Abstract
The potential prebiotic properties of arabino-oligosaccharides (AOS) derived from sugar beet pulp was studied using mixed cultures of human fecal bacteria from patients with ulcerative colitis (UC), in remission or with active disease, and in healthy controls. These results were compared to those for fructo-oligosaccharides (FOS), which are known to have a prebiotic effect. Fermentation studies were carried out using a small-scale static batch system, and changes in the fecal microbial communities and metabolites were monitored after 24 h by quantitative real-time PCR and short-chain fatty acid analysis. With a few minor exceptions, AOS affected the communities similarly to what was seen for FOS. Quantitative real-time PCR revealed that Bifidobacterium spp. and Lactobacillus spp. were selectively increased after fermentation of AOS or FOS by fecal microbiota derived from UC patients. The stimulation of growth of Lactobacillus spp. and Bifidobacterium spp. was accompanied by a high production of acetate and hence a decrease of pH. The fermentation of AOS may help improve the inflammatory conditions in UC patients through stimulation of bacteria eliciting anti-inflammatory responses and through production of acetate. AOS may therefore represent a new prebiotic candidate for reduction of the risk of flare-ups in UC patients. However, human trials are needed to confirm a health-promoting effect.
INTRODUCTION
During the twentieth century a significant increase in the incidence of inflammatory bowel diseases has occurred in Western Europe and North America (33). Ulcerative colitis (UC) is an idiopathic inflammatory bowel disease characterized by chronic inflammation of the colonic mucosa. UC is usually associated with chronic remissions, which are periods in which patients are completely symptom free, and relapse periods, characterized by diarrhea with passage of blood or mucus, occasional abdominal cramping, and pain as well as, in severe cases, systemic symptoms, including fever and weight loss (3, 4). The etiology of inflammatory bowel disease remains unclear, and no causal infectious agent has been identified. The commensal bacterial intestinal community represents the environmental factor most frequently implicated in the development of UC (4, 60). Evidence for this implication is provided by the complete lack of enterocolitis in genetically engineered germfree mice, rats, and guinea pigs, which reproducibly develop intestinal inflammation within 1 to 4 weeks if they are colonized with conventional gut bacteria (61). Thus, a dysbiosis in the composition of the gut microbiota may influence key mechanisms involved in the inflammatory process of the intestinal mucosa (4, 21, 60). Earlier studies have shown differences between the intestinal microbiotas of UC patients and those of healthy subjects (45, 64). In particular, the patients with UC in relapse are reported to have a small amount of bifidobacteria (64) and we have observed that the prevalence of lactobacilli in UC patients is also low compared to that in healthy subjects (L. K. Vigsnæs et al., submitted for publication). Bifidobacteria and lactobacilli are believed to play an important role in maintaining intestinal health due to their effect on maturation and balancing of the immune system (43, 59, 71) and to their inhibition of pathogens (8, 17, 53). Hence, an underrepresentation of bifidobacteria and lactobacilli may compromise the colon health and contribute to a higher risk of flare-up in patients with UC. Maintenance of a healthy gut microbiota and homeostasis can be promoted by the consumption of indigestible carbohydrates or dietary fibers (11, 27, 37). However, a sufficient fiber intake is required for the desired effect (14). Prebiotics are defined as “selectively fermented ingredients that cause specific changes in composition and/or activity in the gastointestinal microbiota, which confer benefits upon host well-being and health” (47). The selective stimulation of specific colonic bacteria is explained by the capability of these bacteria to break down the glycosidic linkages in the prebiotic carbohydrates. These bacteria are able to grow on particular carbon sources, which are less easily fermented by other members of the intestinal community. This provides them with a selective advantage when competing with other bacterial species in a mixed bacterial community such as the human colon (58). Thus, the monosaccharide composition, glycosidic linkage, and length of the prebiotics contribute to the relative increase of beneficial bacteria, including bifidobacteria and lactobacilli (29, 46, 57). Metabolites produced by bacterial fermentation of prebiotics include the short-chain fatty acids (SCFA) acetate, propionate, and butyrate. SCFA have local and systemic biological effects that are proposed to be beneficial to human health (69). Acetate is important for lowering the pH of the intestine, thereby inhibiting enteropathogenic bacteria (16, 17). Butyrate is the preferred energy source for colonocytes, and studies have shown that butyrate can act as an anticarcinogenic agent through selective induction of apoptosis in colon cancer cells (39, 52, 54). With respect to inflammatory bowel diseases, butyrate is important because it stimulates an anti-inflammatory state in the colon by downregulation of proinflammatory cytokine production (62). Ingestion of prebiotic compounds is considered an alternative approach for treatment of UC, since an alteration of the colonic microenvironment toward homeostasis may help decrease the risk of flare-ups.
Sugar beet pulp, originating from industrial production of sugar, is rich in pectins, which are heterogeneous molecules containing regions of homogalacturonan and rhamnogalacturonan I. In sugar beet pectin, the side chains of rhamnogalacturonan I are often rich in arabinan composed of α-1,5-linked backbone with a high degree of α-1,2- and/or 1,3-arabinofuranosyl substitutions along with some β-1,4-linked galactan (40).
Significant potential exists for converting such by-product streams from industrial bioprocesses into new value-added products. Several types of nondigestible fibers and oligosaccharides from plant cell walls have shown the capability to modulate the human gut microbial composition in in vitro studies. Some arabino-oligosaccharides (AOS) from sugar beet pectin (1) and AOS derived from lemon peel (26) are reported to stimulate the growth of bifidobacteria to the same extent as the “gold standard” prebiotic fructo-oligosaccharides (FOS) and inulin, respectively (42, 55, 57). High-molecular-weight rhamnogalacturonan I from potato pulp was superior to FOS in stimulating bifidogenic growth (66).
The aim of this work was to examine the potential prebiotic properties of AOS released from rhamnogalacturonan I during sequential acid extraction of pectin from sugar beet pulp. Fermentation-induced changes in fecal microbial communities obtained from either healthy subjects or patients with UC in remission or relapse were investigated to establish whether fermentation of AOS influenced the fecal communities from UC patients in the same way as known prebiotic FOS.
MATERIALS AND METHODS
Carbohydrates and chemicals.
Sugar beet AOS were obtained from Danisco A/S (Nakskov, Denmark). The AOS were identical with the substrate described as “starting material” by Holck and coworkers (24) and was found as linear and branched arabino-oligosaccharides mainly of 2 to 10 degrees of polymerization (DP). The content of monosaccharides (mainly arabinose, glucose, and fructose) contributed to 125 mg/g dry matter. The arabinose moiety in the total sample accounted for 85 mol%. The ferulic acid content was 36 μg/g dry matter (24). Fructo-oligosaccharides (FOS) (2 to 8 DP) (OraftiR95) were obtained from Beneo-Orafti (Tienen, Belgium). Unless stated otherwise, chemicals were obtained from Sigma-Aldrich (Steinheim, Germany).
Human volunteers and clinical characteristics of the UC patients.
Fecal samples were obtained from 12 patients with UC and six healthy controls (four women and two men). Within the UC group, six patients (three women and three men) were in clinical remission and six patients (three women and three men) had active disease at the time of sampling according to clinical and endoscopic criteria (5). The patients were previously diagnosed with UC according to standardized diagnostic criteria at the Department of Medical Gastroenterology, Herlev Hospital (28). The study was performed in accordance with the Second Helsinki Declaration, reported to the Danish Data Protection Agency, and approved by the Regional Ethics Committee. Four of the six patients with inactive UC received maintenance treatment with oral mesalazine (anti-inflammatory bowel-specific aminosalicylate drug) in a dosage of 1.6 to 2.4 g daily, and one also received azathioprine (purine analogue immunosuppressive drug) 100 mg daily. One patient received oral olsalazine (anti-inflammatory bowel-specific azodisalicylate drug) (1 g daily), and one received no treatment. All six patients with active UC were treated with oral mesalazine in a dosage of 2.4 to 3.2 g daily as well as topical mesalazine 1 g daily either as an enema (n = 5) or as a suppository (n = 1). One patient also received azathioprine 100 mg daily. One patient had active extensive UC, one left-sided colitis, and the rest either active proctitis or proctosigmoiditis. One patient with well-established proctitis refused a new sigmoidoscopy. None of the participants had been treated with antibiotics for at least 2 months before enrollment, and there was no significant difference (P = 0.57) in the mean age of the participants between the three groups (healthy control, 41 ± 9 years; UC patients in remission, 41 ± 8 years; and UC patients in relapse, 45 ± 8 years).
Sample collection and processing.
The stool samples were collected at the home of the participant in airtight containers and stored at 4°C (limited storage time was encouraged [41]) until delivery to the laboratory, where they were processed immediately. Feces (200 mg, wet weight) were collected in triplicates in the middle of each stool sample for DNA extraction and used for the measurement of original bacterial communities. At the same time, feces were prepared for in vitro fermentation in an anaerobic cabinet (Macs Work Station; Don Whitley), containing 10% H2, 10% CO2, and 80% N2. The fecal samples for in vitro fermentation were homogenized in 50% glycerol (1:1 dilution) and stored at −80°C until further analysis, as described below. The use of frozen samples compared to fresh samples in fermentation experiments has been validated previously (49).
Small-scale in vitro fermentation.
A small-scale in vitro fermentation method was used to assess the effect of AOS on the microbial composition in human fecal samples. FOS was applied as a standard with known bifidogenic effect (50). AOS and FOS were sterilely filtrated and added to an autoclaved minimal basal medium to give a final concentration of 5 g/liter in a reaction volume of 2 ml. The minimal basal medium contained 2 g/liter of peptone water, 1 g/liter of yeast extract, 0.1 g/liter of NaCl, 0.04 g/liter of K2HPO4, 0.04 g/liter of KH2PO4, 0.01 g/liter of MgSO4·7H2O, 0.01 g/liter of CaCl2·2H2O, 2 g/liter of NaHCO3, 0.5 g/liter of bile salts, 0.5 g/liter of l-cysteine hydrochloride, 50 mg/liter of hemin, 10 μl/liter of vitamin K1, 2 ml/liter of Tween 80 (VWR, Darmstadt, Germany), and 0.05‰ (wt/vol) resazurin solution. The pH of the minimal medium with added FOS or AOS was adjusted to 7, and the solutions were reduced overnight in an anaerobic cabinet. Fecal samples were defrosted in an anaerobic cabinet, and 10% (wt/vol) fecal slurry was prepared by mixing of the samples with anoxic PBS (Oxoid, Greve, Denmark). The reduced minimal medium with added FOS or AOS was distributed into 3.8-ml sterile screw-cap vials (Nunc) and inoculated to a final concentration of 1% (wt/vol) feces. Caps were loosely placed on the vials to allow gas exchange but avoid evaporation. Each fermentation experiment for the fecal sample of each subject was carried out in triplicates for each carbohydrate source. A parallel incubation with no added substrate was used as a control for the SCFA analysis and pH measurements. The fermentation was non-pH controlled and nonstirred, due to the low reaction volume, and was carried out in an anaerobic cabinet at 37°C. After 24 h of fermentation, the pH of the 2-ml fermentation samples was measured (pH indicator strips, pH 2.0 to 9.0; Merck) in the anaerobic cabinet, and subsequently a 1-ml fermentation sample was taken out for extraction of bacterial DNA and SCFA analysis, respectively.
Extraction of bacterial DNA.
DNA was extracted from each of the triplicate fecal samples and each of the triplicate fermentation samples using the QIAamp DNA stool minikit (Qiagen, Hilden, Germany) with a bead beater step in advance, as previously described (31). The purified DNA was stored at −20°C until use.
Real-time PCR assay conditions.
Amplification and detection of purified bacterial DNA by real-time PCR was performed with the ABI228 Prism 7900 HT from Applied Biosystems using optical grade 384-well plates. Each amplification reaction was done in duplicate for each of the triplicate fecal and triplicate fermentation samples in a final volume of 11 μl containing 5.50 μl EXPRESS SYBR GreenER qPCR SuperMix (Invitrogen A/S, Taastrup, Denmark), 200 nM each of the primers (Eurofins MWG Synthesis GmbH, Ebersberg, Germany), 2 μl template DNA (5 ng/μl), and nuclease-free water purified for PCR (Qiagen). The amplification program consisted of one cycle at 50°C for 2 min, one cycle at 95°C for 10 min, 40 cycles at 95°C for 15 s and 60°C for 1 min, and finally one cycle of melting curve analysis for amplicon specificity at 95°C for 15 s and 60°C for 20 s, increasing the ramp rate by 1.92°C/min until maintaining at 95°C for 15 s. The 16S rRNA-targeting primers used in this study are listed in Table 1. The primers were tested to confirm sensitivity and specificity (see Table S1 in the supplemental material).
Table 1.
16S rRNA gene primers used in this study
| Target taxon | Primer | Sequence (5′-3′) | PCR product (bp) | Efficiency (E)b | Reference |
|---|---|---|---|---|---|
| Faecalibacterium prausnitzii | Fprau 07 | CCA TGA ATT GCC TTC AAA ACT GTT | 140 | 1.90 | 64 |
| Fprau 02 | GAG CCT CAG CGT CAG TTG GT | ||||
| Bifidobacterium spp. | F-bifido | CGC GTC YGG TGT GAA AG | 244 | 2.04 | 9 |
| R-bifido | CCC CAC ATC CAG CAT CCA | ||||
| Lactobacillus spp. | Lacto-F | AGC AGT AGG GAA TCT TCC A | 341 | 1.98 | 22, 70 |
| Lacto-R | CAC CGC TAC ACA TGG AG | ||||
| Clostridium leptum subgroup | sg-Clept-F | GCA CAA GCA GTG GAG T | 239 | 2.03 | 38, 63 |
| sg-Clept-R | CTT CCT CCG TTT TGT CAA | ||||
| Clostridium coccoides group | g-Ccoc-F | AAA TGA CGG TAC CTG ACT AA | 440 | 2.02 | 38 |
| g-Ccoc-R | CTT TGA GTT TCA TTC TTG CGA A | ||||
| Firmicutes phylum | Firm934F | GGA GYA TGT GGT TTA ATT CGA AGC A | 126 | 2.01 | 20 |
| Firm1060R | AGC TGA CGA CAA CCA TGC AC | ||||
| Bacteroidetes phylum | Bact934F | GGA RCA TGT GGT TTA ATT CGA TGA T | 126 | 1.98 | 20 |
| Bact1060R | AGC TGA CGA CAA CCA TGC AG | ||||
| Desulfovibrio spp. | DSV691-F | CCG TAG ATA TCT GGA GGA ACA TCA G | 136 | 1.90 | 15 |
| DSV826-R | ACA TCT AGC ATC CAT CGT TTA CAG C | ||||
| V2-V3 16S rRNA regiona | HDA1 | ACT CCT ACG GGA GGC AGC AGT | 200 | 1.98 | 70 |
| HDA2 | GTA TTA CCG CGG CTG CTG GCA C | ||||
| 16S rRNA regiona | TBA-F | CGG CAA CGA GCG CAA CCC | 130 | 2.04 | 10 |
| TBA-R | CCA TTG TAG CAC GTG TGT AGC C |
The HDA and TBA primers were used as total bacterial DNA targets to normalize for differences in total DNA concentrations between individual samples.
Primer efficiency is calculated from the slope of the standard curve for each primer set, E = 10−1/slope.
Real-time PCR data handling.
The relative quantities and relative ratios of gene targets encoding 16S rRNA sequences of the bacterial taxa were calculated using EΔCT and EΔCT (fermentation sample)/EΔCT (original bacterial community), respectively, where E is the efficiency of the primer calculated from the slope of the standard curve (E = 10−1/slope) and ΔCT is the CT value of the bacterial target normalized against the CT value of the total bacterial population in the sample (23). CT is the threshold cycle calculated by the ABI software (SDS version 2.2; Applied Biosystems, Foster City, CA) as the PCR cycle, where the amplification signal exceeds the selected threshold value, also set by the ABI software. Standard curves were created using serial 10-fold dilutions of bacterial DNA extracted from one of the fermentation samples for all primer sets. Analysis of the standard curves allowed verification of PCR efficiency for the chosen PCR conditions (Table 1). All results were calculated as the means of duplicate determinations, equal values required.
Gas chromatography for SCFA analysis.
Prior to quantification of SCFA, the samples were acidified to pH <2 with 17% H3PO4 and centrifuged at 10,000 × g for 10 min at room temperature to remove particle material. The supernatants were collected and filtered through a 0.45-μm-pore-size filter (Milipore, Copenhagen, Denmark) to remove bacterial cells. SCFA were analyzed using gas chromatography (Perkin Elmer 400 Clarus) with a flame ionization detector and a capillary column (Agilent Technologies, Horsholm, Denmark). The temperatures of the injector and detector were 175°C and 200°C, respectively. The temperature gradient was as follows: 70°C for 1 min, ramped at 8°C/min to 150°C, and after that ramped at 45°C/min to 230°C in 3 min (total time, 15.78 min). The injection volume of the sample was 0.5 μl, and the nitrogen flow was 12 ml/min. The quantified SCFA were analyzed as acids: acetic, propionic, and butyric. The standards used in the SCFA analysis were the same acids as mentioned above with concentrations of 1 to 40 mΜ.
Statistics.
Statistical analysis of the quantitative real-time PCR (qPCR) data was performed with the SAS JMP software (version 6.0.2; SAS Institute Inc., Cary, NC). Data were analyzed by a mixed-model analysis of variance (ANOVA), where the microbiota source (healthy controls, UC patients in remission, and UC patients in relapse) and the incubations (inoculum, FOS, and AOS) were used as the fixed effects, while the individual subjects were entered as random effects. When ANOVA indicated a significant difference, a pairwise multiple comparison of means (Student's t test) was used to determine significant differences among treatment combinations, assuming equal variances. The PCR measurements were log transformed before statistical analysis to obtain normal distribution of the data. Normal distribution was assessed using a D'Agostino and Pearson omnibus normality test with the GraphPad Prism software (version 5.03; GraphPad Software Inc., La Jolla, CA). Statistical analysis of the SCFA data was performed with the SAS JMP software (version 6.0.2) using a two-way ANOVA. Univariate ANOVA was used to analyze differences in the ages of the three groups (GraphPad Prism software, version 5.03). Tests were considered statistically significant if P values lower than 0.05 were obtained. Principal component analysis (PCA) was carried out using the SAS JMP software (version 6.0.2) on the qPCR data to investigate the difference between the carbohydrate incubations and, additionally, which bacterial taxa may be important for fermentation of the tested carbohydrates.
RESULTS
Bacterial communities in the fecal samples.
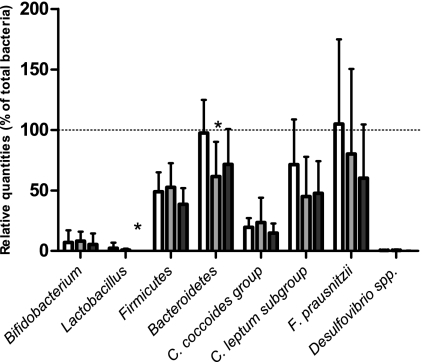
The relative quantification of gene targets encoding 16S rRNA gene sequences present in the original bacterial communities derived from fecal samples (Fig. 1) revealed that densities of Lactobacillus spp. were significantly lower in the UC patients in relapse than in healthy controls (P < 0.05). No significant difference between densities of Lactobacillus spp. present in UC patients in remission and relapse, respectively, was observed. However, there was a trend of lower densities of lactobacilli in the relapse than in the remission group (0.05 < P < 0.10). Densities of the Bacteroidetes phylum were significantly lower in the UC patients in remission than in healthy controls (P < 0.05). The mean relative quantity of Bifidobacterium spp. was lower in the UC patients in relapse than in the other groups, although this was not statistically significant, probably due to high interindividual variation in the three groups. There were no significant differences in the densities of the Firmicutes, the Clostridium coccoides group, the Clostridium leptum subgroup, Faecalibacterium prausnitzii, and Desulfovibrio spp. between the three groups.
Fig. 1.
Relative quantities of gene targets of the original bacterial community in fecal samples from healthy controls (white), ulcerative colitis patients in remission (gray), and ulcerative colitis patients in relapse (black). Target genes encoded 16S rRNA from Lactobacillus spp., Bifidobacterium spp., Firmicutes, Bacteroidetes, Clostridium coccoides group, Clostridium leptum subgroup, Faecalibacterium prausnitzii, and Desulfovibrio spp. The relative quantities (RQ) were calculated as RQ = EΔCT, where E represents the primer efficiency and ΔCT was the CT value of the bacterial target normalized against the CT value of the total bacterial population in the sample. Total bacteria were set to 100% (indicated by dotted line). Error bars represent averages ± standard errors of the means of relative quantities in target species in each of the six samples in each group. Asterisks indicate a significant difference among groups (P < 0.05).
Changes in fecal microbial communities after in vitro fermentation.
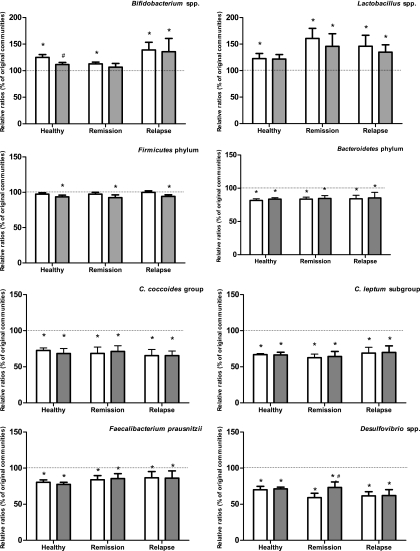
After fermentation of FOS and AOS in fecal slurries obtained from healthy subjects and UC patients in either remission or relapse, qPCR was applied to measure the density of gene targets encoding 16S rRNA genes of selected bacterial taxonomic units (Fig. 2). In fecal communities derived from healthy subjects and UC patients in remission, fermentation of FOS resulted in a significant increase in the content of Bifidobacterium spp. (P < 0.05). However, fermentation of AOS did not similarly increase the proportion of bifidobacteria in these samples, although a trend for higher levels of bifidobacteria in the communities from healthy subjects was observed (P = 0.078). The communities derived from patients with UC in relapse showed a significant increase in the density of bifidobacteria after fermentation with FOS as well as with AOS (P < 0.05). The relative abundances of Lactobacillus spp. in fecal microbiota derived from patients with UC in remission and relapse were significantly increased after fermentation with either FOS or AOS (P < 0.05). The fecal communities from healthy subjects contained a significantly higher density of lactobacilli after fermentation with FOS (P < 0.05). Although this was only borderline significant for the samples fermented with AOS, there was a strong trend of a similar effect (P = 0.057).
Fig. 2.
Relative ratios (RR) of target genes in samples incubated with FOS (white) and AOS (gray) compared to original bacterial communities, calculated as RR = EΔCT (fermentation samples)/EΔCT (original bacterial community). Original communities were set to 100% for each group (indicated by dotted lines). Error bars represent averages ± standard errors of the means of relative changes in target species in each of the six samples in each group. Asterisks (*) indicate significant differences from the original bacterial communities, while pound signs (#) indicate significant differences from FOS-fermented samples.
A significant reduction of the Firmicutes phylum and of taxa belonging to the Firmicutes (C. coccoides group, C. leptum subgroup, and F. prausnitzii) was measured in all three fecal microbiota groups after fermentation with AOS (P < 0.05). Firmicutes phylum levels in the samples fermented with FOS were unaffected, although levels of the C. coccoides group, the C. leptum subgroup, and F. prausnitzii were significantly reduced (P < 0.05). FOS and AOS fermentation both resulted in a decrease of the relative abundance of Bacteroidetes phylum and Desulfovibrio spp. (genus belonging to the Proteobacteria phylum) after fermentation of all three fecal microbiota groups (P < 0.05).
No significant differences between the effects of the two carbohydrate sources AOS and FOS were observed, except in the case of Bifidobacterium spp., which were more abundant (P < 0.05) in the samples from healthy subjects after incubation with FOS than in those incubated with AOS, and Desulfovibrio spp., where the density was significantly lower (P < 0.05) in the samples from UC patients in remission after incubation with FOS than after incubation with AOS.
SCFA analysis and pH measurements after fermentation.
Amounts of SCFA (acetate, propionate, butyrate) produced by fermentation of FOS and AOS and pH measurements are shown in Table 2. For all types of samples (healthy, remission, and relapse), the pH was lower in samples incubated with FOS and AOS than in the samples incubated without added substrate. The amount of acetate was significantly higher (P < 0.05) in all types of samples fermented with FOS than in the samples with no substrate added. Additionally, acetate was higher in samples from patients in relapse incubated with AOS. A significantly lower (P < 0.05) amount of propionate was measured in samples from healthy subjects incubated with FOS than in samples incubated with no substrate added. No significant difference was observed in the amount of butyrate after fermentation, independent of substrate and origin of the microbial community.
Table 2.
pH measurements and short chain fatty acids liberated during fermentation
| Parameter | Result for patient groupa |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Healthy |
Remission |
Relapse |
|||||||
| NS | FOS | AOS | NS | FOS | AOS | NS | FOS | AOS | |
| pH rangeb | 6.5–7.5 | 4.5–5.5 | 5–5.5 | 6.5–7.5 | 4.5–5.5 | 4.5–6 | 6.5–7.5 | 4.5–5.5 | 4.5–5.5 |
| Total SCFA | 36.34 (± 3.5) | 40.97 (± 5.9) | 33.77 (± 4.4) | 24.53 (± 2.6) | 32.79 (± 4.5) | 22.35 (± 2.8) | 14.95 (± 1.2) | 28.95 (± 3.6) | 21.25 (± 2.7) |
| Acetate | 23.06A (± 2.7) | 36.49B (± 5.8) | 27.73A (± 3.3) | 16.96A (± 2.0) | 27.93B (± 1.9) | 17.67A (± 1.8) | 7.74A (± 2.1) | 22.78B (± 4.6) | 17.08B (± 1.6) |
| Propionate | 5.91A (± 2.1) | 1.61B (± 0.18) | 2.54AB (± 1.1) | 2.59 (± 0.82) | 2.24 (± 1.1) | 0.78 (± 0.16) | 2.31 (± 0.7) | 2.40 (± 0.9) | 2.38 (± 1.5) |
| Butyrate | 6.03 (± 1.5) | 1.46 (± 0.41) | 2.97 (± 2.4) | 4.11 (± 1.2) | 2.21 (± 0.9) | 2.16 (± 1.5) | 3.90 (± 1.5) | 3.40 (± 1.8) | 1.51 (± 1.0) |
NS, no substrate added; FOS, fructo-oligosaccharide; AOS, arabino-oligosaccharide. Total SCFA is acetate, butyrate, propionate, iso-butyrate, iso-valate, and valate. All numbers are means of six samples ± standard errors of the means and are expressed as nM. Unlike superscript letters are significantly different (P < 0.05), comparing each disease group.
pH was measured using a pH indicator stick. Due to the uncertainty in the method, a range of pH for the different fermentations is used.
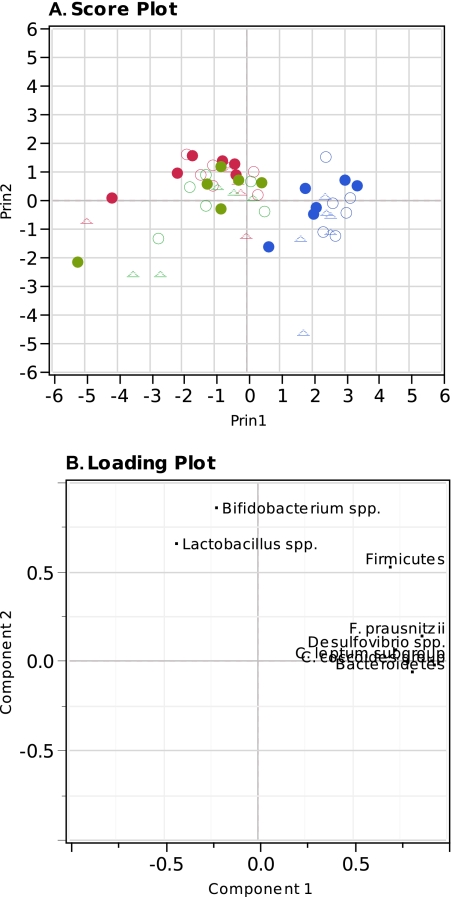
Principal component analysis (PCA) of bacterial composition.
The different bacterial taxonomic units as a function of disease status and incubation with FOS or AOS were subjected to PCA to generate an overview of the variation in the ability of the microbial communities from either healthy subjects, UC patients in remission, or UC patients in relapse to ferment the two carbohydrate sources. Data were visualized in two dimensions using a principal component score and loading plot (Fig. 3A and B, respectively). Two principal components were calculated for the model with a total of 73.60% of variance expressed. The samples before (original bacterial communities) and after incubation with or without either FOS or AOS for all three stages of disease were clearly differentiated by the PC1, with the fermented samples clustering to the left of the PCA score plot (Fig. 3A). However, differentiation between samples incubated with FOS and AOS was not seen. The loading plot revealed that the primary bacterial taxa influencing the differentiation between the original bacterial communities and samples incubated with added substrate were the Bifidobacterium spp. and the Lactobacillus spp. (Fig. 3B).
Fig. 3.
Principal component analysis of the quantitative PCR measurements using the first and second principal component (PC1, 54.94%; PC2, 18.66%). (A) Score plot showing the original bacterial communities (blue), samples incubated with FOS (red) and samples incubated with AOS (green). Sources of the communities are indicated by open circles (○) for healthy subjects, closed circles (•) for UC patients in remission, and triangles (Δ) for UC patients in relapse. (B) Loading plot, indicating each of the measured bacterial taxa as determined by quantitative real-time PCR.
DISCUSSION
In this study, the mean relative quantity of Bifidobacterium spp. in the original bacterial community was lower in UC patients in relapse than in healthy controls (Fig. 1), but this was not statistically significant. A previous study by Sokol and coworkers (64) has shown significantly lower levels of Bifidobacterium spp. in fecal samples of UC patients. Additionally, Lactobacillus spp. was significantly less abundant in the original bacterial community of UC patients in relapse than in that of healthy controls.
It should be acknowledged that differences between the microbial communities may partly be the consequence of the use of anti-inflammatory drugs, since a recent study showed that melsalazine affected the fecal bacterial community composition in patients with inflammatory bowel syndrome (2). In the present study, four out of six UC patients in remission received melsalazine, whereas all the UC patients in relapse received the drug.
Fermentation of FOS resulted in significantly higher levels of bifidobacteria in all types of microbial communities originating from either healthy subjects or UC patients. The bifidogenic effect of FOS using fecal samples from healthy subjects is in accordance with a number of previous in vitro studies (24, 50, 55, 58). In contrast to these observations, a recent study did not reveal any significant difference in the level of bifidobacteria after incubation of fecal microbial communities derived from healthy subjects with FOS and even demonstrated a significant decrease in this bacterial group after FOS incubation of fecal communities from patients with inactive Crohn's disease or active UC (49). The discrepancy in results between the present study and the results reported by Rose and coworkers (49) might be explained by the use of different incubation methods. In the present study, a static batch system was used for each individual fecal community, while Rose and coworkers (49) pooled the individual fecal samples and used the dynamic TIM-2 system. In agreement with the present approach, many in vivo trials suggest a bifidogenic effect of FOS in subjects with inflammatory bowel disease. A recent trial using rats induced with colitis by 2,4,6-trinitrobenzenesulfonic acid (TNBS) showed that administration of FOS had an anti-inflammatory effect and significantly increased concentrations of bifidobacteria in cecum and colon (48). Additionally, a small trial with patients suffering from active Crohn's disease showed that administration of FOS significantly increased the mucosal and fecal counts of bifidobacteria. Furthermore, an increase in the percentage of lamina propria dendritic cells expressing the anti-inflammatory cytokine interleukin 10 (IL-10) as well as a significant upregulation of expression of the Toll-like receptors (TLRs) TLR2 and TLR4 was observed. The authors of reference 32 suggest that this was caused by the bifidogenic effect of FOS and that an increase of bifidobacteria can act via TLRs or other pattern recognition receptors on dendritic cells to stimulate IL-10 production.
Incubation with AOS gave rise to a significantly higher level of bifidobacteria in fecal microbial communities derived from UC patients in relapse, while no difference was seen after AOS incubation of fecal communities from healthy subjects or UC patients in remission. It was surprising to us that no statistically significant increase of bifidobacteria upon AOS fermentation by microbial communities derived from healthy subjects was observed, since previous experiments from our lab have shown that AOS caused a significantly higher level of bifidobacteria in fecal microbial communities derived from healthy subjects (24). However, there was a similar bifidogenic trend in the present study (P = 0.078), and we speculate that the lack of significance is explained by a larger variation between the individual samples used. A previous in vitro fermentation study using feces from healthy subjects reported that Bifidobacterium spp. varied in their ability to hydrolyze arabino-oligosaccharides and that they grew better on low-molecular-weight fractions than on high-molecular-weight fractions of the oligosaccharides (1). Genome sequencing has revealed that carbohydrate-modifying enzymes can vary between species. Bifidobacterium longum, Bifidobacterium adolescentis, Bifidobacterium animalis subsp. lactis, Bifidobacterium bifidum, Bifidobacterium pseudocatenulatum, and Bifidobacterium breve, which are all commonly detected in human adult feces (67), have genes encoding β-fructofuranosidase and α-l-arabinofuranosidase, enabling the bacteria to utilize FOS and AOS, respectively (UniProtKB database, http://www.uniprot.org/uniprot). α-l-Arabinofuranosidase catalyzes the hydrolysis of the glycosidic bond between an arabinose moiety and the backbone, but sequence analysis does not necessarily reveal if the specificity is toward branched arabinan or arabinoxylan. However, B. breve and B. bifidum do not harbor α-l-arabinanase, which catalyzes the hydrolysis of AOS backbone (UniProtKB database, http://www.uniprot.org/uniprot). However, it has been demonstrated that B. adolescentis and B. longum are able to degrade linear arabino-oligosaccharides (8 DP) to the same extent as FOS, whereas B. breve is able only to hydrolyze FOS and B. bifidum is not able to degrade either FOS or AOS. In the same study, B. adolescentis utilized neutral oligosaccharides derived from apple pectin (65). In another study, also based on pure cultures, it was observed that B. breve, B. longum, and B. adolescentis were able to partly ferment a mixture of linear arabino-oligosaccharides (2 to 5 DP) derived from sugar beet pectin; however, FOS was more efficiently fermented (68). The observed differences in the response from the bifidobacterial communities reported here may thus be explained by a different intragenic composition of bifidobacteria in the individual samples. Densities of lactobacilli in all fermentation samples were significantly increased after incubation with FOS of microbial communities from healthy subjects and UC patients. The same was seen after incubation with AOS, although it did not reach statistical significance in healthy subjects (P = 0.057). The importance of the presence of Lactobacillus spp. in the gut was previously highlighted by a study with IL-10-deficient mice with colitis and reduced Lactobacillus spp. levels, which showed that restoration of Lactobacillus spp. to a normal level reduced the development of colitis (35). Additionally, a human trial showed that ingestion of Lactobacillus GG aided the maintenance of remission in patients with UC (72). Our in vitro observations thus suggest that AOS as well as FOS consumption might help patients suffering from UC by increasing their levels of intestinal lactobacilli.
Bacteroidetes and Firmicutes (including C. coccoides group, C. leptum subgroup, F. prausnitzii and Lactobacillus spp.) represent the two dominant bacterial phyla in the human gut and together comprise about 90% of the large intestinal microbiota (13, 45). A changed ratio between these groups has been suggested to affect intestinal inflammation (64). In the present study, no effect on the ratio between the Bacteroidetes and Firmicutes was caused by incubation with either FOS or AOS. Nevertheless, the relative quantity of members of the phylum Bacteroidetes was significantly decreased in samples incubated with FOS and AOS for all fecal communities tested, probably due to the increase in bifidobacteria, which belong to the phylum Actinobacteria. It may, however, be speculated that the observed decrease was the result of the lower level of pH observed after the 24 h of fermentation (Table 2) rather than the inability of species of this phylum to utilize the carbohydrates, since responses to pH vary considerably between different phylogenetic groups of human colonic anaerobes and variation in pH tolerance among the Bacteroidetes phylum has been observed (12). However, in agreement with an effect of the oligosaccharides added, Van Laere and coworkers (68) demonstrated that several species of Bacteroides, including Bacteroides vulgatus, Bacteroides ovatus, and Bacteroides thetaiotaomicron, are not able to utilize a mixture of linear arabino-oligosaccharides (2 to 5 DP) and can only partly degrade FOS. Additionally, pH-controlled in vitro studies also have reported a significant decrease in the population size of Bacteroides after incubation of FOS with fecal communities from UC patients (49) and healthy subjects (36). Taken together, these findings indicate that even though the Bacteriodetes phylum contains genera that can degrade and ferment a wide variety of plant polysaccharides and oligosaccharides (25), they may not be able to utilize FOS and AOS and hence may not compete in the fermentor community when these are the primary carbohydrate sources.
The butyrate-producing species F. prausnitzii and the butyrate-producing bacterial groups C. coccoides (cluster XIVa) and C. leptum (cluster IV) are important for colonic health and have an anti-inflammatory effect due to their production of butyrate (34). However, in the present study their relative abundance was decreased after incubation with FOS as well as AOS in all three types of fecal communities. A similar effect of FOS consumption on the abundance of the C. coccoides group has been observed in mouse trials (44).
The relative density of Desulfovibrio spp. was significantly decreased by incubation with FOS as well as AOS in all three types of samples. Desulfovibrio spp. is the predominant sulfate-reducing bacteria in the human colon, and the principal by-product of sulfate-reducing bacteria is hydrogen sulfide (51). The pH plays an important role for the growth of sulfate-reducing bacteria and their production of hydrogen sulfate. These bacteria prefer a growth environment that is neutral or slightly alkaline (18), hence the low pH in the FOS- and AOS-incubated samples is likely to have caused the relative decrease of Desulfovibrio spp. A similar reduction in pH has been demonstrated in the cecum and colon of animals given FOS compared to control animals (6, 30, 48).
Sulfate-reducing bacteria have previously been linked to UC development, because high concentrations of sulfides inhibit butyrate metabolism in colonocytes and induce hyper-proliferation and metabolic abnormalities in epithelial cells, similar to those observed with UC (7, 15). Additionally, Rowan and coworkers (51) found that higher levels of Desulfovibrio spp. were detected in the colonic mucus of patients with active UC than in healthy controls, indicating a role of this bacterium in the pathogenesis of UC. However, in the present study, no significant difference in the abundance of Desulfovibrio spp. was observed between the bacterial communities originating from each of the three groups, which may be explained by the use of fecal samples as opposed to mucosal samples (73). Still, this does not exclude that a reduction in abundance of Desulfovibrio spp. as caused by FOS and AOS in the present study may be beneficial in the prevention of colitis.
The SCFA profile reflects microbial activity (55). Acetate is the predominant acid produced by bacterial fermentation in fecal communities, and in consistence with the observed increase in the acetate-producing bifidobacteria and lactobacilli, our study revealed a significantly larger amount of acetate in the samples incubated with either FOS or AOS. The higher production of acetate may be the main cause of the observed decrease in pH resulting from FOS and AOS fermentation. Significantly lower levels of propionate were observed after incubation of bacterial communities from healthy subjects with FOS. Similar results have previously been reported by other studies, where FOS fermentation resulted in lower levels of propionate than other substrates, including, e.g., gentio-oligosaccharides (56) and AOS (1). One of the major intestinal producers of propionate is Propionibacterium (19), and we speculate that the population size of Propionibacterium species may be higher (and more easily reduced after fermentation) in healthy subjects than in UC patients, since no difference in propionate level was observed after FOS incubation using fecal samples from UC patients.
PCA of the qPCR data revealed that the bacterial groups separating the fermented samples from the original bacterial communities were Bifidobacterium spp. and Lactobacillus spp., which were both significantly more abundant in the microbiota after fermentation of either FOS or AOS by communities derived from UC patients (Fig. 2 and 3). Communities incubated with FOS and AOS were not differentiated by the PCA (Fig. 3), suggesting that these two oligosaccharides affected the microbiotas similarly.
Our results suggest that the positive effects already observed in human and animal trials using FOS may be applicable also for AOS, which opens the possibility of exploration of, e.g., by-product streams of sugar production for prebiotic preparations. Hence, the presence and fermentation of AOS may help prevent proinflammatory conditions and enhance the resistance to colonization of opportunistic pathogens, e.g., through increased production of acetate (17) and reduced pH in the colonic environment. Considering the reduced levels of Lactobacillus spp. and Bifidobacterium spp. present in UC patients in relapse, a stimulation of these bacterial groups may prevent flare-ups. However, human trials are needed to confirm a health-promoting effect of AOS in UC patients.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Danish Council for Strategic Research (grant no. 2101-06-0067) and the Technical University of Denmark.
We thank Kate Vibefeldt and Bodil Madsen for excellent technical assistance. Additionally, we thank Jørn Brynskov and Casper Steenholdt, Herlev Hospital, for assistance with collection of fecal samples and Peter Westerman and Gitte Hinz-Berg, Aalborg University, for assistance with SCFA measurements.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 7 October 2011.
REFERENCES
- 1. Al-Tamimi M. A. H. M., Palframan R. J., Cooper J. M., Gibson G. R., Rastall R. A. 2006. In vitro fermentation of sugar beet arabinan and arabino-oligosaccharides by the human gut microflora. J. Appl. Microbiol. 100: 407–414 [DOI] [PubMed] [Google Scholar]
- 2. Andrews C., et al. 2011. Mesalazine (5-aminosalicylic acid) alters faecal bacterial profiles, but not mucosal proteolytic activity in diarrhoea-predominant irritable bowel syndrome. Aliment. Pharmacol. Ther. 34: 374–383 [DOI] [PubMed] [Google Scholar]
- 3. Ardizzone S. 2003. Ulcerative colitis. In Orphanet encyclopedia. http://www.orpha.net/data/patho/GB/uk-UC.pdf
- 4. Bamias G., Nyce M. R., De la Rue S. A., Cominelli F. 2005. New concepts in the pathophysiology of inflammatory bowel disease. Ann. Intern. Med. 143: 895–904 [DOI] [PubMed] [Google Scholar]
- 5. Binder V. 1970. A Comparison between clinical state, macroscopic and microscopic appearances of rectal mucosa, and cytologic picture of mucosal exudate in ulcerative colitis. Scand. J. Gastroenterol. 5: 627–632 [PubMed] [Google Scholar]
- 6. Cherbut C., Michel C., Lecannu G. 2003. The prebiotic characteristics of fructooligosaccharides are necessary for reduction of TNBS-induced colitis in rats. J. Nutr. 133: 21–27 [DOI] [PubMed] [Google Scholar]
- 7. Christl S. U., Eisner H. D., Dusel G., Kasper H., Scheppach W. 1996. Antagonistic effects of sulfide and butyrate on proliferation of colonic mucosa—a potential role for these agents in the pathogenesis of ulcerative colitis. Dig. Dis. Sci. 41: 2477–2481 [DOI] [PubMed] [Google Scholar]
- 8. Collado M. C., Meriluoto J., Salminen S. 2007. In vitro analysis of probiotic strain combinations to inhibit pathogen adhesion to human intestinal mucus. Food. Res. Int. 40: 629–636 [Google Scholar]
- 9. Delroisse J. M., et al. 2008. Quantification of Bifidobacterium spp. and Lactobacillus spp. in rat fecal samples by real-time PCR. Microbiol. Res. 163: 663–670 [DOI] [PubMed] [Google Scholar]
- 10. Denman S. E., McSweeney C. S. 2006. Development of a real-time PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. FEMS Microbiol. Ecol. 58: 572–582 [DOI] [PubMed] [Google Scholar]
- 11. De Preter V., Hamer H. M., Windey K., Verbeke K. 2011. The impact of pre- and/or probiotics on human colonic metabolism: does it affect human health? Mol. Nutr. Food. Res. 55: 46–57 [DOI] [PubMed] [Google Scholar]
- 12. Duncan S. H., Louis P., Thomson J. M., Flint H. J. 2009. The role of pH in determining the species composition of the human colonic microbiota. Environ. Microbiol. 11: 2112–2122 [DOI] [PubMed] [Google Scholar]
- 13. Eckburg P. B., et al. 2005. Diversity of the human intestinal microbial flora. Science 308: 1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. European Food Safety Authority 2010. Scientific opinion on dietary reference values for carbohydrates and dietary fibre. EFSA J. 8: 1–77 [Google Scholar]
- 15. Fite A., et al. 2004. Identification and quantitation of mucosal and faecal desulfovibrios using real time polymerase chain reaction. Gut 53: 523–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fooks L. J., Gibson G. R. 2002. In vitro investigations of the effect of probiotics and prebiotics on selected human intestinal pathogens. FEMS Microbiol. Ecol. 39: 67–75 [DOI] [PubMed] [Google Scholar]
- 17. Fukuda S., et al. 2011. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469: 543–U791 [DOI] [PubMed] [Google Scholar]
- 18. Gibson G. R., et al. 1990. Alternative pathways for hydrogen disposal during fermentation in the human colon. Gut 31: 679–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gibson G. R. 1999. Dietary modulation of the human gut microflora using the prebiotics oligofructose and inulin. J. Nutr. 129: 1438S–1441S [DOI] [PubMed] [Google Scholar]
- 20. Guo X., et al. 2008. Development of a real-time PCR method for Firmicutes and Bacteroidetes in faeces and its application to quantify intestinal population of obese and lean pigs. Lett. Appl. Microbiol. 47: 367–373 [DOI] [PubMed] [Google Scholar]
- 21. Hart A. L., et al. 2005. Characteristics of intestinal dendritic cells in inflammatory bowel diseases. Gastroenterology 129: 50–65 [DOI] [PubMed] [Google Scholar]
- 22. Heilig H. G. H. J., et al. 2002. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl. Environ. Microbiol. 68: 114–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. 2007. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biology. 8(2): R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Holck J., et al. 2011. Feruloylated and nonferuloylated arabino-oligosaccharides from sugar beet pectin selectively stimulate the growth of Bifidobacterium spp. in human fecal in vitro fermentations. J. Agric. Food. Chem. 59: 6511–6519 [DOI] [PubMed] [Google Scholar]
- 25. Hooper L. V., Midtvedt T., Gordon J. I. 2002. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22: 283–307 [DOI] [PubMed] [Google Scholar]
- 26. Hotchkiss A. T., Nunez A., Rastall R. A., Gibson G. R. December 2010. Growth promotion of beneficial bacteria in gut of human comprises administering composition comprising arabino oligosaccharide as prebiotic. U.S. patent 2010316766-A1
- 27. Jacobs D. M., Gaudier E., van Duynhoven J., Vaughan E. E. 2009. Non-digestible food ingredients, colonic microbiota and the impact on gut health and immunity: a role for metabolomics. Curr. Drug Metab. 10: 41–54 [DOI] [PubMed] [Google Scholar]
- 28. Langholz E., Munkholm P., Davidsen M., Binder V. 1994. Course of ulcerative-colitis—analysis of changes in disease-activity over years. Gastroenterology 107: 3–11 [DOI] [PubMed] [Google Scholar]
- 29. Langlands S. J., Hopkins M. J., Coleman N., Cummings J. H. 2004. Prebiotic carbohydrates modify the mucosa associated microflora of the human large bowel. Gut 53: 1610–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lara-Villoslada F., et al. 2006. Short-chain fructooligosaccharides, in spite of being fermented in the upper part of the large intestine, have anti-inflammatory activity in the TNBS model of colitis. Eur. J. Nutr. 45: 418–425 [DOI] [PubMed] [Google Scholar]
- 31. Leser T. D., Lindecrona R. H., Jensen T. K., Jensen B. B., Moller K. 2000. Changes in bacterial community structure in the colon of pigs fed different experimental diets and after infection with Brachyspira hyodysenteriae. Appl. Environ. Microbiol. 66: 3290–3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lindsay J. O., et al. 2006. Clinical, microbiological, and immunological effects of fructo-oligosaccharide in patients with Crohn's disease. Gut 55: 348–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lloyd D. A. J., et al. 2002. Crohn's disease and ulcerative colitis: divergent trends in hospital admission rates. Health Stat. Q. 16: 19–24 [Google Scholar]
- 34. Louis P., Flint H. J. 2009. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 294: 1–8 [DOI] [PubMed] [Google Scholar]
- 35. Madsen K. L., Doyle J. S., Jewell L. D., Tavernini M. M., Fedorak R. N. 1999. Lactobacillus species prevents colitis in interleukin 10 gene-deficient mice. Gastroenterology 116: 1107–1114 [DOI] [PubMed] [Google Scholar]
- 36. Mandalari G., et al. 2007. In vitro evaluation of the prebiotic activity of a pectic oligosaccharide-rich extract enzymatically derived from bergamot peel. Appl. Microbiol. Biotechnol. 73: 1173–1179 [DOI] [PubMed] [Google Scholar]
- 37. Manning T. S., Gibson G. R. 2004. Microbial-gut interactions in health and disease. Prebiotics. Best Pract. Res. Clin. Gastroenterol. 18: 287–298 [DOI] [PubMed] [Google Scholar]
- 38. Matsuki T., Watanabe K., Fujimoto J., Takada T., Tanaka R. 2004. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Appl. Environ. Microbiol. 70: 7220–7228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Medina V., Afonso J. J., Alvarez-Arguelles H., Hernandez C., Gonzalez F. 1998. Sodium butyrate inhibits carcinoma development in a 1,2-dimethylhydrazine-induced rat colon cancer. JPEN J. Parenter. Enteral. Nutr. 22: 14–17 [DOI] [PubMed] [Google Scholar]
- 40. Oosterveld A., Beldman G., Schols H. A., Voragen A. G. J. 2000. Characterization of arabinose and ferulic acid rich pectic polysaccharides and hemicelluloses from sugar beet pulp. Carbohydr. Res. 328: 185–197 [DOI] [PubMed] [Google Scholar]
- 41. Ott S. J., et al. 2004. In vitro alterations of intestinal bacterial microbiota in fecal samples during storage. Diagn. Microbiol. Infect. Dis. 50: 237–245 [DOI] [PubMed] [Google Scholar]
- 42. Palframan R. J., Gibson G. R., Rastall R. A. 2002. Effect of pH and dose on the growth of gut bacteria on prebiotic carbohydrates in vitro. Anaerobe 8: 287–292 [DOI] [PubMed] [Google Scholar]
- 43. Peran L., et al. 2007. A comparative study of the preventative effects exerted by three probiotics, Bifidobacterium lactis, Lactobacillus casei and Lactobacillus acidophilus, in the TNBS model of rat colitis. J. Appl. Microbiol. 103: 836–844 [DOI] [PubMed] [Google Scholar]
- 44. Petersen A., et al. 2010. Analysis of the intestinal microbiota of oligosaccharide fed mice exhibiting reduced resistance to Salmonella infection. Benef. Microbes 3: 271–281 [DOI] [PubMed] [Google Scholar]
- 45. Qin J. J., et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rastall R. A., Maitin V. 2002. Prebiotics and synbiotics: towards the next generation. Curr. Opin. Biotechnol. 13: 490–496 [DOI] [PubMed] [Google Scholar]
- 47. Roberfroid M. 2007. Prebiotics: the concept revisited. J. Nutr. 137: 830S–837S [DOI] [PubMed] [Google Scholar]
- 48. Rodriguez-Cabezas M. E., et al. 2010. The combination of fructooligosaccharides and resistant starch shows prebiotic additive effects in rats. Clin. Nutr. 29: 832–839 [DOI] [PubMed] [Google Scholar]
- 49. Rose D. J., Venema K., Keshavarzian A., Hamaker B. R. 2010. Starch-entrapped microspheres show a beneficial fermentation profile and decrease in potentially harmful bacteria during in vitro fermentation in faecal microbiota obtained from patients with inflammatory bowel disease. Br. J. Nutr. 103: 1514–1524 [DOI] [PubMed] [Google Scholar]
- 50. Rossi M., et al. 2005. Fermentation of fructooligosaccharides and inulin by bifidobacteria: a comparative study of pure and fecal cultures. Appl. Environ. Microbiol. 71: 6150–6158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rowan F., et al. 2010. Desulfovibrio bacterial species are increased in ulcerative colitis. Dis. Colon Rectum 53: 1530–1536 [DOI] [PubMed] [Google Scholar]
- 52. Roy M. J., et al. 2009. In vitro studies on the inhibition of colon cancer by butyrate and carnitine. Nutrition 25: 1193–1201 [DOI] [PubMed] [Google Scholar]
- 53. Ruas-Madiedo P., et al. 2010. Exopolysaccharides produced by Lactobacillus and Bifidobacterium strains abrogate in vitro the cytotoxic effect of bacterial toxins on eukaryotic cells. J. Appl. Microbiol. 109: 2079–2086 [DOI] [PubMed] [Google Scholar]
- 54. Ruemmele F. M., et al. 2003. Butyrate induced Caco-2 cell apoptosis is mediated via the mitochondrial pathway. Gut 52: 94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rycroft C. E., Jones M. R., Gibson G. R., Rastall R. A. 2001. A comparative in vitro evaluation of the fermentation properties of prebiotic oligosaccharides. J. Appl. Microbiol. 91: 878–887 [DOI] [PubMed] [Google Scholar]
- 56. Rycroft C. E., Jones M. R., Gibson G. R., Rastall R. A. 2001. Fermentation properties of gentio-oligosaccharides. Lett. Appl. Microbiol. 32: 156–161 [DOI] [PubMed] [Google Scholar]
- 57. Sanz M. L., Gibson G. R., Rastall R. A. 2005. Influence of disaccharide structure on prebiotic selectivity in vitro. J. Agric. Food Chem. 53: 5192–5199 [DOI] [PubMed] [Google Scholar]
- 58. Sanz M. L., et al. 2005. In vitro investigation into the potential prebiotic activity of honey oligosaccharides. J. Agric. Food Chem. 53: 2914–2921 [DOI] [PubMed] [Google Scholar]
- 59. Sartor R. B. 2008. Therapeutic correction of bacterial dysbiosis discovered by molecular techniques. Proc. Natl. Acad. Sci. U. S. A. 105: 16413–16414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sartor R. B. 2006. Mechanisms of disease: pathogenesis of Crohn's disease and ulcerative colitis. Nat. Clin. Pract. Gastroenterol. Hepatol. 3: 390–407 [DOI] [PubMed] [Google Scholar]
- 61. Sartor R. B. 2004. Therapeutic manipulation of the enteric microflora in inflammatory bowel diseases: antibiotics, probiotics, and prebiotics. Gastroenterol. 126: 1620–1633 [DOI] [PubMed] [Google Scholar]
- 62. Segain J. P., et al. 2000. Butyrate inhibits inflammatory responses through NF kappa B inhibition: implications for Crohn's disease. Gut 47: 397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shen J., et al. 2006. Molecular profiling of the Clostridium leptum subgroup in human fecal microflora by PCR-denaturing gradient gel electrophoresis and clone library analysis. Appl. Environ. Microbiol. 72: 5232–5238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sokol H., et al. 2009. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel Dis. 15: 1183–1189 [DOI] [PubMed] [Google Scholar]
- 65. Suzuki Y., Tanaka K., Nano T., Asakura T., Muramatsu N. 2004. Utilization by intestinal bacteria and digestibility of arabino-oligosaccharides in vitro. J. Jpn. Soc. Hortic. Sci. 73: 574–579 [Google Scholar]
- 66. Thomassen L. V., Vigsnaes L. K., Licht T. R., Mikkelsen J. D., Meyer A. S. 2011. Maximal release of highly bifidogenic soluble dietary fibers from industrial potato pulp by minimal enzymatic treatment. Appl. Microbiol. Biotechnol. 90: 873–884 [DOI] [PubMed] [Google Scholar]
- 67. Turroni F., et al. 2009. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl. Environ. Microbiol. 75: 1534–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Van Laere K. M. J., Hartemink R., Bosveld M., Schols H. A., Voragen A. G. J. 2000. Fermentation of plant cell wall derived polysaccharides and their corresponding oligosaccharides by intestinal bacteria. J. Agric. Food Chem. 48: 1644–1652 [DOI] [PubMed] [Google Scholar]
- 69. Vernazza C. L., Rabiu B. A., Gibson G. R. 2006. Human colonic microbiology and the role of dietary intervention: introduction to prebiotics, p. 1–28 In Gibson G. R., Rastall R. A.(ed.), Prebiotics: development and application. John Wiley & Sons, Ltd., Chichester, England [Google Scholar]
- 70. Walter J., et al. 2000. Detection and identification of gastrointestinal Lactobacillus species by using denaturing gradient gel electrophoresis and species-specific PCR primers. Appl. Environ. Microbiol. 66: 297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zeuthen L. H., et al. 2010. Lactobacillus acidophilus induces a slow but more sustained chemokine and cytokine response in naive foetal enterocytes compared to commensal Escherichia coli. BMC Immunol. 11: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zocco M. A., et al. 2006. Efficacy of Lactobacillus GG in maintaining remission of ulcerative colitis. Aliment. Pharmacol. Ther. 23: 1567–1574 [DOI] [PubMed] [Google Scholar]
- 73. Zoetendal E. G., et al. 2002. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl. Environ. Microbiol. 68: 3401–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.