Abstract
The metabolically versatile Rhodococcus sp. strain DK17 is able to grow on tetralin and indan but cannot use their respective desaturated counterparts, 1,2-dihydronaphthalene and indene, as sole carbon and energy sources. Metabolite analyses by gas chromatography-mass spectrometry and nuclear magnetic resonance spectrometry clearly show that (i) the meta-cleavage dioxygenase mutant strain DK180 accumulates 5,6,7,8-tetrahydro-1,2-naphthalene diol, 1,2-indene diol, and 3,4-dihydro-naphthalene-1,2-diol from tetralin, indene, and 1,2-dihydronaphthalene, respectively, and (ii) when expressed in Escherichia coli, the DK17 o-xylene dioxygenase transforms tetralin, indene, and 1,2-dihydronaphthalene into tetralin cis-dihydrodiol, indan-1,2-diol, and cis-1,2-dihydroxy-1,2,3,4-tetrahydronaphthalene, respectively. Tetralin, which is activated by aromatic hydroxylation, is degraded successfully via the ring cleavage pathway to support growth of DK17. Indene and 1,2-dihydronaphthalene do not serve as growth substrates because DK17 hydroxylates them on the alicyclic ring and further metabolism results in a dead-end metabolite. This study reveals that aromatic hydroxylation is a prerequisite for proper degradation of bicyclics with aromatic and alicyclic rings by DK17 and confirms the unique ability of the DK17 o-xylene dioxygenase to perform distinct regioselective hydroxylations.
INTRODUCTION
The ability of bacteria to use aromatic hydrocarbons for growth was first demonstrated over a century ago by isolating Bacillus hexacarborum on the basis of its ability to grow with toluene and xylene (22). Up until now, much research has centered on elucidating the bacterial metabolism of monocyclic and polycyclic aromatic hydrocarbons at the biochemical and molecular level, and the details of these metabolic pathways have been well documented (3, 5, 6, 15, 24). In contrast, little in-depth work has been reported regarding the degradation of bicyclics containing both aromatic and alicyclic moieties. Two different catabolic pathways, through either initial alicyclic or aromatic ring oxidation, have been proposed in several different bacteria, although the alicyclic ring oxidation route seems more common (1, 4, 11, 12, 13, 16, 18, 23).
Recently, we reported that Rhodococcus sp. strain DK17 uses indan as a growth substrate via the o-xylene pathway (9). Specifically, indan degradation by DK17 is initiated by o-xylene dioxygenase, leading to the formation of 4,5-indandiol, which then undergoes ring cleavage by a meta-cleavage dioxygenase (methylcatechol 2,3-dioxygenase, encoded by akbC). The fact that DK17 is the first example of a bacterial strain that metabolizes indan by aromatic oxidation led us to examine its ability to metabolize indan-related compounds such as indene, tetralin, and 1,2-dihydronaphthalene. Our preliminary growth tests showed that DK17 is able to grow on tetralin, while 1,2-dihydronaphthalene and indene failed to serve as sole carbon and energy sources.
Considering their structural similarities to tetralin and indan, respectively, the inability of DK17 to grow on 1,2-dihydronaphthalene and indene is unexpected. The present study was initiated to investigate the observed variations in the ability of DK17 to degrade indan derivatives and gain a comprehensive understanding of the degradation of tetralin, 1,2-dihydronaphthalene, and indene by DK17.
MATERIALS AND METHODS
Bioconversion experiments with Rhodococcus sp. strain DK180.
Rhodococcus sp. strain DK180, derived from DK17, is unable to grow on alkylbenzenes due to the loss of meta-cleavage enzyme activity (7). To analyze the metabolites accumulated during the degradation of tetralin, indene, or 1,2-dihydronaphthalene, DK180 was inoculated in MSB (21) containing 20 mM glucose and incubated overnight at 30°C in shaking incubator (200 rpm). A 15-ml sample of the overnight culture was used to inoculate 200 ml of fresh MSB medium containing 20 mM glucose in a 1-liter Erlenmeyer flask. Then, 200 μl of tetralin (Sigma-Aldrich Korea, Seoul, Korea), 1,2-dihydronaphthalene (TCI, Tokyo, Japan), and indene (Sigma-Aldrich Korea) were placed in the inlet cup with a hole 0.6 mm in diameter connected to a rubber stopper, respectively. Subsequently, the culture flasks were incubated for 24 h at the same conditions.
Bioconversion experiments with the recombinant o-xylene dioxygenase.
Chemically competent cells of E. coli BL21(DE3) were transformed with recombinant pKEB051 containing DK17 o-xylene dioxygenase genes (akbA1A2A3), which encode the large and small subunits and ferredoxin reductase, respectively (8). Subsequently, the resultant BL21(DE3) cells were cotransformed with a chaperon plasmid pKJE7 (TaKaRa Bio, Inc., Ohtsu, Japan) for higher expression and stable maintenance of AkbA1A2A3. Preculture of the resulting transformants was prepared by inoculating one colony into 50 ml of Luria-Bertani (LB) medium supplemented with carbenicillin (100 μg/ml) and chloramphenicol (34 μg/ml), followed by incubation overnight at 37°C. Then, portions (4 ml) of the overnight culture were transferred into 200 ml of LB plus carbenicillin-chloramphenicol, followed by incubation under the same conditions. The culture was induced by adding IPTG (isopropyl-β-d-thiogalactopyranoside; 1.0 mM) and l-arabinose (0.002%) when the bacterial cells reached an optical density at 600 nm of 0.4 to 0.6, followed by further incubation for 2 h. Subsequently, the culture was harvested by centrifugation at 10,000 × g for 15 min, washed twice with 50 mM potassium phosphate buffer (pH 7.4), and resuspended in 30 ml of the same solution supplemented with 20 mM glucose and carbenicillin-chloramphenicol. Tetralin, indene, or 1,2-dihydronaphthalene was provided in the vapor phase from the inlet-cup and the cells were incubated at 30°C for 12 h. A previously developed E. coli BL21(DE3) harboring pCR T7/CT-TOPO vector which was used to construct pKEB051 (10) was used as a negative control strain.
Structural identification of metabolites.
Cells of DK180 or the recombinant E. coli were sedimented by centrifugation (10,000 × g, 20 min, 4°C), and the supernatant was extracted twice with an equal volume of ethyl acetate, followed by concentration in a rotary evaporator. Gas chromatography-mass spectrometry (GC-MS) analysis of tetralin and indene metabolites was carried out with a HP 5972 mass detector (electron impact ionization, 70 eV) connected to a HP 5890 gas chromatograph fitted with a fused silica capillary column (HP-5MS; 0.25 mm by 30 m, 0.25-μm film thickness). The following conditions were used for the GC: 1 ml of He/min; on-column injection mode (injector temperature, 250°C); oven temperature of 100°C for 1 min, increased to 300°C at a rate of 10°C/min, and then held at 300°C for 10 min. The 1,2-dihydronaphthalene metabolites were analyzed by GC-MS with a Perkin-Elmer Clarus 500 MS (70 eV) connected to a Clarus 500 GC with an Elite-5 capillary column (0.25 mm by 30 m, 0.25-μm film thickness). The following conditions were used for the GC: 1.5 ml of He/min; conventional split/splitless injector (CAP, injector temperature, 250°C); oven temperature was 100°C for 1 min, increased to 250°C at a rate of 10°C/min, and then held at 250°C for 10 min. The GC interface inlet line and ionization temperatures were 200 and 180°C, respectively.
Prior to nuclear magnetic resonance (NMR) spectroscopy analysis, the ethyl acetate extracts of tetralin, indene, and 1,2-dihydronaphthalene metabolites were dissolved in methanol and purified through a preparative high-performance liquid chromatography (HPLC) system (Agilent 1100 series) equipped with a YMC-Pack ODS-A column (120 Å, 5 μm, 250 by 10 mm). The mobile phase was a 55-min linear gradient of acetonitrile-water (from 5:95 to 95:5) at a flow rate of 1.5 ml/min. The purified metabolite was dissolved in perdeuterated methanol (CD3OD) and analyzed by one-dimensional 1H and 13C (Varian Unity-300 or -500) and two-dimensional 1H-1H COSY (Varian Unity-500) NMR techniques.
Molecular modeling.
The molecular structure of the large subunit of the Rieske oxygenase component of the DK17 o-xylene dioxygenase was predicted using the Swiss-Model Server (19) based on the crystal structure of biphenyl oxygenase from Rhodococcus sp. strain RHA1 (PDB ID 1ULI).
RESULTS
Comparison of metabolism of tetralin, indene, and 1,2-dihydronaphthalene by the wild-type strain DK17 and the mutant strain DK180.
Rhodococcus sp. strain DK17 grows on tetralin, forming 1-mm-sized colonies in 5 days, while no growth was observable on indene or 1,2-dihydronapthalene under the same conditions. In addition, DK17 excreted a yellow-colored compound into the medium when growing on tetralin. While no growth was observed on indene and 1,2-dihydronaphthalene, a yellowish hue was visible in the first streaked sections for these two substrates.
The yellow pigment is indicative of a meta-ring cleavage product of the catecholic metabolite of tetralin. We thus examined the ability of the mutant strain DK180, which lacks AkbC meta-cleavage enzyme activity and is therefore unable to use alkylbenzenes including o-xylene and indan as a growth substrate (7, 8), to grow with tetralin as the sole carbon source and found that it is unable to grow on tetralin. It should also be noted that a dark brown color developed around DK180 colonies when grown on glucose in the presence of tetralin, indene, or 1,2-dihydronaphthalene. Since all of the results obtained thus far implicate the AkbC enzyme in the degradation of tetralin, indene, or 1,2-dihydronaphthalene, attempts were made to identify the corresponding metabolites in each catabolic pathway.
GC-MS analysis of tetralin, indene, and 1,2-dihydronaphthalene metabolites formed by DK180.
DK180 was grown on glucose in the presence of tetralin, indene, or 1,2-dihydronaphthalene, and metabolites were extracted from the culture supernatant and analyzed by GC-MS as described in Materials and Methods. Table 1 summarizes the GC-MS analysis results of the three extracts. One significant tetralin (molecular weight [MW], 132.09) metabolite peak was detected at 10.20 min on the total ion chromatogram (TIC) and had the molecular ion M+ at m/z 164. When this metabolite was fragmented, a base peak ion at m/z 136 [M-28]+ and a series of prominent ions at m/z 147 [M-OH]+, 146 [M-(OH, H)]+, and 145 [M-(OH, 2H)]+ were produced, which matched well with the mass fragment patterns of a vicinally dihydroxylated metabolite of tetralin, 5,6,7,8-tetrahydro-1,2-naphthalene diol (20).
Table 1.
GC-MS data for tetralin, 1,2-dihydronaphthalene, and indene metabolites by DK180
| Compound | Retention time (min) on GC | Prominent ions (m/z, % relative intensity) |
|---|---|---|
| Tetralin metabolite | 10.20 | 164 (M+, 95), 163 (14), 147 (20), 146 (19), 145 (25), 136 (100), 131 (15), 123 (15), 117 (22), 115 (20), 107 (18), 91 (15), 77 (17), 51 (14) |
| Indene metabolite I | 7.28 | 150 (M+, 21), 132 (50), 131 (33), 115 (13), 107 (54), 104 (100), 103 (44), 91 (37), 79 (28), 78 (30), 77 (48), 51 (23) |
| Indene metabolite II | 9.48 | 148 (M+, 100), 120 (63), 119 (30), 92 (29), 91 (92), 65 (24), 63 (20), 51 (12), 44 (23) |
| 1,2-Dihydronaphthalene metabolite | 9.19 | 162 (M+, 100), 161 (22), 147 (28), 145 (15), 144 (37), 143 (24), 116 (42), 115 (74), 77 (18), 58 (20) |
In the case of indene, GC-MS analysis showed two major indene (MW, 116.16) metabolites, I (7.28 min) and II (9.48 min), on the TIC. Metabolite I produced a molecular ion M+ at m/z 150, prominent ions at m/z 132 [M-(OH, H)]+ and 131 [M-(OH, 2H)]+, and a base peak ion at m/z 104 [M-46]+. The fragmentation pattern of metabolite I was in perfect concordance with that of indan-1,2-diol (cis-1,2-indandiol) in the GC-MS library database (Wiley 7N edition) and the published mass spectrum of indan-1,2-diol (25). Metabolite II produced a molecular ion at m/z 148 (base peak ion) and prominent ions at m/z 120 [M-28]+ and 91 [M-28-29]+. The molecular mass and the mass fragmentation pattern are consistent with those of 1,2-indene diol, which is derived from indan-1,2-diol by a dehydrogenation reaction (17).
GC-MS analysis revealed one major peak for the 1,2-dihydronaphthalene (MW, 130.08) metabolite that eluted at 9.19 min. When this metabolite was fragmented, a molecular ion M+ at m/z 162 (base peak ion) and a series of prominent ions due to the fission of M+ at m/z 145 [M-OH]+, 144 [M-(OH, H)]+, and 143 [M-(OH, 2H)]+ were produced, suggesting that this metabolite is a dihydroxylated 1,2-dihydronaphthalene.
NMR spectroscopy analysis of tetralin, indene, and 1,2-dihydronaphthalene metabolites formed by DK180.
For more rigorous structural determination, each of the above-mentioned metabolites was purified by a preparative HPLC system and subjected to NMR studies. Tetralin and indene metabolites were analyzed by one-dimensional NMR (1H NMR) and two-dimensional NMR (1H-1H COSY) techniques, while 1H and 13C NMR techniques were used in the structure elucidation of the 1,2-dihydronaphthalene metabolites.
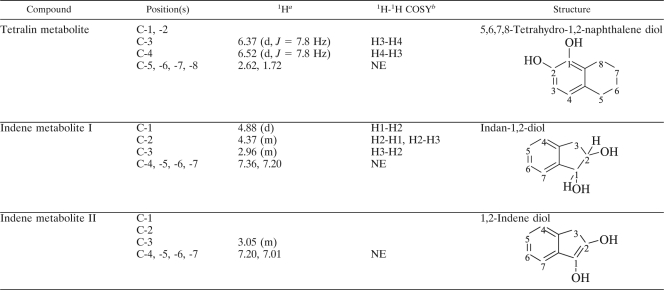
As summarized in Table 2, the protons on carbons 5, 6, 7, and 8 of the tetralin metabolite, which were easily assigned on the basis of their chemical shifts and multiplicities, enabled the assignment of the carbons to which they are attached. The eight protons on the alicyclic ring were detected at δ 1.72 (m, 4H) and δ 2.62 (m, 4H). Two aromatic protons on C-3 and C-4, reciprocally coupled, were absorbed at δ 6.37 (d, J = 7.8 Hz) and δ 6.52 (d, J = 7.8 Hz) with no proton signals on C-1 and C-2, proving that these aromatic protons are attached at C-3 and C-4. Based on the GC-MS and NMR data, the tetralin metabolite was rigorously confirmed to be 5,6,7,8-tetrahydro-1,2-naphthalene diol.
Table 2.
1H (CD3OD, 300 MHz) chemical shifts and 1H-1H COSY (CD3OD, 500 MHz) correlations of tetralin and indene metabolites
1H chemical shifts δ in ppm (multiplicity, J in Hz).
1H-1H COSY data are cross-peaks between signals. NE, not estimated.
In the case of indene metabolite I (putative indan-1,2-diol), two protons on C-1 and C-2 were assigned on the basis of their chemical shifts and multiplicities, and the assignments were further confirmed by 1H-1H COSY signals between these two protons (Table 2). Based on the GC-MS and NMR data, the structure of indene metabolite I was rigorously confirmed to be indan-1,2-diol (cis-1,2-indandiol). The structure of indene metabolite II (putative 1,2-indene diol) was analyzed by comparing to that of indan-1,2-diol. The signals of the two protons on C-1 and C-2 of metabolite II disappeared, although the 1H signal of C-3 remained. This clearly indicates that the two protons of metabolite II are detached during double bond formation between C-1 and C-2, resulting in the molecular ion value m/z 148 [M (150)-2]+. Accordingly, indene metabolite II was rigorously identified as 1,2-indene diol.
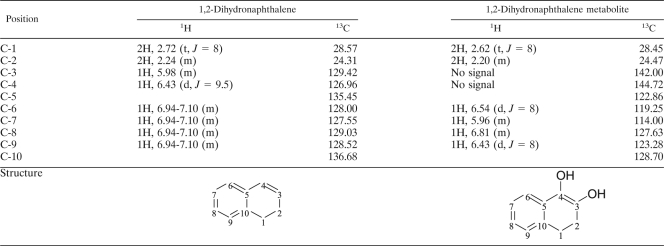
The 1H and 13C chemical shifts and coupling constants for the substrate 1,2-dihydronaphthalene and its dihydroxylated metabolite are given in Table 3. The aromatic protons on C-6, -7, -8, and -9 in 1,2-dihydronaphthalene were absorbed at δ 6.94 to 7.10 (m), but the protons in the metabolite were more widely shifted. The two protons attached to the double bond between C-3 and C-4 in 1,2-dihydronaphthalene were assigned on the basis of their chemical shifts and multiplicities, enabling the assignment of the carbons to which they are attached. The protons were absorbed at δ 5.98 (m) and δ 6.43 (d, J = 9.5 Hz). However, in contrast to 1,2-dihydronaphthalene, the signals for the two protons on the C-3 and C-4 double bond in the metabolite were not detected. This observation indicates that the two protons on C-3 and C-4 are replaced by hydroxyl (-OH) groups. The carbon chemical shifts of the metabolite were also altered. The C-3 and C-4 carbons on 1,2-dihydronaphthalene were absorbed at δ 129.42 and 126.96, respectively, while the chemical shifts of these two carbons on the metabolite were shifted downfield because of the two hydroxyl groups at the C-3 and C-4 positions. Thus, based on GC-MS and NMR data, the dihydroxylated metabolite was clearly identified as 3,4-dihydro-naphthalene-1,2-diol.
Table 3.
Data (500 MHz 1H and 13C NMR) for 1,2-dihydronaphthalene metabolitea
The chemical structures for 1,2-dihydronaphthalene (left) and 3,4-dihydro-naphthalene-1,2-diol (right) are presented at the bottom of the table. 1H, chemical shifts δ in ppm (multiplicity, J in Hz); 13C, chemical shifts δ in ppm.
Hydroxylation of tetralin, 1,2-dihydronaphthalene, and indene by o-xylene dioxygenase.
The rigorous structural identification of a tetralin metabolite as 5,6,7,8-tetrahydro-1,2-naphthalene diol indicates that the initial oxidation of tetralin occurs at the 1,2 positions on the aromatic ring carbon. Thus, to determine whether the DK17 o-xylene dioxygenase, which catalyzes the initial dioxygenation of o-xylene ring, is responsible for the hydroxylation of tetralin, biotransformation experiments were performed using recombinant E. coli cells expressing the DK17 o-xylene dioxygenase. Prior to injection into the GC, the dried residue from the tetralin reaction was treated with methaneboronic acid in pyridine at 80°C for 20 min to produce a methaneboronate derivative as described previously (7). When tetralin metabolites were separated by GC, three significant metabolite peaks were detected at 6.55, 6.75, and 6.97 min on the TIC. The metabolite at 6.55 min has a molecular ion M+ at m/z 190 and a base ion at m/z 147 [M-(CH3, B, OH)]+ (Table 4), suggesting that the metabolite is a methaneboronate derivative and the original form is a vicinally dihydroxylated tetralin (tetralin cis-dihydrodiol). In addition, two other metabolites having an identical molecular ion at m/z 148 eluted at 6.75 and 6.97 min, the mass fragmentation patterns of which matched well with 5,6,7,8-tetrahydro-1-naphthol and 5,6,7,8-tetrahydro-2-naphthol, respectively, in the GC-MS library database (NIST MS Search 2.0). One plausible explanation for the formation of these two phenolics is that tetralin cis-dihydrodiol would spontaneously dehydrate to 5,6,7,8-tetrahydro-1-naphthol and 5,6,7,8-tetrahydro-2-naphthol, with the latter produced in greater quantity due to the chemically preferred rearrangement of tetralin cis-dihydrodiol to this compound (20). In fact, we have previously observed that, when expressed in E. coli, o-xylene dioxygenase transformed o-xylene into 2,3- and 3,4-dimethylphenol, which were derived from an unstable o-xylene cis-3,4-dihydrodiol for the same reason (8).
Table 4.
GC-MS data for tetralin, indene, and 1,2-dihydronaphthalene metabolites by E. coli BL21(DE3) expressing o-xylene dioxygenase
| Compound | Retention time(s) (min) on GC | Prominent ions (m/z, % relative intensity) |
|---|---|---|
| Compound | Retention time(s) (min) on GC | Prominent ions (m/z, % relative intensity) |
| Tetralin metabolitea (tetralin cis-dihydrodiol) | 6.55 | 190 (M+, 62), 175 (6), 161 (15), 147 (100), 133 (27), 120 (52), 105 (12), 91 (85), 77 (35), 65 (17) |
| Indene metabolite (indan-1,2-diol) | 5.97, 6.06 | 150 (M+, 29), 132 (53), 115 (31), 107 (53), 104 (100), 91 (47), 77 (42), 65 (21), 51 (29) |
| 1,2-Dihydronaphthalene metabolite (cis-1,2-dihydroxy-1,2,3,4-tetrahydronaphthalene) | 7.56 | 164 (M+, 8), 146 (44), 128 (4), 120 (82), 119 (100), 115 (21), 104 (8), 91 (54), 77 (17), 65 (19), 51 (11) |
The original sample was derivatized to a methaneboronate.
The bioconversion products of 1,2-dihydronaphthalene by o-xylene dioxygenase were analyzed under the same conditions as described above. A 1,2-dihydronaphthalene metabolite exhibiting a molecular ion at m/z 164 was eluted at 7.56 min (Table 4), suggesting that the metabolite is a vicinally dihydroxylated 1,2-dihydronaphthalene. The mass fragmentation patterns of this metabolite matched well with cis-1,2-dihydroxy-1,2,3,4-tetrahydronaphthalene in the GC-MS library database (NIST MS Search 2.0). In addition, when the original sample was derivatized to a methaneboronate, the vicinally dihydroxylated metabolite peak disappeared, and a compound exhibiting a molecular ion at m/z 188 [M-2H+(BCH3)]+ was newly detected, reinforcing that the metabolite is cis-1,2-dihydroxy-1,2,3,4-tetrahydronaphthalene (data not shown). In addition, two significant 1,2-dihydronaphthalene metabolite peaks were detected at 6.69 and 6.77 min from the original sample. The two metabolites showed an identical molecular ion at m/z 146 [M-H+OH]+ and similar fragmentation patterns. They showed the highest hit value with 5,8-dihydronaphthalen-1-ol (m/z 146), a monohydroxylated derivative from 1,4-dihydronaphtalene, in the GC-MS library database. These two phenolic compounds are also thought to be spontaneously derived from unstable cis-1,2-dihydroxy-1,2,3,4-tetrahydronaphthalene due to the electron-donating nature of the hydrogens bound to carbons 1 and 2. The apparent biotransformation of 1,2-dihydronaphthalene to cis-1,2-dihydroxy-1,2,3,4-tetrahydronaphthalene proves that the o-xylene dioxygenase initiates the 1,2-dihydronaphthalene breakdown pathway in DK17.
When indene metabolites were separated by GC, two significant metabolite peaks were detected at 5.97 and 6.06 min on the TIC. Both of them have a molecular ion M+ at m/z 150 and two decisive ions at m/z 132 [M-(OH, H)]+ and 115 [M-(2OH, H)]+ (Table 4), demonstrating that the metabolites are from a dihydro-dihydroxylated indene. The mass fragmentation patterns of these metabolites matched well with that of indan-1,2-diol in the GC-MS library database (NIST MS Search 2.0). Two other major metabolites were also detected at 5.16 and 5.21 min, respectively, and shown to have an identical molecular ion at m/z 132, indicating that they are likely monohydroxylated indenes derived from indan-1,2-diol. It is noteworthy that none of the above metabolites were detectable from the culture of the negative control strain and the cell-free medium, which were incubated with tetralin, 1,2-dihydronaphthalene, or indene under the same conditions.
DISCUSSION
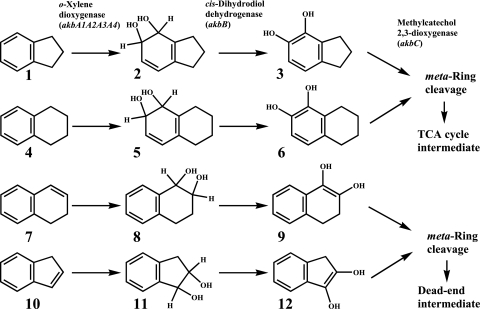
Metabolite analyses by GC-MS and NMR spectrometry clearly show that (i) the meta-cleavage dioxygenase mutant strain DK180 accumulates 5,6,7,8-tetrahydro-1,2-naphthalene diol, 1,2-indene diol, and 3,4-dihydro-naphthalene-1,2-diol from tetralin, indene, and 1,2-dihydronaphthalene, respectively, and (ii) when expressed in E. coli, the DK17 o-xylene dioxygenase transforms tetralin, indene, and 1,2-dihydronaphthalene into tetralin cis-dihydrodiol, indan-1,2-diol, and cis-1,2-dihydroxy-1,2,3,4-tetrahydronaphthalene, respectively. It is noteworthy that all three metabolites formed by DK180 are dehydrogenated forms of the corresponding metabolites produced from tetralin, indene, and 1,2-dihydronaphthalene by the DK17 o-xylene dioxygenase. These observations strongly suggest that, as in the case in indan degradation (9), the three initial o-xylene-degrading enzymes are implicated in the initial degradation of tetralin, indene, and 1,2-dihydronaphthalene by DK17 (Fig. 1).
Fig. 1.
Proposed pathways for early steps in the catabolism of bicyclics by Rhodococcus sp. strain DK17. Gene designations are shown in parentheses. Compounds: 1, indan; 2, cis-indan-4,5-dihydrodiol; 3, 4,5-indandiol; 4, tetralin; 5, tetralin cis-dihydrodiol; 6, 5,6,7,8-tetrahydro-1,2-naphthalene diol; 7, 1,2-dihydronaphthalene; 8, cis-1,2-dihydroxy-1,2,3,4-tetrahydronaphthalene; 9, 3,4-dihydro-naphthalene-1,2-diol; 10, indene; 11, indan-1,2-diol; 12, 1,2-indene diol.
Comparison of the pathways depicted in Fig. 1 reveals the interesting finding that indan and tetralin are subjected to dioxygenation on their aromatic rings by the DK17 o-xylene dioxygenase, while the same enzyme catalyzes the saturation of the alicyclic double bonds of indene and 1,2-dihydronaphthalene. These differential initial hydroxylations place the resulting metabolites into contrasting metabolic fates. The former two bicyclics, which are activated by aromatic hydroxylation, are channeled into the ring-cleavage pathway and are further degraded to support growth of DK17. In contrast, the vicinally dihydroxylated metabolites derived from the latter two bicyclics by alicyclic hydroxylation fail to serve as growth substrates because, although they are destined for degradation through the same ring-cleavage pathway, further degradation of these metabolites is not possible. A fundamental question that arises from these observations is why the DK17 o-xylene dioxygenase fails to attack the aromatic rings of indene and 1,2-dihydronaphthalene. A molecular modeling approach was used to initially address this question.
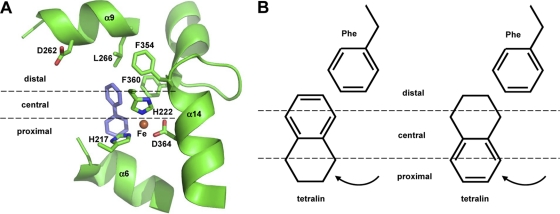
The DK17 o-xylene dioxygenase, which belongs to the Rieske oxygenase class of enzymes (2), consists of a reductase (AkbA4) component, a ferredoxin (AkbA3) component, and a Rieske oxygenase component that contains large and small subunits (AkbA1A2) (8). In order to elucidate the catalytic mechanism of the DK17 o-xylene dioxygenase, we have recently performed molecular functional studies on AkbA1, which contains the catalytic and substrate-binding domains (10, 26). As shown in Fig. 2A, the substrate-binding pocket of AkbA1 is large enough to accommodate bicyclics and can be divided into three regions (distal, central, and proximal), depending on the distance to the mononuclear iron atom. In case of a substrate containing two ring moieties, the one located in the proximal site is to be attacked for oxidation. Further, hydrophobic interactions in the distal position apparently play an important role in substrate binding, largely via the stacking of benzene ring structures from the substrate and phenylalanines at positions 354 and 360. In theory, the substrates indan, indene, tetralin, and 1,2-dihydronaphthalene can be positioned in two ways. In the first position, each molecule exposes its alicyclic moiety to the catalytic motif (Fig. 2B). Alternatively, each molecule can rotate 180° from the first position to expose its aromatic moiety to the catalytic motif. Based on the role of hydrophobic interactions in substrate binding, the first position is likely to be much more favored than the second. In the first configuration, neither indan nor tetralin can be dioxygenated by the DK17 o-xylene dioxygenase due to their lack of double bonds, while indan-1,2-diol, and cis-1,2-dihydroxy-1,2,3,4-tetrahydronaphthalene would be formed as products from indene and 1,2-dihydronaphthalene, respectively. In contrast, the second configuration would allow all of the substrates to be dioxygenated at their aromatic rings, although this is a less favorable configuration. These explanations are not only in good agreement with the observation that indan and tetralin are degraded exclusively via aromatic hydroxylation but also address the possibility of aromatic hydroxylation on indene and 1,2-dihydronaphthalene. We thoroughly examined all of the significant peaks of the TIC of metabolites formed during the incubation of DK180 or the cloned o-xylene dioxygenase with indene or 1,2-dihydronaphthalene. However, no indene and 1,2-dihydronaphthalene metabolites other than those with alicyclic hydroxylation were detected, implicating other unknown factors in the positioning of indene and 1,2-dihydronaphthalene.
Fig. 2.
Molecular modeling and proposed substrate positioning in the active site of o-xylene dioxygenase from Rhodococcus sp. strain DK17. (A) Modeling of biphenyl interaction at the active site. A brown circle indicates the mononuclear iron atom, which is coordinated by three amino acid residues (H217, H222, and D364). (B) Graphic illustration of two alternative binding orientations of bicyclic substrates using tetralin as a representative example. For simplicity, only one aromatic ring from phenylalanines is shown above the substrate. Curved arrows indicate hydroxylation sites.
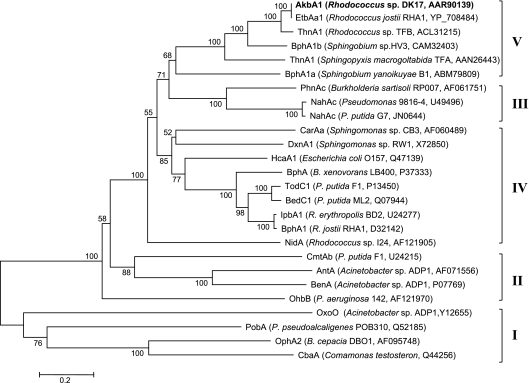
These results reinforce our previous reports that the DK17 o-xylene dioxygenase catalyzes unique oxidations of aromatic compounds, producing metabolites that are not seen in oxidations by other dioxygenases. The novelty of the DK17 o-xylene dioxygenase led us to further investigate phylogenetic relationships among aromatic dioxygenase enzymes. The amino acid sequence of the DK17 AkbA1 was aligned with those of closely related enzymes retrieved from the GenBank database by BLAST search, and a phylogenetic tree was constructed. As shown in Fig. 3, AkbA1/EtbA1, along with four other sequences, forms a monophyletic group, which is clearly separated from the other groups defined by Nam et al. (14). From these results, we propose that AkbA1/EtbA1 forms a fifth phylogenetic lineage (designated group V) of large subunits of oxygenase components in aromatic ring-hydroxylating dioxygenase systems, with AkbA1 the only member with a confirmed function. Overall, the present results demonstrate that aromatic hydroxylation is a prerequisite for the proper degradation of bicyclics with aromatic and alicyclic rings by DK17 and sheds more light on the unique ability of the DK17 o-xylene dioxygenase to perform distinct regioselective hydroxylations.
Fig. 3.
Phylogeny of large subunits (amino acid sequences) of oxygenase components in aromatic ring-hydroxylating dioxygenase systems. The names of bacterial strains and GenBank or Swiss-Prot accession numbers are indicated in parentheses after the protein names. The scale bar denotes 0.2 substitution per site and unit time. The phylogenetic tree was constructed by using the MEGA software (version 4.0) with the neighbor-joining method, and bootstrap values (>50) from 1,000 replicates are shown for each node. Putative monophyletic groups are indicated by the Roman numerals I to V.
ACKNOWLEDGMENTS
This study was supported by the 21C Frontier Microbial Genomics and Applications Center Program and by the Basic Science Research Program (grant 2011-0001111) through the National Research Foundation of Korea. Both are funded by the Ministry of Education, Science, and Technology of the Republic of Korea. D.K. acknowledges the support of the Korea Polar Research Institute under projects PE10050 and PE11060. G.J.Z. acknowledges the support of NIEHS grant P42-ES004911 under the Superfund Research Program. M.Y. and K.Y.C. are recipients of the Brain Korea 21 scholarship.
Footnotes
Published ahead of print on 30 September 2011.
REFERENCES
- 1. Andújar E., Hernáez M. J., Kaschabek S. R., Reineke W., Santero E. 2000. Identification of an extradiol dioxygenase involved in tetralin biodegradation: gene sequence analysis and purification and characterization of the gene product. J. Bacteriol. 182: 789–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ferraro D. J., Gakhar L., Ramaswamy S. 2005. Rieske business: structure-function of Rieske non-heme oxygenases. Biochem. Biophys. Res. Commun. 338: 175–190 [DOI] [PubMed] [Google Scholar]
- 3. Habe H., Omori T. 2003. Genetics of polycyclic aromatic hydrocarbon metabolism in diverse aerobic bacteria. Biosci. Biotechnol. Biochem. 67: 225–243 [DOI] [PubMed] [Google Scholar]
- 4. Hernáez M. J., et al. 2000. Identification of a serine hydrolase which cleaves the alicyclic ring of tetralin. J. Bacteriol. 182: 5448–5453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ju K.-S., Parales R. E. 2010. Nitroaromatic compounds, from synthesis to biodegradation. Microbiol. Mol. Biol. Rev. 74: 250–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Karegoudar T. B., Kim C. K. 2000. Microbial degradation of monohydroxybenzoic acids. J. Microbiol. 38: 53–61 [Google Scholar]
- 7. Kim D., et al. 2002. Monocyclic aromatic hydrocarbon degradation by Rhodococcus sp. strain DK17. Appl. Environ. Microbiol. 68: 3270–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim D., et al. 2004. Identification of a novel dioxygenase involved in metabolism of o-xylene, toluene, and ethylbenzene by Rhodococcus sp. strain DK17. Appl. Environ. Microbiol. 70: 7086–7092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim D., et al. 2010. Aromatic hydroxylation of indan by o-xylene-degrading Rhodococcus sp. strain DK17. Appl. Environ. Microbiol. 76: 375–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim D., et al. 2010. Benzylic and aryl hydroxylations of m-xylene by o-xylene dioxygenase from Rhodococcus sp. strain DK17. Appl. Microbiol. Biotechnol. 86: 1841–1847 [DOI] [PubMed] [Google Scholar]
- 11. Lie F., Chen Y., Wang Z., Li Z. 2009. Enantioselective benzylic hydroxylation of indan and tetralin with Pseudomonas monteilii TA-5. Tetrahedron Asymmetry 20: 1206–1211 [Google Scholar]
- 12. Limberger R. P., Ursini C. V., Moran P. J. S., Rodrigues J. A. R. 2007. Enantioselective benzylic microbial hydroxylation of indan and tetralin. J. Mol. Catal. B Enzym. 46: 37–42 [Google Scholar]
- 13. Moreno-Ruiz E., Hernáez M. J., Martínez-Pérez O., Santero E. 2003. Identification and functional characterization of Sphingomonas macrogolitabida strain TFA genes involved in the first two steps of the tetralin catabolic pathway. J. Bacteriol. 185: 2026–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nam J. W., et al. 2001. New classification system for oxygenase components involved in ring-hydroxylating oxygenations. Biosci. Biotechnol. Biochem. 65: 254–263 [DOI] [PubMed] [Google Scholar]
- 15. Peng R. H., et al. 2008. Microbial biodegradation of polyaromatic hydrocarbons. FEMS Microbiol. Rev. 32: 927–955 [DOI] [PubMed] [Google Scholar]
- 16. Priefert H., et al. 2004. Indene bioconversion by a toluene inducible dioxygenase of Rhodococcus sp. I24. Appl. Microbiol. Biotechnol. 65: 168–176 [DOI] [PubMed] [Google Scholar]
- 17. Raschke H., Fleischmann T., Van der Meer J. R., Kohler H. P. 1999. cis-Chlorobenzene dihydrodiol dehydrogenase (TcbB) from Pseudomonas sp. strain P51, expressed in Escherichia coli DH5α(pTCB149), catalyzes enantioselective dehydrogenase reactions. Appl. Environ. Microbiol. 65: 5242–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schreiber A. F., Winkler U. K. 1983. Transformation of tetralin by whole cells of Pseudomonas stutzeri AS39. Appl. Microbiol. Biotechnol. 18: 6–10 [Google Scholar]
- 19. Schwede T., Kopp J., Guex N., Peitsch M. C. 2003. Swiss-Model: an automated protein homology-modeling server. Nucleic Acids Res. 31: 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sikkema J., de Bont J. A. M. 1993. Metabolism of tetralin (1,2,3,4-tetrahydronaphthalene) in Corynebacterium sp. strain C125. Appl. Environ. Microbiol. 59: 567–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stanier R. Y., Palleroni N. J., Doudoroff M. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43: 159–271 [DOI] [PubMed] [Google Scholar]
- 22. Störmer K. 1908. ÜUber die Wirkung des Schwefelkohlenstoffs und ähnlicher Stoffe auf den Boden. Centr. Bakteriol. Parasitenk. Infekt. 11: 282–286 [Google Scholar]
- 23. Treadway S. L., et al. 1999. Isolation and characterization of indene bioconversion genes from Rhodococcus strain I24. Appl. Microbiol. Biotechnol. 51: 786–793 [DOI] [PubMed] [Google Scholar]
- 24. Ullrich R., Hofrichter M. 2007. Enzymatic hydroxylation of aromatic compounds. Cell. Mol. Life Sci. 64: 271–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wackett L. P., Kwart L. D., Gibson D. T. 1988. Benzylic monooxygenation catalyzed by toluene dioxygenase from Pseudomonas putida. Biochemistry 27: 1360–1367 [DOI] [PubMed] [Google Scholar]
- 26. Yoo M., Kim D., Zylstra G. J., Kang B. S., Kim E. 2011. Biphenyl hydroxylation enhanced by an engineered o-xylene dioxygenase from Rhodococcus sp. strain DK17. Res. Microbiol. 162: 724–728 [DOI] [PubMed] [Google Scholar]