett. Appl. Microbiol. 47: 405–409 [DOI] [PubMed] [Google Scholar]
- 56. Wobus C., et al. 2004. Replication of norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2: e432. [DOI] [PMC free article] [PubMed] [Google Scholar]