Abstract
Aphids (Hemiptera: Aphididae) have been the focus of several studies with respect to their interactions with inherited symbionts, but bacterial communities of most aphid species are still poorly characterized. In this research, we used bar-coded pyrosequencing to characterize bacterial communities in aphids. Specifically, we examined the diversity of bacteria in two obligately parthenogenetic aphid species (the melon aphid, Aphis gossypii, and the cardamom aphid, Pentalonia caladii) cocolonizing two plant species (taro, Colocasia esculenta, and ginger, Alpinia purpurata) across four Hawaiian Islands (Hawaii, Kauai, Maui, and Oahu). Results from this study revealed that heritable symbionts dominated the bacterial communities for both aphid species. The bacterial communities differed significantly between the two species, and A. gossypii harbored a more diverse bacterial community than P. caladii. The bacterial communities also differed across aphid populations sampled from the different islands; however, communities did not differ between aphids collected from the two host plants.
INTRODUCTION
Aphids (Hemiptera: Aphididae) compose a diversified taxon of phytophagous insects specialized to feed on plant sap through a piercing sucking apparatus. This group of insects has been extensively studied with respect to their association with bacteria, which display diverse phenotypic effects on the aphid hosts and range from detrimental to beneficial. While a few bacteria, typically from the family Enterobacteriaceae, have been shown to be pathogenic (11, 12), the majority of characterized bacteria of aphids are mutualists and maternally inherited (i.e., vertically transmitted) symbionts (6, 13, 27). For instance, nearly all aphids are infected with Buchnera aphidicola (5), an obligate endosymbiont localized within specific cells derived from the host, the bacteriocytes (5). As a result of an ancestral infection and vertical transmission along the host lineages, Buchnera shows a pattern of cospeciation with the aphid hosts. The bacteria have been shown to produce essential amino acids scarcely found in phloem sap of plants upon which aphids feed and develop (5).
In addition to Buchnera, aphids may harbor facultative symbionts that are not essential for host survival. These are diversified bacterial lineages that, in addition to being vertically inherited, are also horizontally transferred within and across host species and sometimes lost within certain lineages (27). Under specific environmental conditions, facultative symbionts provide the hosts with net fitness benefits. For instance, the bacterium Serratia symbiotica, a member of the gammaproteobacteria, has been implicated in the tolerance of aphids to heat stress (21), whereas the bacteria “Candidatus Hamiltonella defensa” and “Candidatus Regiella insecticola,” both of which belong to the gammaproteobacteria, display protective functions against aphid parasitoids and entomopathogenic fungi, respectively (1, 28). “Candidatus Regiella insecticola,” has also being implicated in host plant use (32), and another aphid secondary endosymbiont from the genus Rickettsia has been shown to be responsible for determining chromatic changes in the host body (33). In one case, a consortium of bacteria provided tryptophan to the aphid host with some necessary genes in the Buchnera genome and others in the Serratia genome (10). Although facultative symbionts provide net fitness benefits when hosts are exposed to specific stresses, they also incur some reproductive costs to the hosts; therefore, balancing selection maintains these bacteria at intermediate frequencies in aphid populations (26).
The pea aphid, Acyrthosyphon pisum, has been used as a model system to address several questions related to the diversity and interaction of symbiotic bacteria with aphids (27). However, only scattered information is available on the bacteria associated with other aphid species (13, 24), and little is known about the entire bacterial communities of aphids. High-throughput DNA sequencing approaches have opened up new venues to characterize bacterial communities, and this approach permits the investigation of ecological questions that are not possible to address with more traditional microbial genotyping procedures (2, 3, 8, 18).
To determine the effects of aphid species, host plant, and geographic isolation on aphid-associated bacterial communities, we explored nonnative aphid species on the Hawaiian Islands. The Hawaiian Archipelago hosts approximately 100 nonnative aphid species (20), all of which are believed to reproduce entirely parthenogenetically. The melon aphid, Aphis gossypii, and the cardamom aphid, Pentalonia caladii, are among the most widespread species across the Hawaiian Islands and can coexist on the same plants, like red ginger, Alpinia purpurata, and taro, Colocasia esculenta. We sampled individuals of A. gossypii and P. caladii cocolonizing red ginger and taro across four Hawaiian Islands (Hawaii, Kauai, Maui, and Oahu) and characterized their bacterial communities by using high-throughput DNA sequencing. By sampling two species of aphids from the same host plant species across four islands, we were able to concurrently test three hypotheses: (i) the aphid host governs bacterial community assemblages, (ii) the aphid diet governs bacterial community assemblages, and (iii) physical isolation or environmental differences among localities govern bacterial community assemblages. (These three hypotheses are not necessarily mutually exclusive of each other.)
MATERIALS AND METHODS
Apterous adults were collected with a paintbrush from aphid colonies from December 2008 through February 2010 and stored directly in 70% ethanol. DNA from 46 whole insects (one aphid per colony) was extracted by using the MoBIO (Carlsbad, CA) PowerSoil-htp 96-well DNA isolation kit. Bacterial communities were characterized by using a bar-coded pyrosequencing-based approach described previously (3, 8). Briefly, each sample (n = 46) was amplified in triplicate by using 5 Prime (Gaithersburg, MD) MasterMix under the following conditions: 94°C for 5 min; 40 cycles of 94°C for 45 s, 50°C for 30 s, and 72°C for 90 s; and 72°C for 10 min with bacterial 16S rRNA gene primers 27f and 338r containing 454 adaptors and a 12-bp error-correcting bar code. PCR products from each sample were combined and cleaned by using the MoBIO (Carlsbad, CA) UltraClean-htp 96-well PCR clean-up kit. DNA concentrations were estimated by using the Invitrogen (Carlsbad, CA) Quant-IT PicoGreen dsDNA assay kit. Sample concentrations were normalized and pooled, with sequencing conducted by using GS FLX titanium reagents on an FLX genome sequencer by EnGenCore (Columbia, SC).
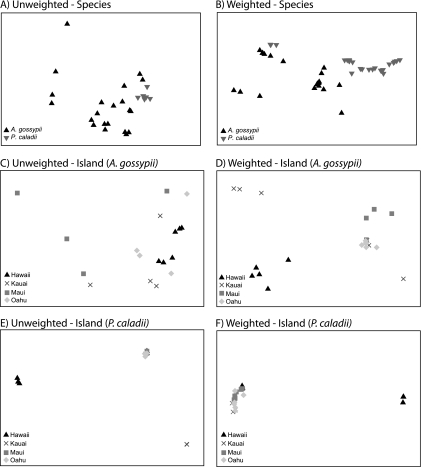
DNA sequence data were processed by using QIIME v1.2 (2). After removing low-quality sequences with the default settings in QIIME, we obtained 111,110 DNA sequences from 46 aphid samples. We binned sequences into 2,657 phylotypes using the uclust algorithm and a 97% similarity threshold. Sequencing errors and limitations of clustering algorithms create spurious phylotypes (16); to eliminate the majority of these, we removed phylotypes from each sample that represented less than 1% of sequences from a given sample. One drawback of this approach is that very rare members of the community will not be detected, but it should minimize the effects of spurious phylotypes on observed diversity patterns. We aligned representative sequences from each phylotype with the NAST aligner (4), used Greengene's lane mask to filter the alignment of highly variable regions, and constructed a phylogenetic tree using fasttree (30). We used both weighted (relative abundances of phylotypes included) and unweighted (presence or absence of phylotypes) UniFrac metrics; UniFrac is a phylogenetic measure of beta diversity that can be used to determine pairwise distances between all samples (18, 19). We used nonmetric multidimensional scaling of UniFrac distance values to visualize the bacterial communities based on aphid species and sampling location (Fig. 1). We also used an analysis of similarity (ANOSIM) procedure to test the hypothesis that bacterial communities differed based on aphid species, host plant, or sampling location (PRIMER6; PRIMER-E Ltd., Ivybridge, United Kingdom).
Fig. 1.
Nonmetric multidimensional scaling plots of individual aphid-associated bacterial communities determined by using unweighted (includes presence and absence data) and weighted (includes relative abundance data) UniFrac. Communities are displayed according to aphid host species (A and B) and according to sampling location for A. gossypii (C and D) and for P. caladii (E and F). Analyses of similarity between aphid species, islands, and host plants are presented in Tables 2 and 3.
Nucleotide sequence accession numbers.
Our final data set included 91,984 sequences binned into 28 phylotypes, deposited as GenBank accession numbers FR832349 to FR832376.
RESULTS AND DISCUSSION
The phylotypes were further classified into eight groups: Buchnera aphidicola (16 phylotypes; 59,293 sequences), Serratia symbiotica (2 phylotypes; 15,345 sequences), Wolbachia (2 phylotypes; 10,236 sequences), a novel lineage of the Enterobacteriaceae (2 phylotypes; 5,576 sequences), Arsenophonus sp. (1 phylotype; 485 sequences), Rickettsia bellii (1 phylotype; 320 sequences), a gammaproteobacterial lineage previously detected in a honey bee (1 phylotype; 117 sequences), and three lineages commonly found in environmental samples (Lactococcus sp. [22 sequences], Stenotrophomonas sp. [23 sequences], and a Brevundimonas sp. [567 sequences]). The prevalences and relative abundances of these eight bacterial groups across aphid species and Hawaiian Islands are presented in Table 1.
Table 1.
Prevalences and average relative abundances of eight bacterial groups in aphid species and Hawaiian Islands
| Bacterium | Aphid species |
Island |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
A. gossypii (sample size = 22) |
P. caladii (sample size = 24) |
Hawaii (sample size = 12) |
Kauai (sample size = 12) |
Maui (sample size = 10) |
Oahu (sample size = 12) |
|||||||
| Prevalence (no. of individuals) | Relative abundance (%) | Prevalence (no. of individuals) | Relative abundance (%) | Prevalence (no. of individuals) | Relative abundance (%) | Prevalence (no. of individuals) | Relative abundance (%) | Prevalence (no. of individuals) | Relative abundance (%) | Prevalence (no. of individuals) | Relative abundance (%) | |
| Arsenophonus | 7 | 1.3 | 2 | 0.3 | 5 | 2.0 | ||||||
| Buchnera | 22 | 63.4 | 24 | 67.9 | 12 | 37.0 | 12 | 60.4 | 10 | 87.6 | 12 | 81.5 |
| Enterobacteriaceae | 3 | 11.0 | 3 | 20.1 | ||||||||
| Gammaproteobacteria | 1 | 0.4 | 1 | 0.7 | ||||||||
| Serratia | 6 | 21.5 | 3 | 10.4 | 9 | 60.3 | ||||||
| Rickettsia | 3 | 0.5 | 3 | 1.0 | ||||||||
| Wolbachia | 2 | 0.4 | 21 | 21.5 | 3 | 1.4 | 6 | 17.0 | 7 | 12.0 | 7 | 15.4 |
| Other/environmentala | 4 | 1.6 | 2 | 0.2 | 4 | 2.6 | 1 | 0.4 | 1 | 0.4 | ||
Includes a Lactococcus sp., a Stenotrophomonas sp., and a Brevundimonas sp.
Most of the phylotypes detected through pyrosequencing are likely to be inherited symbionts; in fact, in addition to Buchnera, the obligate endosymbiont, the majority of other bacterial phylotypes detected (namely, Serratia, Wolbachia, Arsenophonus, and Rickettsia) were previously described as being facultative and inherited endosymbionts of aphids (27) or other arthropods (22), and this may also be the case for the novel lineages of the Enterobacteriaceae detected. The three other bacteria species (Lactococcus sp., Stenotrophomonas sp., and Brevundimonas sp.) represent fewer than 1% of detected sequences, have not been previously described for aphids, and are likely not ecologically important to aphids. These results indicate that bacterial communities of aphids are largely dominated by inherited symbionts.
Three of the 16 Buchnera phylotypes accounted for 99.5% of the total sequences of Buchnera detected. We also detected 13 other Buchnera phylotypes and suggest that these sequences may be rare mutants detected by the high-throughput approach but unlikely to be fixed in the host population. Buchnera has a reduced genome, which lacks, among other genes, the genetic coding for a functional DNA-repairing system and, as a consequence, tends to accumulate mutations at much higher rates than related free-living bacteria and facultative symbionts (22). Thus, the high mutation rate of Buchnera may increase the generation of new lineages, but the reduced fitness of these new lineages likely limits their abundance and distribution. Alternatively, since the aphids are relatively new colonists to the Hawaiian Islands, the detection of uncommon Buchnera lineages may be a result of an incomplete lineage sorting of a polymorphic population.
The high prevalence of Wolbachia in Pentalonia caladii (21/24 individuals) is surprising. It has been thought that aphids are not permissive for hosting Wolbachia, which has been rarely found on those arthropods (9, 25, 34). Wolbachia is typically associated with the manipulation of the reproduction of several arthropod hosts, inducing a number of reproductive disorders, such as cytoplasmic incompatibility and sex ratio distortion (35). However, aphids such as Pentalonia caladii living in subtropical and tropical climates reproduce exclusively parthenogenetically, suggesting that the symbiont may have other effects on these hosts. Some strains of Wolbachia (but not others) associated with Drosophila were shown to have antiviral properties (29), and this antiviral effect is a possible explanation for the presence of Wolbachia in the parthenogenetic aphid.
Beta diversity patterns provide insight into how ecological factors (e.g., aphid host, plant host, and spatial isolation) govern the assembly of entire bacterial communities. Bacterial communities were significantly different between the two aphid species (weighted ANOSIM R = 0.513 [P < 0.001]; unweighted ANOSIM R = 0.487 [P < 0.001]) (Table 2 and Fig. 1A and B). These results suggest that the identity of the host species may affect bacterial composition directly through interactions with the bacterial community. For instance, long periods of coevolution between insects and their vertically transmitted symbionts can lead to cocladogenesis, as is likely the case with the Buchnera bacteria that we detected in A. gossypii and P. caladii. On the other hand, host-specific symbionts may affect community composition via interactions with other bacteria. For example, in Wolbachia-infected P. caladii (21/24 individuals), Buchnera spp. and Wolbachia spp. accounted for 99.74% of the detected bacteria, whereas Wolbachia was largely absent in A. gossypii (only 2 individuals of 22 tested were infected at a low abundance) (Table 1). A. gossypii also harbored greater bacterial diversity than P. caladii, with 7 symbiont genera detected in A. gossypii and 3 genera detected in P. caladii (Table 1). These data may suggest that Wolbachia excludes other secondary symbionts from invading the host, an effect that was demonstrated previously for insect-vectored prokaryotes (23) and insect viruses (29, 31). However, Wolbachia coexists with secondary symbionts in other insects, suggesting that this effect is not universal (7).
Table 2.
ANOSIM of UniFrac distances for aphid-associated bacterial communities across aphid species, Hawaiian islands, and host planta
| Parameter | Unweighted UniFrac |
Weighted UniFrac |
||
|---|---|---|---|---|
| R | P value | R | P value | |
| Species | 0.487 | 0.001 | 0.513 | 0.001 |
| Islands | 0.121 | 0.004 | 0.257 | 0.001 |
| Hawaii, Kauai | 0.148 | 0.037 | 0.294 | 0.007 |
| Hawaii, Maui | 0.235 | 0.008 | 0.539 | 0.001 |
| Hawaii, Oahu | 0.228 | 0.012 | 0.476 | 0.001 |
| Kauai, Maui | 0.047 | 0.150 | 0.067 | 0.130 |
| Kauai, Oahu | 0.028 | 0.202 | 0.028 | 0.227 |
| Maui, Oahu | 0.097 | 0.055 | 0.040 | 0.208 |
| Plant | −0.003 | 0.445 | 0.059 | 0.102 |
Results with significance values less than 0.05 are in boldface.
The composition of aphid-associated bacterial communities also differed among islands (weighted ANOSIM R = 0.257 [P < 0.001]; unweighted ANOSIM R = 0.121 [P = 0.004]) (Table 2). This effect was greater within individual aphid species and was more pronounced in A. gossypii (weighted ANOSIM R = 0.384 [P < 0.001]; unweighted ANOSIM R = 0.537 [P < 0.001]) than in P. caladii (weighted ANOSIM R = 0.143 [P = 0.008]; unweighted ANOSIM R = 0.337 [P < 0.001]) (Fig. 1C to F and Table 3). While Buchnera was detected in every individual and Wolbachia was widespread among the islands, lineages such as Arsenophonus, Serratia, and Rickettsia were limited to one or two islands (Table 1). The geographic variability may be due to selection for specific symbionts in response to differences in environmental conditions or parasite exposure between islands. Symbionts that confer resistance to a threat provide a faster means for insects to adapt than via host evolution and may rapidly spread through insect populations (15). Alternatively, differences between islands may be due to stochastic forces, such as ecological drift, acting on communities of aphid-associated bacteria (14).
Table 3.
ANOSIM of UniFrac distances within aphid species and across the Hawaiian Islandsb
| Parametera |
Aphis gossypii |
Pentalonia caladii |
||||||
|---|---|---|---|---|---|---|---|---|
| Unweighted |
Weighted |
Unweighted |
Weighted |
|||||
| R | P value | R | P value | R | P value | R | P value | |
| Global | 0.384 | 0.001 | 0.537 | 0.001 | 0.142 | 0.008 | 0.337 | 0.001 |
| H, K | 0.265 | 0.048 | 0.489 | 0.002 | 0.207 | 0.095 | 0.546 | 0.002 |
| H, M | 0.619 | 0.014 | 1 | 0.005 | 0.151 | 0.182 | 0.461 | 0.002 |
| H, O | 0.524 | 0.006 | 1 | 0.002 | 0.235 | 0.045 | 0.576 | 0.002 |
| K, M | 0.488 | 0.010 | 0.175 | 0.110 | 0.069 | 0.303 | 0.365 | 0.028 |
| K, O | 0.122 | 0.126 | 0.200 | 0.071 | 0.104 | 0.123 | −0.070 | 0.682 |
| M, O | 0.532 | 0.014 | 0.528 | 0.019 | 0.220 | 0.061 | 0.185 | 0.102 |
H, Hawaii; K, Kauai; M, Maui; O, Oahu.
Results with significance values less than 0.05 are in boldface.
Bacterial communities did not differ based on the host plant (Table 2), and our results corroborate the finding that geography has a greater effect on bacterial community assemblages than plant use in A. gossypii (24). This result is somewhat unexpected in light of previous work that found facultative symbionts to increase aphid fitness on certain plants (17, 32). However, there is also evidence that there may be no effect of secondary symbionts on nutrient use (6), suggesting that symbionts may protect aphids from plant-produced defense rather than providing a dietary advantage. Perhaps, the defenses of taro and ginger do not affect bacterial composition or do not substantially differ between the two plant species. It is also possible, though less likely due to the high dispersal rates of bacteria, that not enough time has passed since the aphids' colonization of Hawaii for the bacterial communities to adapt to differences among host plants.
Overall, our study finds that bacterial communities differ markedly between aphid species but that communities also differ among localities, especially within a species, due to either deterministic or stochastic factors. Because many aphid symbionts are vertically inherited, bacterial communities may differ among species and locations due to the localized proliferation of certain aphid lineages (along with their symbionts). This work suggests that in addition to host effects, biogeographic and stochastic effects may also strongly influence insect-associated bacterial communities and should be considered when characterizing host-associated bacteria.
ACKNOWLEDGMENTS
We thank Jia Hu, who provided funds for R.T.J.'s travel to and accommodations in Hawaii.
We thank members of the University of Hawaii's Department of Plant and Environmental Protection Services and Insect Museum for hosting R.T.J. during his visit and for valuable discussions.
Footnotes
Published ahead of print on 30 September 2011.
REFERENCES
- 1. Brownlie J. C., Johnson K. N. 2009. Symbiont-mediated protection in insect hosts. Trends Microbiol. 17: 348–354 . [DOI] [PubMed] [Google Scholar]
- 2. Caporaso J. G., et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7: 335–336 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Costello E. K., et al. 2009. Bacterial community variation in human body habitats across space and time. Science 326: 1694–1697 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. DeSantis T. Z., et al. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34: W394–W399 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Douglas A. E. 1998. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 43: 17–37 . [DOI] [PubMed] [Google Scholar]
- 6. Douglas A. E., Francois C., Minto L. B. 2006. Facultative ‘secondary’ bacterial symbionts and the nutrition of the pea aphid, Acyrthosiphon pisum. Physiol. Entomol. 31: 262–269 . [Google Scholar]
- 7. Duron O., et al. 2008. The diversity of reproductive parasites among arthropods: Wolbachia do not walk alone. BMC Biol. 6: 27 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fierer N., Hamady M., Lauber C. L., Knight R. 2008. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc. Natl. Acad. Sci. U. S. A. 105: 17994–17999 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gomez-Valero L., et al. 2004. Coexistence of Wolbachia with Buchnera aphidicola and a secondary symbiont in the aphid Cinara cedri. J. Bacteriol. 186: 6626–6633 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gosalbes M., Lamelas A., Moya A., Latorre A. 2008. The striking case of tryptophan provision in the cedar aphid, Cinara cedri. J. Bacteriol. 190: 6026–6029 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grenier A. M., Duport G., Pages S., Condemine G., Rahbe Y. 2006. The phytopathogen Dickeya dadantii (Erwinia chrysanthemi 3937) is a pathogen of the pea aphid. Appl. Environ. Microbiol. 72: 1956–1965 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harada H., Ishikawa H. 1997. Experimental pathogenicity of Erwinia aphidicola to pea aphid, Acyrthosiphon pisum. J. Gen. Appl. Microbiol. 43: 363–367 . [DOI] [PubMed] [Google Scholar]
- 13. Haynes S., et al. 2003. Diversity of bacteria associated with natural aphid populations. Appl. Environ. Microbiol. 69: 7216–7223 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hubbell S. 2001. The unified neutral theory of biodiversity and biogeography. Princeton University Press, Princeton, NJ: . [Google Scholar]
- 15. Jaenike J., Unckless R., Cockburn S. N., Boelio L. M., Perlman S. J. 2010. Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science 329: 212–215 . [DOI] [PubMed] [Google Scholar]
- 16. Kunin V., Engelbrektson A., Ochman H., Hugenholtz P. 2010. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ. Microbiol. 12: 118–123 . [DOI] [PubMed] [Google Scholar]
- 17. Leonardo T. E., Muiru G. T. 2003. Facultative symbionts are associated with host plant specialization in pea aphid populations. Proc. R. Soc. Lond. B Biol. Sci. 270: S209–S212 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lozupone C., Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71: 8228–8235 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lozupone C. A., Hamady M., Kelley S. T., Knight R. 2007. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73: 1576–1585 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Messing R. H., Tremblay M. N., Mondor E. B., Foottit R. G., Pike K. S. 2007. Invasive aphids attack native Hawaiian plants. Biol. Invasions 9: 601–607 . [Google Scholar]
- 21. Montllor C. B., Maxmen A., Purcell A. H. 2002. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol. Entomol. 27: 189–195 . [Google Scholar]
- 22. Moran N. A., McCutcheon J. P., Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42: 165–190 . [DOI] [PubMed] [Google Scholar]
- 23. Moreira L. A., et al. 2009. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell 139: 1268–1278 . [DOI] [PubMed] [Google Scholar]
- 24. Najar-Rodriguez A. L., McGraw E. A., Mensah R. K., Pittman G. W., Walter G. H. 2009. The microbial flora of Aphis gossypii: patterns across host plants and geographical space. J. Invertebr. Pathol. 100: 123–126 . [DOI] [PubMed] [Google Scholar]
- 25. Nirgianaki A., et al. 2003. Wolbachia infections of the whitefly Bemisia tabaci. Curr. Microbiol. 47: 93–101 . [DOI] [PubMed] [Google Scholar]
- 26. Oliver K. M., Campos J., Moran N. A., Hunter M. S. 2008. Population dynamics of defensive symbionts in aphids. Proc. R. Soc. Lond. B Biol. Sci. 275: 293–299 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Oliver K. M., Degnan P. H., Burke G. R., Moran N. A. 2010. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu. Rev. Entomol. 55: 247–266 . [DOI] [PubMed] [Google Scholar]
- 28. Oliver K. M., Russell J. A., Moran N. A., Hunter M. S. 2003. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl. Acad. Sci. U. S. A. 100: 1803–1807 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Osborne S., Leong Y., O'Neill S., Johnson K. 2009. Variation in antiviral protection mediated by different Wolbachia strains in Drosophila simulans. PLoS Pathog. 5: e1000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Price M. N., Dehal P. S., Arkin A. P. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26: 1641–1650 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Teixeira L., Ferreira A., Ashburner M. 2008. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 6: 2753–2763 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tsuchida T., Koga R., Fukatsu T. 2004. Host plant specialization governed by facultative symbiont. Science 303: 1989. [DOI] [PubMed] [Google Scholar]
- 33. Tsuchida T., et al. 2010. Symbiotic bacterium modifies aphid body color. Science 330: 1102–1104 . [DOI] [PubMed] [Google Scholar]
- 34. Wang Z., Shen Z. R., Song Y., Liu H. Y., Li Z. X. 2009. Distribution and diversity of Wolbachia in different populations of the wheat aphid Sitobion miscanthi (Hemiptera: Aphididae) in China. Eur. J. Entomol. 106: 49–55 . [Google Scholar]
- 35. Werren J. H., Baldo L., Clark M. E. 2008. Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6: 741–751 . [DOI] [PubMed] [Google Scholar]