Abstract
Studies on the effect of environmental conditions on plants and microorganisms are a central issue in ecology, and they require an adequate experimental setup. A strategy often applied in geobotanical studies is based on the reciprocal transplantation of plant species at different sites. We adopted a similar approach as a field-based tool to investigate the relationships of soil bacterial communities with the environment. Soil samples from two different (calcareous and siliceous) unvegetated glacier forefields were reciprocally transplanted and incubated for 15 months between 2009 and 2010. Controls containing local soils were included. The sites were characterized over time in terms of geographical (bedrock, exposition, sunlight, temperature, and precipitation) and physicochemical (texture, water content, soluble and nutrients) features. The incubating local (“home”) and transplanted (“away”) soils were monitored for changes in extractable nutrients and in the bacterial community structure, defined through terminal restriction fragment length polymorphism (T-RFLP) of the 16S rRNA gene. Concentrations of soluble ions in most samples were more significantly affected by seasons than by the transplantation. For example, NO3− showed a seasonal pattern, increasing from 1 to 3 μg NO3− (g soil dry weight)−1 after the melting of snow but decreasing to <1 μg NO3− (g soil dry weight)−1 in autumn. Seasons, and in particular strong precipitation events occurring in the summer of 2010 (200 to 300 mm of rain monthly), were also related to changes of bacterial community structures. Our results show the suitability of this approach to compare responses of bacterial communities to different environmental conditions directly in the field.
INTRODUCTION
A traditional concept in microbial ecology says that “everything is everywhere, but the environment selects” (14). This principle is a useful basis to study the biogeographies and structures of natural communities. It describes the influence of the environment on microorganisms, which could be potentially cosmopolitan, as they can be limitlessly dispersed (44).
Recent findings suggest that microbial community structures are shaped by factors related to both geographical and specific physicochemical characteristics of their home site (12, 18, 20). However, there is still uncertainty about whether and where one or the other mechanism prevails (3, 38). The debate on whether microbial communities are cosmopolitan (affected primarily by geographical factors such as climate, migration, and spatial isolation) or locally adapted (influenced primarily by the local abiotic and biotic factors) is still open (14).
The use of advanced high-resolution methodologies based mainly on sequence analyses and DNA-based community profiling permitted the comparison of spatially distant microbial communities (47, 48). Particularly, the latest improvements in the terminal restriction fragment length polymorphism (T-RFLP) profiling technique (1, 3, 19, 53) allow detailed characterizations of microbial community changes.
In addition to community characterization, there is an increasing interest in understanding the main factors driving microbial community structure in different natural environments, (19, 60). However, most of these studies are limited to surveys and comparisons of community structures or to laboratory-based incubations of extracted microbial consortia under different conditions (9, 15, 23, 26, 30). While these approaches have advantages in their simplicity and are useful to understand the effects of isolated factors, they may not represent the real situation in nature, where there is a wide array of geographical, physical, and chemical factors interplaying and influencing community structure. A current challenge is therefore to perform these kinds of studies directly in the field (61).
Reciprocal transplantation experiments have been commonly applied in geobotany to study the local adaptation of plants to their habitat (50, 52), the effects of the adaptation capacity of plant pathogens and invasive species, or plant performance along environmental gradients (5, 33, 34). The advantage of these experiments lies in the elimination of biases related to the growth of seedlings in the laboratory and in avoiding an erroneous simulation of natural conditions. In microbial ecology, field transplantation experiments have been often performed to assess responses of specific microbial functional groups and microbially mediated processes (6, 8) to changes in vegetation and microclimate (25). Currently, there are only few studies that have used the reciprocal transplantation approach to investigate relationships of total microbial communities to different factors, such as vegetation (2, 24), soil properties (41), and temperature (58, 59), or also to test ecological theories (3). Due to the experimental design, which often involves the growth of plants or the use of sealed incubation vessels (3, 24), in many of these studies sampling is limited to a few time points (2, 41, 58). Gradual dynamics of responses of microbial communities to environmental changes may therefore be overlooked. The present study was conceived after we noticed that the bacterial community structures in various alpine glacier forefields posing on lithologically different bedrocks (calcareous and siliceous) were different (31). The structures of these communities are generally relatively simple, being characterized by dominance of distinct operational taxonomic units (OTUs). This feature facilitates the detailed analysis of community changes. We therefore evaluated the feasibility of a field-based approach which permits long-term incubation of transplanted soil samples, regular monitoring of bacterial community structures, and characterization of the relevant geographical (e.g., climate) and physicochemical (e.g., nutrient availability) factors.
MATERIALS AND METHODS
Field sites and soil sampling.
For the experiment we selected 2 unvegetated glacier forefields, one posing on a siliceous (Tiefen, Canton Uri) and one on a calcareous (Griessen, Canton Obwalden) bedrock. According to data provided from the Swiss Glacier Monitoring Network (http://glaciology.ethz.ch/messnetz/), the soils have an estimated age of <30 years of development. General characteristics of the sites are shown in Table 1. The physicochemical properties of these glacier forefields and their bacterial communities have been previously compared (31). At each site, the soil used for the incubation experiment was collected randomly from approximately the first 5 cm at 5 spots, sieved through a 2-mm sieve, and pooled by gentle mixing.
Table 1.
General characteristics of the sampling sites and of conditions during the sampling perioda
| Glacier | Altitude (m above sea level) | Exposition | Bedrock | Avg pHb | Avg TC [mg (g soil dry wt)−1]b | Avg TN [mg (g soil dry wt)−1]b | Avg recession (m)c | Slope (%) | Mean sunshine (h)d | Mean precipitation (mm)d | Dominant plant speciese |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tiefen | 2,510 | Southeast | Siliceous | 5.50 | <0.08 | <0.001 | −15 | 11.5 | 150.13 | 100.75 | Leodonton montanum |
| Griessen | 2,450 | West | Calcareous | 7.07 | 2.16 ± 0.79 | <0.010 | −12.5 | 25 | 110.28 | 128.75 | Saxifraga sp. |
Geographical data represent the area where the incubation pots were placed.
The values represent averages in the incubation pots measured at the different time points.
Estimated from recession data since 1990.
Data averaged from the CLIMAP database.
The characterization was based on our own observations in the vicinity of the sampling sites.
Experimental setup.
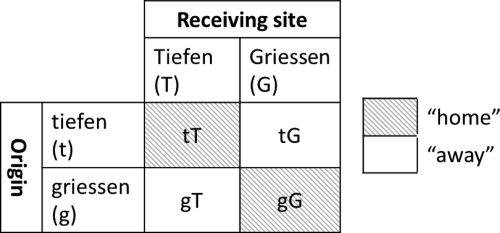
The experimental setup is schematized in Fig. 1. Briefly, at each site (T [Tiefen] and G [Griessen]), we buried pots containing soil originating from Tiefen (t) or from Griessen (g) in order to obtain all possible combinations of “home” and “away” samples. At each site, all the pots were repeated in quadruplicates, for a total of 16 pots (8 pots at each glacier forefield).
Fig. 1.
Conceptual representation of the experimental setup. The soils from the different locations were reciprocally incubated in their home sites or in the reciprocal locations.
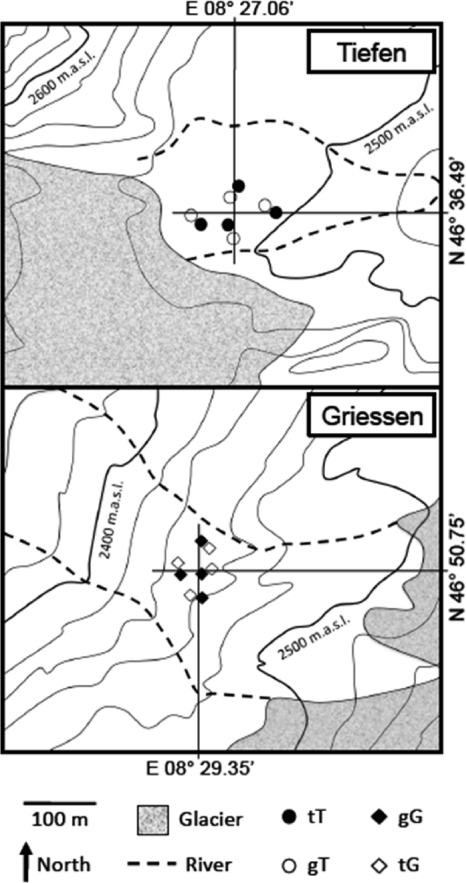
The field experiment was based on in situ incubations of soil samples. Stainless steel pots of 12 cm in diameter and 13 cm in height (IKEA, catalog no. 300.118.32) were set up in quadruplicates at 4 random, unvegetated sites in both forefields (Fig. 2). It was important to select appropriate pots, which were perforated on all sides, to allow exchange of gases and water with the surroundings while at the same time preventing waterlogging. The pots were buried completely except from a rim of approximately 1 cm to avoid surface runoff into the pots. For the same reason, pots were slightly overfilled. The rim was covered with pebbles to avoid direct sunlight and heating of the metal. After burial, the direct surroundings were reestablished as before.
Fig. 2.
Schematic representation of the experimental setup. The Tiefen and the Griessen sites harbor 4 pairs of pots, each containing 1 “home” and 1 “away” sample.
Sampling was performed regularly (Table 2) except between October 2009 and June 2010, when the experimental sites were inaccessible due to heavy snow cover. In total, we could sample 6 different time points (D0, D1, D2, D3, D4, and D5) at the Tiefen glacier for a total of 15 months of incubation. Due to heavy snow cover in June 2010 at Griessen, sampling at D3 was not possible.
Table 2.
Sampling dates for the pot transplantation experiment at the two different incubation sites (Tiefen and Griessen)
| Designation | Date of sampling |
|
|---|---|---|
| Tiefen site | Griessen site | |
| D0 | 21 August 2009 | 20 August 2009 |
| D1 | 10 September 2009 | 9 September 2009 |
| D2 | 30 September 2009 | 29 September 2009 |
| D3 | 29 June 2010 | —a |
| D4 | 3 August 2010 | 4 August 2010 |
| D5 | 2 October 2010 | 4 October 2010 |
—, sampling in Griessen at this time point was incomplete due to the inaccessibility of most of the pots and therefore was not considered.
At each time point, two soil samples per pot were taken. For molecular analysis, approximately 2 g of soil was taken at a depth of approximately 5 cm and placed in a 2-ml reaction tube. For chemical analysis, approximately 20 g of soil was taken at the same depth and placed in plastic bags. Samples were transported on ice and stored in the lab at 4°C (plastic bags) or −20°C (reaction tubes).
Chemical analysis and physical parameters.
Total carbon (TC) and total nitrogen (TN) were measured through combustion with a CN 1500 analyzer (CE Instruments, Wigan, United Kingdom). Soluble ions were extracted from 1 g of fresh, sieved soil in 5 ml of 0.01 M CaCl2 (for anions) and 2 M KCl (for cations). The ion concentrations were measured with an IC Dionex 2300 separation system (Dionex, Sunnyvale, CA). NH4+ concentrations were determined colorimetrically according to the protocol of Mulvaney (42) within 1 week after sampling.
Dissolved organic carbon (DOC) was measured in water extracts. One gram of fresh soil was added to 10 ml Milli-Q water and shaken in a rotary shaker for 24 h. The extracts were filtered through a 0.45-μm filter and acidified with 5 μl 37% HCl to eliminate inorganic carbon. Finally, DOC was measured with a TOC-5000 analyzer (Shimadzu SSI, Columbia, MD) with standards ranging from 0.5 to 25 ppm DOC. Water extracts were also used to measure pH with a XMP225 pH meter (Mettler-Toledo, Columbus, OH).
The soil water content in all the pots at all time points was estimated by converting weight loss of oven-dried soil (60°C for 24 h) to percent water. The soil temperature in selected pots was recorded at approximately a 5-cm depth with temperature data loggers (Ibuttons; Maxim Integrated Products, Inc., Sunnyvale, CA). The measured temperature was recorded every 8 h for the entire sampling period. General climatic data at the Tiefen glacier were obtained by the automatic meteorological station operated by the WSL Institute for Snow and Avalanche Research SLF (www.slf.ch). Parameters for the Griessen glacier are taken from CLIMAP (www.meteoswiss.ch).
Cell counts.
To estimate cell numbers in the soil samples, DAPI (4′,6′-diamidino-2-phenylindole) staining was performed (4). Shortly after sampling, 1 g of fresh soil was fixed with 1 ml of 4% paraformaldehyde overnight at 4°C and washed twice with 900 μl of phosphate-buffered saline (PBS). Prior to analysis, the sample was stored at −20°C in 200 μl of a 1:1 mixture of PBS and ethanol.
For cell extraction, fixed samples were first shaken for 15 min on a horizontal vortex plate. Sixty microliters of a 60% Nycodenz solution (Sigma-Aldrich Chemie GmbH, Buchs, Switzerland) was added to 100 μl of the soil slurry and centrifuged for 90 min at 10,000 × g. Ten microliters of cell-containing supernatant was pipetted onto an 8-well glass slide and dried over a gas flame. The samples were stained with 10 μl of 25-ng/μl DAPI solution, and then the slide was mounted with Citifluor AFI (Citifluor Ltd., Leicester, United Kingdom) and examined microscopically (Leica CTR 6000; Leica Microsystems GmbH, Wetzlar, Germany). At least 10 fields of view per well were counted.
DNA extraction and T-RFLP profiling.
DNA was extracted with the UltraClean soil DNA isolation kit (Mobio Laboratories Inc., Carlsbad, CA) starting from 0.25 g of fresh soil, according to the manufacturer's instructions. Quantification was done with the Sybr green I colorimetric method (39). PCR of the 16S rRNA bacterial gene with primers 27F (labeled with 6-carboxyfluorescein [FAM] at the 5′ end) and 1401 Rev (29) and T-RFLP analysis of MspI-digested PCR fragments were performed as described by Lazzaro et al. (31).
Statistical analysis of soil chemical parameters.
The data on water content and chemical analysis were processed with multivariate analysis of variance (MANOVA) for repeated measurements to explore the question of whether time, substrates, or treatments (transplantation) of the soil samples have significant effects on a certain chemical property. Calculations were run with Systat V.12.
Statistical analysis of T-RFLP profiles.
In the T-RFLP profiles, peaks within a range of ±0.5 bp were defined as operational taxonomic units (OTU) and were considered in the subsequent analysis if present in at least 2 out of 4 replicates. Relative bacterial 16S rRNA gene fingerprints were created by transforming each peak height into a percentage of the total peak heights. The height threshold for a peak to be taken into account was set to 0.3% of total fluorescence (18).
Pairwise similarities between the soils at the different sites and, among each set of soils, between D0 and the subsequent time points, were performed by averaging the 4 replicate profiles for each soils. On the basis of the average T-RFLP profiles, we estimated the Simpson dominance index (l) as , where s is the total number of OTUs in the profile, ni is the abundance of each OTU, and N is the total abundance in the profile. From this equation, we then could derive Morisita's index of community similarity (Im) (16): Im = (2Σn1in2i)/(l1 + l2)N1N2. This index describes the similarity between two profiles and ranges from 0 (no similarity) to 1 (complete similarity). Significances of the pairwise comparisons were further tested by ADONIS with the R package vegan (43).
Log normalized T-RFLP profiles were further investigated using principal-component analysis (PCA) (57). Results were visualized in 2-dimensional ordination plots, which show the distribution of the samples along the first and second principal components. With these plots, similarities and differences between the samples could be assessed.
In order to identify potential driving forces responsible for observed population changes, redundancy analysis (RDA) was conducted, combining the T-RFLP profiles with the log-normalized geographical and physicochemical data obtained. The goal of this ordination method is to clarify how much of the variability of the community data can be explained by a certain environmental variable. The parameter describing geographical exposition was represented by number classes. Temperature data for each time point were averaged over a 3-week period before the actual sampling date. Liquid precipitation was summed up over the same time span. RDA and DCA were conducted with the software CANOCO V. 4.5.
RESULTS
Meteorological data and water content.
At Tiefen and Griessen, temperature and precipitation followed similar patterns (see Fig. S1 in the supplemental material). Generally, Griessen tended to be subjected to slightly higher mean temperatures and more intense precipitation than Tiefen. Heavy rainfall at this site was registered in June and July 2010. The variation in the meteorological conditions was reflected in the water content of the incubation pots, which appeared to be significantly influenced by both time and incubation site (Table 3). Generally, the water contents of the replicates were similar. The samples incubating at Tiefen were generally drier than those at Griessen. However, water content tended to increase with time, from average starting values of 1.4% ± 0.2% water (Tiefen soil) and 1.3% ± 0.2% water (Griessen soil) to maximum values of 28.2% ± 1.9% water (tG5 samples) and 30.3% ± 2.8% water (gG5 samples).
Table 3.
MANOVA for repeated measures of soil parameters measured in this study
| Soil | Significance of difference ina: |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Water content |
DOC |
NH4+ |
NO3− |
SO42− |
|||||||||||
| Between | Within | Factor interaction | Between | Within | Factor interaction | Between | Within | Factor interaction | Between | Within | Factor interaction | Between | Within | Factor interaction | |
| Tiefen | ** | * | NS | ** | ** | ** | NS | ** | NS | NS | * | NS | NS | NS | NS |
| Griessen | ** | ** | ** | NS | ** | * | NS | NS | NS | NS | * | * | NS | ** | * |
Significances were tested within one soil at its home site in comparison to the transplanted soil (“away”). The analysis was performed on the basis of 5 time points (D0, D1, D2, D4, and D5). D3, which is not available for the Griessen site, was excluded from the analysis. “Between” significances indicate differences between “home” and “away” samples; “within” significances indicate effects of time. “Factor interactions” indicates the effects of time × transplantation interactions. Asterisks indicate significant differences at the 0.05 (*) or at the 0.01 (**) level.
Soil temperature measurements performed with temperature data loggers in summer were in good agreement with the air temperature data derived from the Meteoswiss database (data not shown). However, during the winter months, air temperature fell many degrees below 0°C. Although we do not have our own soil temperature measurements for these months due to the several meters of snow cover, soil temperatures must have remained close to 0°C (56).
pH and extractable nutrients.
While pH, TC, and TN values remained stable during the course of the experiment (Table 1), DOC and soluble ion concentrations appeared to be affected by the transplantation or by the course of time in a different way (Table 4). DOC tended to increase over time, by 4-fold (gG samples) or even 20-fold (gT samples) from D0 to D5.
Table 4.
Physicochemical and biological parameters measured in the two soils
| Samplea | Value (mean ±SD) for parameterb: |
|||||
|---|---|---|---|---|---|---|
| NH4+ [μg (g soil dry wt)−1] | NO3− [μg (g soil dry wt)−1] | SO42− [μg (g soil dry wt)−1] | DOC [μg (g soil dry wt)−1] | DNA [ng (g soil dry wt)−1] | Cell number [106 DAPI-stained cells (g soil dry wt)−1] | |
| tT0a | 0.64 ± 0.27 | 0.38 ± 0.21 | 0.41 ± 0.29 | 16.25 ± 9.83 | 152.03 ± 49.81 | 0.95 ± 0.90 |
| tT1 | 0.45 ± 0.34 | 0.44 ± 0.29 | 0.48 ± 0.15 | 16.58 ± 8.48 | 163.35 ± 11.89 | 0.66 ± 0.20 |
| tT2 | 0.21 ± 0.10 | 0.59 ± 0.34 | 0.33 ± 0.06 | 3.71 ± 1.05 | 138.41 ± 54.72 | 0.67 ± 0.20 |
| tT3 | 0.77 ± 0.08 | 0.44 ± 0.27 | 0.37 ± 0.03 | 35.05 ± 20.33 | 134.27 ± 107.03 | NA |
| tT4 | 0.32 ± 0.06 | 1.97 ± 0.89 | 0.43 ± 0.09 | 46.93 ± 20.42 | 247.02 ± 89.10 | 2.81 ± 0.89 |
| tT5 | 0.27 ± 0.03 | 0.25 ± 0.08 | 0.43 ± 0.18 | 106.16 ± 31.34** | 508.04 ± 382.57 | 0.83 ± 0.55 |
| tG0 | 0.85 ± 0.17 | 0.91 ± 0.29 | 0.50 ± 0.39 | 7.70 ± 2.97 | 163.37 ± 33.62 | 1.98 ± 1.50 |
| tG1 | 0.17 ± 0.06** | 0.01 ± 0.02** | 0.41 ± 0.08 | 7.46 ± 4.67 | 80.13 ± 43.44 | 1.14 ± 1.13 |
| tG2 | 0.28 ± 0.15** | 0.47 ± 0.22** | 0.97 ± 0.90 | 20.01 ± 19.12 | 149.03 ± 48.44 | 0.94 ± 1.21 |
| tG4 | 0.24 ± 0.05** | 3.67 ± 0.23** | 0.57 ± 0.18 | 31.59 ± 23.17 | 206.71 ± 153.50 | 0.93 ± 0.66 |
| tG5 | 0.41 ± 0.18** | <0.01** | 0.16 ± 0.13 | 21.53 ± 13.06 | 115.74 ± 17.25 | 2.58 ± 1.29 |
| gG0 | 0.35 ± 0.22 | 0.80 ± 0.36 | 0.48 ± 0.02 | 8.90 ± 3.96 | 179.13 ± 18.58 | 3.42 ± 2.00 |
| gG1 | 0.34 ± 0.02 | 0.58 ± 0.61 | 0.55 ± 0.12 | 12.08 ± 10.79 | 187.64 ± 53.19 | 0.91 ± 0.70 |
| gG2 | 0.25 ± 0.12 | 0.93 ± 0.33 | 0.78 ± 0.52 | 3.38 ± 2.26 | 199.04 ± 200.45 | 0.69 ± 0.59 |
| gG4 | 0.31 ± 0.17 | 3.67 ± 0.49** | 1.12 ± 0.24 | 42.89 ± 23.00* | 864.27 ± 643.20 | 1.90 ± 0.62 |
| gG5 | 0.78 ± 0.42 | 0.28 ± 0.33 | 0.13 ± 0.13 | 45.81 ± 34.91* | 363.53 ± 68.14 | 1.02 ± 0.65 |
| gT0 | 0.47 ± 0.03 | 0.56 ± 0.39 | 0.49 ± 0.12 | 8.15 ± 3.46 | 187.10 ± 22.41 | 1.68 ± 1.48 |
| gT1 | 0.40 ± 0.08 | 2.41 ± 2.58 | 0.75 ± 0.13 | 9.90 ± 4.59 | 194.26 ± 29.47 | 2.83 ± 1.77 |
| gT2 | 0.35 ± 0.23 | 1.76 ± 0.90 | 0.49 ± 0.18 | 2.02 ± 0.35 | 106.59 ± 10.78 | 2.81 ± 1.75 |
| gT3 | 1.05 ± 0.52 | 1.56 ± 1.09 | 0.45 ± 0.10 | 36.27 ± 20.76 | 411.96 ± 142.79 | NA |
| gT4 | 0.38 ± 0.11 | 2.81 ± 0.58 | 0.46 ± 0.04 | 42.60 ± 20.36* | 145.78 ± 26.84 | 2.53 ± 0.82 |
| gT5 | 0.25 ± 0.12 | 0.72 ± 0.15 | 0.19 ± 0.01 | 115.13 ± 49.82* | 457.52 ± 380.13 | 1.13 ± 0.58 |
Measurements at D0 were performed on the initial homogenized soil partitioned into 4 subsamples.
Asterisks indicate significant differences at the 0.05 (*) or 0.01 (**) level between D0 and the subsequent time points within each transplantation set, calculated with the Tukey test for pairwise comparisons. NA, not available.
NH4+ measured in both soils at both sites tended to decrease in September and October of both sampling years (Table 4) to values below 0.85 μg (g soil dry weight)−1. Only in the Griessen “home” samples did we not observe this decline. NO3− and SO42− concentrations remained constant over time, although generally higher concentrations of NO3− were measured in the Griessen “home” and “away” samples. At the Tiefen site, NO3− concentrations tended to increase significantly at D4, as peaks of 1.97 ± 0.89 and 2.81 ± 0.58 μg NO3− (g soil dry weight)−1 were detected in the tT and gT samples, respectively (Table 4). MANOVA analysis (Table 3) showed a strongly significant within-treatment (“time”) effect for the DOC amounts in both soils and for the NH4+ concentrations in the Tiefen soil. Time also appeared to be significantly related to NO3− amounts at both sites. The effect of transplantation (“between” effect) appeared to be significant only in the DOC concentrations measured at Tiefen and in the SO42− concentrations measured at Griessen.
Microbial biomass and cell counts.
DNA amounts in all samples ranged from 80.13 ± 43.44 to 864.27 ± 643.20 ng DNA (g soil dry weight)−1. The largest amounts, which were above 400 ng DNA (g soil dry weight)−1, were detectable only in tT5, gG4, gT3, and gT5. The counts of DAPI-stained cells were highly variable, ranging from 0.69 × 106 ± 0.59 × 106 to 3.42 × 106 ± 2.00 × 106 (g soil dry weight)−1, and did not show any clear trend (Table 4). However, a reduction in cell numbers was noticed in both the Tiefen and Griessen soils incubating at the Griessen site (tG and gG).
Bacterial community structure.
Overall, we distinguished and aligned 161 OTUs in the T-RFLP profiles, which represent only the detectable fraction of the total microbial community in the soils (3). The Morisita index (Table 5) was used to assess similarities between the two soils at D0 and, subsequently, within the replicates of each setup (Tiefen “home,” Tiefen “away,” Griessen “home,” and Griessen “away”) over time. The index showed a divergence of the T-RFLP profiles from Tiefen in comparison with those from Griessen at D0. In all the samples, the index was equal to or above 0.79 at the beginning of the experiment and then tended to decline (indicating increasing dissimilarity) with the sampling time. In the gT samples, however, the index appeared to fall to 0.45 at D4 and then rose again to 0.64 at D5. ADONIS showed that both communities changed significantly (P < 0.05) after the transplantation. In addition, significant changes (P < 0.05) were noticeable within each group (Tiefen “home,” Tiefen “away,” Griessen “home,” and Griessen “away”) with time, from D2 to D5 (Table 5).
Table 5.
Morisita community similarity indices based on pairwise comparison of the T-RFLP profiles between D0 and the subsequent time points of each transplantation seta
| Group | Reference time pointb | Morisita index at time pointc: |
|||||
|---|---|---|---|---|---|---|---|
| D0 | D1 | D2 | D3 | D4 | D5 | ||
| Home | tT0 | 1 | 0.79 | 0.80 | 0.42* | 0.42* | 0.52* |
| gG0 | 1 | 0.90 | 0.88 | 0.70* | 0.49* | ||
| Away | tG0 | 1 | 0.85 | 0.71* | 0.52* | 0.34* | |
| gT0 | 1 | 0.89 | 0.75* | 0.65* | 0.45* | 0.64* | |
The pairwise comparisons were performed on T-RFLP profiles generated from the average of the 4 replicates per sample/time point.
The pairwise comparison between tT0 and gG0 gave a Morisita index of 0.47 (ADONIS P < 0.05).
Asterisks indicate significant (P < 0.05) differences of the pairwise comparisons according to ADONIS.
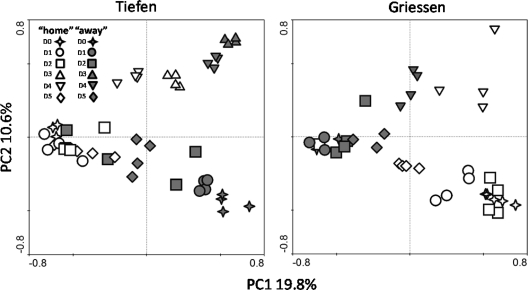
PCA permitted further visualization of similarities between all the T-RFLP profiles at the different sites and during the incubation period. Dynamics and changes of the transplanted communities in comparison with the local communities are illustrated in Fig. 3. First, the T-RFLP profiles generated from the replicate samples appear to be very homogeneous and always cluster together. The communities of the 2 different sites appear to be different (Fig. 3). However, at Tiefen, the “away” community gT (Griessen soil) tended to shift with time along PC1 (Fig. 3), partially overlapping the “home” community tT (Tiefen soil), which clustered more closely throughout the sampling points. The tendency was not as pronounced in the samples incubating at Griessen. We also noticed, at both sites, a clustering of the “home” and “away” samples from D3 and D4 along PC2. However, at D5, the T-RFLP profiles were again positioned along PC1.
Fig. 3.
PCA of the 16S rRNA gene T-RFLP profiles. The data were analyzed together, but for clarity of presentation they are divided into “home” and “away” samples incubating at Tiefen and Griessen.
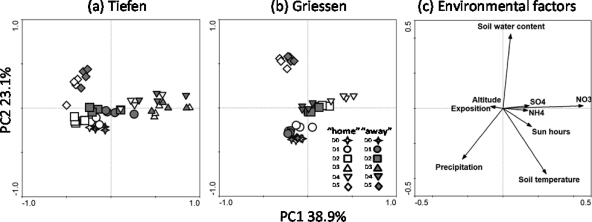
Redundancy analysis (RDA) showed possible environmental conditions that could be related to the T-RFLP profiles. PC1 and PC2 accounted for 38.9% and 23.1% of the total variance, respectively. Soluble ions, and in particular NO3−, were positively correlated with the samples from D3 and D4 at Tiefen (Fig. 4a) along PC1. Factors such as soil water content, total precipitation, and average temperature were placed instead along PC2 (Fig. 4c). Soil water content appeared to be strongly correlated with samples from D5, taken in October 2010, especially with those at Griessen (Fig. 4b). These samples were negatively correlated with total precipitation and average soil temperature.
Fig. 4.
RDA of the 16S rRNA gene T-RFLP profiles and different environmental factors. The data were analyzed together but for clarity are divided into samples incubating at Tiefen (a) and Griessen (b) and the related environmental factors (c).
DISCUSSION
With our field transplantation experiment, we achieved our primary objective of establishing a simple experimental setup which would allow the long-term monitoring of structural changes in soil bacterial communities. Moreover, the experiment provided some preliminary information on important environmental factors influencing glacier forefield bacterial communities.
Ideally, field incubations permit assessment of ecological parameters under conditions as close to the natural state as possible and take into account the great multitude of factors which may affect a microbial community (49, 51). However, it is of vital importance that the microbial communities are kept stable and in equilibrium with the surroundings. All the replicate T-RFLP profiles clustered together (Fig. 3), indicating that the incubation pots maintained similar conditions throughout the course of the experiment. In fact, the pots did not show any sign of rust or degradation over time, and due to the perforation, water saturation and the development of an anoxic bottom layer could be avoided.
The comparison of the T-RFLP profiles of the “home” soils (retransplanted at their home site) with those of the “away” soils showed that there was no major effect of the transplantation procedure on either of the soils, aside from a divergence at D3 and D4. The community structure shift at D3 and D4 was temporary, as at D5 the communities tended to return to the original composition, and may be an indication of seasonal effects. An impact of seasonal changes on microbial community structures was expected, as previous studies in similar environments (35, 36) found changes in the abundance and diversity of several bacterial taxa in samples taken at different times of the year. In particular, in a related study, Lipson (34) reported a significant relationship between bacterial community structures and temperature fluctuations. In our study, the main differences we observed between the climates of the summer seasons of 2009 and 2010 (D1 and D4) appeared to be related to the strong precipitation events of 2010. As it appears also in the redundancy analysis (Fig. 4), it is likely that the heavy rainfall events in the summer of 2010 had a strong influence on the water content of the incubating soil (Table 3). At D4 and D5, the soils were close to saturation (30%), while values determined from the previous samples rarely exceeded 10%.
Our findings are in accordance with the results reported by Schwartz et al. (54) for parameters along a precipitation gradient and by Clark et al. (13) on precipitation effects in arid zones, as well as with the outcomes of the field-based humidity manipulations performed by Castro et al. (11). These results generally highlight the importance of precipitation events for bacterial community structures. However, a possible effect of precipitation on microbial biomass remains unclear, as seen, for example, in the case of contrasting outcomes observed in two different desert environments (13, 32). In agreement with Papatheodorou et al. (46) and Clark et al. (13), we did not notice any significant change in our biomass estimations (represented by DAPI cell counts and DNA amounts) from the different time points sampled. As our soils do not experience drought like the desert environments previously mentioned, precipitation may not have a strong impact on microbial biomass but rather may cause a replacement of sensitive bacterial groups with other, more opportunistic species (13).
In terms of soil chemical properties, generally the concentrations of the extractable ions remained within the normal range detected for unvegetated glacier forefields (31) and followed similar trends in spite of the different soil bedrock types. A significant effect of time was noticeable for the NO3− concentrations, which tended to increase in the August 2010 sampling. At the Tiefen site the increase was coupled with a decrease in NH4+, suggesting nitrification during the summer. Some nitrification activity has been detected at the early stages of a glacier chronosequence in the summer season (10). Moreover, several studies demonstrated the effect of precipitation and dry-wet cycles on nitrification processes in soil (21, 22, 40). Although the glacier forefields which were object of this study do not usually experience full drought, they are exposed to short-term dry-wet cycles, which could influence the bacterial community processes.
We could also observe a tendency of an autumn decline of NO3− and NH4+ concentrations in all the samples, which would suggest leaching. Leaching also is strongly influenced by precipitation, which can flush the soluble ions at depths beyond the point where capillary forces can draw water back toward the surface. It is therefore possible that the heavy rain events before our autumn sampling caused the lower concentrations observed.
DOC concentrations in soil depend on different biotic and abiotic factors; in particular, seasonality and precipitation regimen have been identified as key drivers for DOC amounts in grassland soils (17). Moreover, a clayey texture of the soil is believed to increase absorption and retention of DOC, which would otherwise easily leach through the soil profile (27). In our soils, we could not find any evidence of seasonality in the DOC patterns, although a significant increase with time was noticeable in the Griessen samples, which may indicate absorption events and accumulation of DOC.
With this experimental setup we could establish a strategy to monitor the effects of similar environmental conditions on different bacterial communities. In general, allochthonous soil microbial communities may respond in different ways when transferred to a new environment. For example, Bottomley et al. (6) found positive responses of potential nitrification in forest communities transferred to meadows. Bottomley et al. (7) also observed microbial community shifts which could be related to changes in N and C availability. In contrast, in a reciprocal transplantation study between two different forest stands, microbial community composition did not change (24). Bacterial communities originating from hostile or extreme environments may be able to adapt rapidly to changing conditions (55), and therefore they may efficiently establish and develop in a new environment. This has been shown, for example, with experiments involving transfer of gut microflora to different hosts. These communities are able to rapidly and dynamically adapt to new conditions, such as the transfer from human to porcine hosts (45) or to hosts with a modified gut community (28). In our work with unvegetated glacier forefield communities, we also observed that certain OTUs were not apparently affected, in terms of number and of relative abundance, by the transfer to a different site. For example, the number of dominant OTUs in the profiles remained generally stable in all the Tiefen (22.23 to 33.83%) and Griessen (22.51 to 33.9%) “home” and “away” samples (data not shown). Our results, however, also point to local adaptation of some members of the “away” calcareous soil (Griessen), as seen from a close examination of the T-RFLP profiles. For example, the number of unique OTUs in the “away” Griessen samples declined from 13.06% (D0) to 2.44% (D2) and then rose again at D4 and D5 (data not shown). These OTUs may represent the more sensitive members of the Griessen community and may have responded negatively to a certain factor (e.g., precipitation) which varied above their “home” regimen at only a certain moment during the course of the experiment. However, seasonal climatic variations generally had a stronger effect than the transplantation itself. Further analysis is needed to better understand the roles of environmental factors such as precipitation, independently and in the frame of cyclical environmental variations. In addition, an investigation on the long-term dynamics of microbial functions would provide a more detailed insight on the adaptation mechanisms of microbial communities.
Conclusions.
Bacterial communities inhabiting unvegetated forefields of receding glaciers have a relatively simple composition but are also able to respond dynamically to environmental changes. They therefore represent good study systems for bacterial adaptation. In this work we conceived and validated the reciprocal transplantation approach, which permitted us to monitor long-term changes of these bacterial communities in the field. T-RFLP profiling proved to be a sensitive and powerful tool to compare bacterial community structures. Overall, the reciprocal transplantation of soils was successful in evidencing structural changes of the “home” and “away” bacterial communities and indicating their relationships to environmental factors. However, the establishment of clear-cut cause-effect relationships requires further investigation. Our results generally suggest that this approach can be applied to assess microbial adaptation in other simple natural systems.
Supplementary Material
ACKNOWLEDGMENTS
We are particularly grateful to Karl Schroff, Climate Physics, Institute for Atmospheric and Climate Science, for the preparation of the portable rain collector. T-RFLP analysis was performed at the Genetic Diversity Center of ETH Zürich. We also thank Alessandro Franchini, Ruth Henneberger, Beat Frey, Martin Vogt, Isolde Erny, and Annika Vössner for help in the field and laboratory.
This project was funded by ETH Zürich.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 30 September 2011.
REFERENCES
- 1. Angel R., Soares M. I. M., Ungar E. D., Gillor O. 2010. Biogeography of soil archaea and bacteria along a steep precipitation gradient. ISME J. 4: 553–563 [DOI] [PubMed] [Google Scholar]
- 2. Balser T. C., Firestone M. K. 2005. Linking microbial community composition and soil processes in a California annual grassland and mixed-conifer forest. Biogeochemistry 73: 395–415 [Google Scholar]
- 3. Bell T. 2010. Experimental tests of the bacterial distance-decay relationship. ISME J. 4: 1357–1365 [DOI] [PubMed] [Google Scholar]
- 4. Bennett P. C., Summers Engel A., Roberts J. A. 2006. Counting and imaging bacteria on mineral surfaces, p. 37–78 In Maurice J. P. A., Warren L. A. (ed.), Methods of investigating microbial-mineral interactions. CMS workshop lectures, vol. 14 The Clay Mineral Society, Chantilly, VA [Google Scholar]
- 5. Bischoff A., et al. 2006. Detecting local adaptation in widespread grassland species—the importance of scale and local plant community. J. Ecol. 94: 1130–1142 [Google Scholar]
- 6. Bottomley P. J., et al. 2004. Responses of nitrification and ammonia-oxidizing bacteria to reciprocal transfers of soil between adjacent coniferous forest and meadow vegetation in the Cascade Mountains of Oregon. Microb. Ecol. 48: 500–508 [DOI] [PubMed] [Google Scholar]
- 7. Bottomley P. J., et al. 2006. Responses of soil bacterial and fungal communities to reciprocal transfers of soil between adjacent coniferous forest and meadow vegetation in the Cascade Mountains of Oregon. Plant Soil 289: 35–45 [DOI] [PubMed] [Google Scholar]
- 8. Boyle S. A., Rich J. J., Bottomoley P. J., Cromack K., Jr., Myrold D. D. 2006. Reciprocal transfer effects on denitrifying community composition and activity at forest and meadow sites in the Cascade Mountains of Oregon. Soil Biol. Biochem. 38: 870–878 [Google Scholar]
- 9. Bradford M. A., Watts B. W., Davies C. A. 2010. Thermal adaptation of heterotrophic soil respiration in laboratory microcosms. Global Change Biol. doi:10.1111/j.1365-2486.2009.02040.x [Google Scholar]
- 10. Brankatschk R., Töwe S., Kleineidam K., Schloter M., Zeyer J. 2011. Abundances and potential activities of nitrogen cycling microbial communities along a chronosequence of a glacier forefield. ISME J. 5: 1025–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Castro H. F., Classen A. T., Austin E. E., Norby R. J., Schadt C. W. 2010. Soil microbial community responses to multiple experimental climate change drivers. Appl. Environ. Microbiol. 76: 999–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cho J. C., Tiedje J. M. 2000. Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Appl. Environ. Microbiol. 66: 5448–5456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Clark J., Campbell J., Grizzle H., Acosta-Martinez V., Zak J. C. 2009. Soil microbial community response to drought and precipitation variability in the Chihuahuan desert. Microb. Ecol. 57: 248–260 [DOI] [PubMed] [Google Scholar]
- 14. de Wit R., Bouvier T. 2006. ‘Everything is everywhere, but, the environment selects’; what did Baas Becking and Beijerinck really say? Environ. Microbiol. 8: 755–758 [DOI] [PubMed] [Google Scholar]
- 15. Dimitriu P. A., Lee D., Grayston S. J. 2010. An evaluation of the functional significance of peat microorganisms using a reciprocal transplant approach. Soil Biol. Biochem. 42: 65–71 [Google Scholar]
- 16. Dollhopf S. L., Hashsham S. A., Tiedje J. M. 2001. Interpreting 16S rDNA T-RFLP data: application of self-organizing maps and principal component analysis to describe community dynamics and convergence. Microb. Ecol. 42: 495–505 [DOI] [PubMed] [Google Scholar]
- 17. Don A., Schulze E. D. 2008. Controls on fluxes and export of dissolved organic carbon in grasslands with contrasting soil types. Biogeochemistry 91: 117–131 [Google Scholar]
- 18. Dunbar J., Ticknor L. O., Kuske C. R. 2000. Assessment of microbial diversity in four southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl. Environ. Microbiol. 66: 2943–2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fierer N., Jackson R. B. 2006. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U. S. A. 103: 626–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fulthorpe R. R., Roesch L. F. W., Riva A., Triplett E. W. 2008. Distantly samples soils carry few species in common. ISME J. 2: 901–910 [DOI] [PubMed] [Google Scholar]
- 21. Gelfand I., Yakir D. 2008. Influence of nitrite accumulation in association with seasonal patterns and mineralization of soil nitrogen in a semi-arid pine forest. Soil Biol. Biochem. 40: 415–424 [Google Scholar]
- 22. Hamer U., Makeschin F., Stadler J., Klotz S. 2008. Soil organic matter and microbial community structure in set-aside and intensively managed arable soils in NE-Saxony, Germany. Appl. Soil Ecol. 40: 465–475 [Google Scholar]
- 23. Hämmerli A., Waldhuber S., Miniaci C., Zeyer J., Bunge M. 2007. Local expansion and selection of soil bacteria in a glacier forefield. Eur. J. Soil Sci. 58: 1437–1445 [Google Scholar]
- 24. Hannam K. D., Quideau S. A., Kishchuk B. E. 2007. The microbial communities of aspen and spruce forest floors are resistant to changes in litter inputs and microclimate. Appl. Soil Ecol. 35: 635–647 [Google Scholar]
- 25. Hart S. C., Perry D. A. 1999. Transferring soils from high- to low-elevation forests increases nitrogen cycling rates: climate change implications. Global Change Biol. 5: 23–32 [Google Scholar]
- 26. Hosoi-Tanabe S., et al. 2010. Comprehensive analysis of an Antarctic bacterial community with the adaptability of growth at higher temperatures than those in Antarctica. Biocontrol Sci. 15: 57–62 [DOI] [PubMed] [Google Scholar]
- 27. Kalbitz, Solinger K. S., Park J. H., Michalzik B., Matzner E. 2000. Controls on the dynamics of dissolved organic matter in soils: a review. Soil Sci. 165: 277–304 [Google Scholar]
- 28. Landy J., et al. 2011. Faecal transplantation therapy for gastrointestinal disease. Aliment. Pharmacol. Ter. 34: 409–415 [DOI] [PubMed] [Google Scholar]
- 29. Lane D. 1991. 16S/23S rRNA sequencing. p. 131–175 In Strackebrandt E., Goodfellow M. (ed.), Nucleic acid techniques in bacterial systematics. Wiley, New York, NY [Google Scholar]
- 30. Langenheder S., Lindström E. S., Tranvik L. J. 2006. Communities emerging from different sources under identical conditions. Appl. Environ. Microbiol. 72: 212–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lazzaro A., Abegg C., Zeyer J. 2009. Bacterial community structure in glacier forefields on calcareous and siliceous bedrock. Eur. J. Soil Sci. 60: 860–870 [Google Scholar]
- 32. Li X., Chen Z. 2004. Soil microbial biomass C and N along a climatic transect in the Mongolian steppe. Biol. Fertil. Soils 39: 344–351 [Google Scholar]
- 33. Link S. O., Smith J. L., Halvorson J. J., Bolton H. 2003. A reciprocal transplantation experiment within a climatic gradient in a semiarid shrub-steppe ecosystem: effects on bunchgrass growth and reproduction, soil carbon and soil nitrogen. Global Change Biol. 9: 1097–1105 [Google Scholar]
- 34. Lipson D. A. 2007. Relationships between temperature responses and bacterial community structure along seasonal and altitudinal gradients. FEMS Microbiol. Ecol. 59: 418–427 [DOI] [PubMed] [Google Scholar]
- 35. Lipson D. A., Schadt C. W., Schmidt S. K. 2002. Changes in soil microbial community structure and function in an alpine dry meadow following spring snow melt. Microb. Ecol. 43: 307–314 [DOI] [PubMed] [Google Scholar]
- 36. Lipson D. A., Schmidt S. K. 2004. Seasonal changes in an alpine soil bacterial community in the Colorado Rocky Mountains. Appl. Environ. Microbiol. 70: 2867–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reference deleted.
- 38. Martiny J. B. H., et al. 2006. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4: 102–112 [DOI] [PubMed] [Google Scholar]
- 39. Matsui K., Ishii N., Honjo M., Kawabata Z. 2004. Use of the SYBR green I fluorescent dye and a centrifugal filter device for rapid determination of dissolved DNA concentration in fresh water. Aquat. Microb. Ecol. 36: 99–105 [Google Scholar]
- 40. McIntyre R. E. S., Adams M. A., Grierson P. E. 2009. Nitrogen mineralization potential in rewetted soils from a semi-arid stream landscape, north-west Australia. J. Arid Environ. 73: 48–54 [Google Scholar]
- 41. Miglia K. J., et al. 2007. Genotype, soil type, and locale effects on reciprocal transplant vigor, endophyte growth, and microbial functional diversity of a narrow sagebrush hybrid zone in Salt Creek Canyon, Utah. Am. J. Bot. 94: 425–436 [DOI] [PubMed] [Google Scholar]
- 42. Mulvaney R. L. 1996. Nitrogen-inorganic forms, p. 1123–1184 In Sparks D. L. (ed.), Methods of soil analysis, part 3. American Society of Agronomy-Soil Science Society of America, Madison, WI [Google Scholar]
- 43. Oksanen J., et al. 2011. vegan: community ecology package. R package version 1.17-6. http://CRAN.R-project.org/package=vegan
- 44. O'Malley M. A. 2008. “Everything is everywhere: but the environment selects”: ubiquitous distribution and ecological determinism in microbial biogeography. Stud. Hist. Philos. Biol. Biomed. Sci. 39: 314–325 [DOI] [PubMed] [Google Scholar]
- 45. Pang X., et al. 2007. Inter-species transplantation of gut microbiota from human to pigs. ISME J. 1: 156–162 [DOI] [PubMed] [Google Scholar]
- 46. Papatheodorou E. M., Stamou G. P., Giannotaki A. 2004. Response of soil chemical and biological variables to small and large scale changes in climatic factors. Pedobiologia 48: 329–338 [Google Scholar]
- 47. Ranjard L., et al. 2010. Biogeography of soil microbial communities: a review and a description of the ongoing French national initiative. Agron. Sustain. Dev. 30: 359–365 [Google Scholar]
- 48. Rappè M. S., Giovannoni S. J. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57: 369–394 [DOI] [PubMed] [Google Scholar]
- 49. Risk D., Kellmann L., Beltrami H., Diochon A. 2008. In situ incubations highlight the environmental constraints on soil organic carbon decomposition. Environ. Res. Lett. 3: 044004 [Google Scholar]
- 50. Ross C. A., Faust D., Auge H. 2009. Mahonia invasions in different habitats: local adaptation or general-purpose genotypes? Biol. Invest. 11: 441–452 [Google Scholar]
- 51. Savin M. C., Gorres J. H., Neher D. A., Amador J. A. 2001. Biogeophysical factors influencing soil respiration and mineral nitrogen content in an old field soil. Soil Biol. Biochem. 33: 429–438 [Google Scholar]
- 52. Scheepens J. F., Frei E. S., Stöcklin J. 2010. Genotypic and environmental variation in specific leaf area in a widespread Alpine plant after transplantation to different altitudes. Oecology 164: 141–150 [DOI] [PubMed] [Google Scholar]
- 53. Schütte U. M. E., et al. 2008. Advances in the use of terminal restriction fragment length polymorphism (T-RFLP) analysis of 16S rRNA genes to characterize microbial communities. Appl. Microbiol. Biotechnol. 80: 365–380 [DOI] [PubMed] [Google Scholar]
- 54. Schwartz E., Adair K. L., Schuur E. A. 2007. Bacterial community structure correlates with decomposition parameters along a Hawaiian precipitation gradient. Soil Biol. Biochem. 39: 2164–2167 [Google Scholar]
- 55. Sigler W. V., Zeyer J. 2004. Colony-forming analysis of bacterial community succession in deglaciated soils indicates pioneer stress-tolerant opportunists Microb. Ecol. 48: 316–323 [DOI] [PubMed] [Google Scholar]
- 56. Taras B., Sturm M., Liston G. E. 2002. Snow-ground interface temperatures in the Kuparuk River Basin, Arctic Alaska: measurements and model. J. Hydrometeorol. 3: 377–394 [Google Scholar]
- 57. Ter Braak C. J. F. 1986. Canonical correspondence analysis: a new eigenvector technique for multivariate direct gradient analysis. Ecology 67: 1167–1179 [Google Scholar]
- 58. Vanhala P., et al. 2009. Transplantation of organic surface horizons of boreal soils into warmer regions alters microbiology but not the temperature sensitivity of decomposition. Global Change Biol. 17: 538–550 [Google Scholar]
- 59. Waldrop M. P., Firestone M. K. 2006. Response of microbial community composition and function to soil climate change. Microb. Ecol. 52: 716–724 [DOI] [PubMed] [Google Scholar]
- 60. Yannarell A. C., Triplett E. W. 2004. Within- and between-lake variability in the composition of bakterioplankton communities: investigations using multiple spatial scales. Appl. Environ. Microbiol. 70: 214–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zvomuya F., Janzen H. H., Larney F. J., Olson B. M. 2008. A long-term field bioassay of soil quality indicators in a semiarid environment. Soil Sci. Soc. Am. J. 72: 683–692 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.