Abstract
Listeria monocytogenes is among the most important food-borne pathogens and is well adapted to persist in the environment. To gain insight into the genetic relatedness and potential virulence of L. monocytogenes strains causing central nervous system (CNS) infections, we used multilocus variable-number tandem-repeat analysis (MLVA) to subtype 183 L. monocytogenes isolates, most from ruminant rhombencephalitis and some from human patients, food, and the environment. Allelic-profile-based comparisons grouped L. monocytogenes strains mainly into three clonal complexes and linked single-locus variants (SLVs). Clonal complex A essentially consisted of isolates from human and ruminant brain samples. All but one rhombencephalitis isolate from cattle were located in clonal complex A. In contrast, food and environmental isolates mainly clustered into clonal complex C, and none was classified as clonal complex A. Isolates of the two main clonal complexes (A and C) obtained by MLVA were analyzed by PCR for the presence of 11 virulence-associated genes (prfA, actA, inlA, inlB, inlC, inlD, inlE, inlF, inlG, inlJ, and inlC2H). Virulence gene analysis revealed significant differences in the actA, inlF, inlG, and inlJ allelic profiles between clinical isolates (complex A) and nonclinical isolates (complex C). The association of particular alleles of actA, inlF, and newly described alleles of inlJ with isolates from CNS infections (particularly rhombencephalitis) suggests that these virulence genes participate in neurovirulence of L. monocytogenes. The overall absence of inlG in clinical complex A and its presence in complex C isolates suggests that the InlG protein is more relevant for the survival of L. monocytogenes in the environment.
INTRODUCTION
Listeria monocytogenes is a food- and feed-borne pathogen of major concern with regard to public and animal health. This Gram-positive bacterium affects a wide range of mammalian species, most commonly humans and domestic ruminants (15). Clinical manifestations are similar in all susceptible hosts and include septicemia, abortion, severe gastroenteritis, and central nervous system (CNS) infections, such as meningitis, meningoencephalitis, and rhombencephalitis (2, 55). CNS involvement is a characteristic feature in humans and ruminants and accounts for the high fatality rates (20 to 50%) associated with listeriosis compared to other food-borne infections (9, 43, 55). Rhombencephalitis is the most commonly encountered form of listeriosis in farm ruminants and is of importance not only because it results in significant economic losses in livestock production due to morbidity and high mortality in animals but also because of its impact on food safety and public health, as infected animals represent a possible reservoir for human infection (4, 38, 44, 61). Clinically affected animals and healthy carriers may shed the bacteria in their feces and milk, thus contaminating pastures, vegetables, surface water, and milk products (5, 14, 28, 62).
L. monocytogenes is widespread in the environment due to its eurythermal and osmotolerant characteristics as well as its ability to grow in a wide pH range (1, 15), which hampers surveillance and infection control. This species encompasses numerous strains with a high genetic diversity (13, 21), and several studies have suggested that there are interstrain differences in virulence and transmission (38, 61).
Because of the high frequency of fatal outcomes of L. monocytogenes infections and the ubiquity of the agent, high-resolution subtyping methods that accurately discriminate between strains are very important tools in routine surveillance, outbreak investigation, and control. The standard method is serotyping, which allows the differentiation of at least 13 serotypes (53), of which only three (1/2a, 1/2b, and 4b) are responsible for the vast majority of human and animal infections (30, 35, 46). Genomic subtyping methods divide L. monocytogenes into two major evolutionary lineages (lineages I and II) and one minor lineage (lineage III) (45, 46, 49, 61), which apparently differ in host specificity and pathogenic potential (3, 7, 27).
Most studies have focused on human clinical isolates and outbreak-related food isolates, including the epidemic clones, which is a group of related, highly clonal LM subtypes with apparently high pathogenic potential. In contrast, information on the subtypes of isolates from ruminants is rare (38, 61).
This study had two objectives. The first was to investigate the genotypic subtypes of ruminant clinical isolates and compare them to human clinical isolates and food and environmental isolates to determine whether ruminant isolates are commonly found in food and the environment. For this purpose, a multilocus variable-number tandem-repeat analysis (MLVA) method that allows the clustering of L. monocytogenes strains with a strong epidemiological concordance (57) was utilized. This method is based on the analysis of the number of small repeated units present in eight different variable-number tandem-repeat (VNTR) loci, which mostly encode extracellular invasion-associated proteins (57). Hence, the method not only clusters strains purely according to their genetic relatedness, but being based on potential virulence markers, it also indirectly separates strains based on their ecological niche and pathogenic potential (10). The second objective was to determine whether polymorphisms of virulence genes are associated with a putative MLVA cluster or origin of the isolate. A wide variety of virulence genes are involved in the intracellular life cycle of L. monocytogenes within the host (11, 12, 15, 17, 54, 60). These include Listeria pathogenicity island 1 (LIPI-1), which contains six key virulence genes (i.e., prfA, plcA, hly, mpl, actA, and plcB) (26, 31, 40, 60), and various internalin genes scattered over the L. monocytogenes genome (18, 19, 51, 52).
To these ends, MLVA was used to type 183 isolates of L. monocytogenes from two main sources: (i) clinical rhombencephalitis isolates from small ruminants and cattle and human clinical isolates and (ii) L. monocytogenes isolated from food and the environment, collected from two geographically and climatically different countries. Subsequently, representative isolates belonging or linked to the MLVA clonal complexes were screened for polymorphisms in nine internalin genes and two additional virulence-associated genes by PCR and sequencing.
MATERIALS AND METHODS
Bacterial isolates and growth conditions.
A collection of 183 L. monocytogenes isolates were investigated in this study (see Table S1 in the supplemental material). The collection contained 109 isolates from the brains of ruminants with listeric rhombencephalitis that were collected in Switzerland within the framework of surveillance programs and routine diagnostics from 1996 to 2010. Furthermore, the collection included 4 isolates from the placentas of bovine abortions, 10 isolates from human neonatal infections, 10 from human cerebrospinal fluid (CSF), 40 isolates from non-outbreak-related food from Switzerland and Greece (16), 10 isolates from the environment, and 14 isolates of other origins (see Table S1 in the supplemental material). Isolates from human patients and environmental isolates were obtained from the Swiss National Centre for Listeriosis (CHUV, Lausanne, Switzerland). Other isolates were obtained from fresh or frozen tissues (brainstem, placenta, and endocardium) and cultured directly on polymyxin acriflavine lithium chloride ceftazidime esculin mannitol (Palcam) agar (Oxoid, Basel, Switzerland). Potential Listeria sp. colonies were subcultivated on tryptic soy agar (TSA) containing 5% sheep blood (Oxoid, Basingstoke, United Kingdom) and identified using a Vitek 2 compact system (bioMérieux, Geneva, Switzerland) according to the operating instructions. Subsequently, the isolates were confirmed by an in-house-validated real-time PCR using specific primers and probes for the detection of the hly gene of L. monocytogenes (50). All isolates are deposited in the ZOBA strain collection of the Institute of Veterinary Bacteriology, University of Bern, Switzerland.
Lysates.
Bacterial DNA templates for PCR were obtained by a direct lysis method. Isolates were grown on TSA at 37°C for 24 h. Five bacterial colonies were lysed in 450 μl lysate buffer containing 0.1 M Tris-HCl (pH 8.5), 0.05% Tween 20, and 0.24 mg/ml of proteinase K for 1 h at 60°C. This was followed by inactivation of the proteinase K for 15 min at 97°C. Lysed samples were kept at −20°C until further analysis.
Characterization of L. monocytogenes isolates by MLVA.
Eight reference loci (Lm-2, Lm-3, Lm-8, Lm-10, Lm-11, Lm-15, Lm-23, and Lm-32) and eight corresponding primer pairs, previously described by Sperry et al. (57), were used to type L. monocytogenes isolates by MLVA. All selected loci have relatively small repeat units, 6 to 15 bp, and are able to distinguish serotypes 1/2a, 1/2b, and 4b (57). All selected primers were manufactured by Microsynth AG (Balgach, Switzerland). The specificity of all primers was verified using the Basic Local Alignment Search Tool (BLAST) (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
The eight loci were tested in a single PCR before being combined in a multiplex PCR assay. The multiplex PCR was a modification of that described by Sperry et al. (57) and consisted of four multiplex PCRs with two primer loci in each reaction: R1 (Lm-2, Lm-8), R2 (Lm-10, Lm-11), R3 (Lm-3, Lm-23), and R4 (Lm-15, Lm-32). PCR products were amplified using a Qiagen multiplex PCR kit (Qiagen AG, Basel, Switzerland) as per the manufacturer's instructions. Amplification was performed using a Gene Amp 9600 DNA thermal cycler (Applied Biosystems, Foster City, CA), with the following settings: initial denaturation step at 95°C 15 min, followed by 35 cycles of three-step cycling at 94°C for 30 s, 52°C for 90 s, and 72°C for 90 s, and one final extension step at 72°C for 10 min.
DNA fragments were separated on an Agilent 2100 bioanalyzer using an Agilent DNA 1000 kit (Agilent Technologies, Waldbronn, Germany) according to the manufacturer's standard protocol. The number of repeats in each locus was calculated as described by Sperry et al. (57). The fragment size with no tandem repeats (a) was subtracted from the amplicon size (x) and then divided by the repeat unit length (b) [(x-a)/b]. A categorical minimum spanning tree (MST) was created using BioNumerics software (version 5.1; Applied Math, Sint-Martens-Latem, Belgium) from MLVA data. Discriminatory power was calculated with an internet-based tool (http://insilico.ehu.es/mini_tools/discriminatory_power/) that applies the Simpson's index of diversity formula (22).
Serogroup-related PCR typing.
Molecular serovar group type was determined for 165 isolates using protocols described by Doumith et al. (13).
DNA polymorphism analysis in 12 virulence-associated genes by PCR and DNA sequence analysis.
The presence and possible allelic variations of nine internalin genes (inlA, inlB, inlC, inlD, inlE, inlF, inlG, inlJ, and inlC2H) and two additional virulence-associated genes (prfA and actA) in L. monocytogenes were analyzed by PCR amplification and DNA sequencing, using the primers listed in Table 1 . Four isolates (two ovine, one caprine, and one bovine) from complex A (L2, L55, L11, and L81), and four isolates (one ovine, one caprine, and two environmental) from complex C or linked to complex C (L86, L84, CHUV 206/2005 [1/2a], and CHUV 031/2005 [1/2a]) were selected for this screening. All isolates were screened with primers targeting the actA and inlJ genes. The presence or absence of the inlF and inlG genes was investigated in 150 and 131 isolates, respectively. PCR with 35 cycles and an annealing temperature of 54°C was used to amplify DNA fragments. DNA sequence analysis of PCR fragments was performed with an ABI Prism 3100 genetic analyzer (Applied Biosystems) and a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems) following the manufacturer's instructions. Sequences were edited with Sequencer 4.6 software (GeneCodes, Ann Arbor, MI). The entire genes of the actA alleles actA3 and actA4 (isolates L86/2007 and L11/2007) and of the inlJ alleles inlJ1, inlJ2, and inlJ3 (isolates L11/2007, L41/2007, and CHUV 206/2005) were amplified as overlapping fragments using the primers shown in Table 1. They were subsequently sequenced.
Table 1.
Primers used to amplify virulence-associated genes actA, prfA, inlA, inlB, inlC, inlD, inlE, inlF, inlG, inlJ, and inlC2H
| Primer use and gene or primer name | Sequence (5′–3′) |
|
|---|---|---|
| Forward primer | Reverse primer | |
| Determination of gene size variations | ||
| actA | GTTGATAAAAGTGCAGGGTTAATTG | GCAATTAGTTTTCCTTCTTCAATGC |
| prfA | ATGAACGCTCAAGCAGAAGAATTC | TTAATTTAATTTTCCCCAAGTAGCA |
| inlA-a | TAACAAAGCTTCAAAGATTATTTTTCTATAATAAC | TCAGTCAATAAATTCCCAGCTTCC |
| inlA-b | AAGCTGGGAATTTATTGACTGAACC | TGAATTATAAGGGTCATAAGCGTTCATT |
| inlB | TGGTTTTCGGACTATATCTAGCTTTT | TAAAATCGTTTCCGGACATATAATCC |
| inlC | TGCTAACATATAAGTATACAAAGGGACA | AACTGTTCCATCAAATATAGCCTCA |
| inlD | AACTCTGGCTGTAGTAATGGCAAT | CGCCAGTTTTCGCATCATAC |
| inlE | TAAATTCATCTAAACGTCACAACTAAA | TGACTGATTGGTTAAATGTGTAACTAA |
| inlF | AGAAGCGGAAATTTGCATATTAA | ACCAACCATCAAATGTATAACCTTG |
| inlG | AAACCTCAGTACTACATGTTTTACTTGTAG | TCCTACAAATGTGTAGCCATCTTTC |
| inlJ | GACATCCCAGATTATACATTAACGACTACT | TTTGCCATTAGCATCCACGTAATTCA |
| inlC2H | GTTTTAATGGTAACTGCTATTCTCGG | AAAACCGCAGTGTAAGCTTCTG |
| Amplification of actA and inlJ | ||
| inlJ-a | AGTCATTAAACCGACACAATCGAGT | CAGTCTAGCTCAGTTAATTGTGTATTGTGG |
| inlJ-b | AACTTATTTAAACTGCGCACGCA | TTAAAGTTTCTTTAGGCATCGTGATTG |
| inlJ-c | TTGCGTAAATGCTCACATCCAA | TGTACACGTACGTTACGGACTGGC |
| inlJ | GACATCCCAGATTATACATTAACGACTACT | TTTGCCATTAGCATCCACGTAATTCA |
| inlJ-d | GCTAACAACTACACCAGATAACGCAACC | ACTGGATGTGGGTGTTTCTATCCAAGT |
| actA-a | TTTCCTTGTTCTAAAAAGGTTGTATTAGC | GTGGAAAGTCCGAAGCATTTACC |
| actA | GTTGATAAAAGTGCAGGGTTAATTG | GCAATTAGTTTTCCTTCTTCAATGC |
| actA-b | TGAAACCGCAAACCGAGG | ATTTACCTCAGTATTTGTCGGATTATCC |
The complete actA from strains L86/2007 and L11/2007 and inlJ from strains L11/2007, L41/2007, and CHUV 206/2005 were sequenced.
Plaque-forming assays.
The in vitro virulence of 99 randomly chosen isolates was assessed in a slightly modified plaque-forming assay as previously described (59). A bovine macrophage cell line (58) (kindly provided by D. Dobbelaere, Department of Clinical Research and Veterinary Public Health, Vetsuisse Faculty Bern) was used between passages 6 and 25. Cells were grown in Dulbecco's modified Eagle medium (DMEM) plus Glutamax (61965-026; GIBCO) supplemented with 10% fetal calf serum and 2 mM l-glutamine (Bioconcept, Allschwil, Switzerland) and kept in a humidified incubator at 37°C in the presence of 5% CO2. Penicillin (100 IU ml−1) and streptomycin (100 μg ml−1) were routinely added to the culture medium, except for the virulence assays. For the plaque-forming assay, cells were grown to confluence in 24-well tissue culture plates (2 × 106 per well) for 24 h. Monolayers were washed with phosphate-buffered saline (PBS) and inoculated with 104 bacteria per well as determined by the McFarland turbidity standard. Bacterial suspensions were prepared in DMEM, and infected cultures were incubated at 37°C. After 1 h of incubation, wells were washed three times with 500 μl PBS and covered with 700 μl DMEM–1% agarose containing 10 μg of gentamicin per ml. Wells were incubated at 37°C for 3 days to allow plaque formation. In each experiment, L. monocytogenes strain L114, which causes large plaques in the bovine macrophage cell line, was used as a positive control, and the reference strain, L. innocua strain L136, was used as a negative control. Additionally, mock infection of cell monolayers was included. Experiments were carried out in duplicate for each strain. The sizes of 10 plaques were determined using an inverted microscope. The mean plaque size of a given strain was expressed as a percentage of that of the reference strain, L114.
Statistical analysis.
Contingency tables were used to measure the statistical significance of the difference between clonal complexes, ActA protein, InlJ protein, and the origin of the strain using Fisher's exact test (GraphPad Software). Variation in plaque size was compared between groups (origin, MLVA complex, actA allele, and inlJ allele) with analysis of variance (Kruskal Wallis ANOVA followed by Bonferroni correction for multiple comparisons).
Nucleotide sequence accession numbers.
The sequences of the actA and inlJ alleles were deposited in the GenBank/EMBL database under the following accession numbers: actA4 (strain L86/2007), FR686458; actA3 (strain L11/2007), FR686459; inlJ1 (strain L11/2007), FR686501; inlJ2 (strain L41/2007), FR686503; and inl3 (strain CHUV 206/2005), FR686502.
RESULTS
MLVA clusters L. monocytogenes into clonal complexes correlating with the origin of isolates.
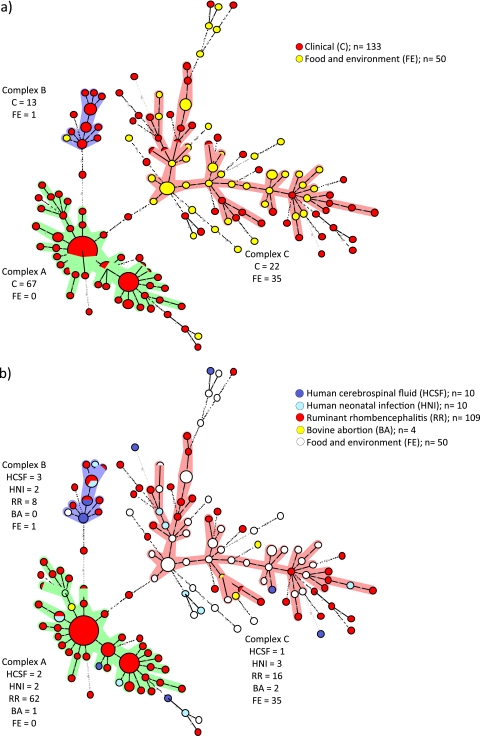
One hundred forty-three MLVA types were detected, corresponding to a discriminatory power of 0.987. Allelic profile-based comparisons of MLVA data clustered L. monocytogenes isolates from this study into two major clonal complexes (arbitrarily designated A and C), one minor complex (B), and numerous single-locus variants (SLV) (Fig. 1a). Clonal complexes A and C contained the majority of isolates (67 and 56 of 197, respectively), whereas only 14 isolates were assigned to complex B (Fig. 1a). There was a strong tendency of L. monocytogenes isolates to cluster based on their origin. Complex A contained uniquely human and ruminant clinical isolates, and except for the Swiss food isolate CHUV 005/2007 (serovar 4b), all isolates of complex B were of clinical origin (Fig. 1a). In contrast, complex C (n = 57) appeared to be more heterogeneous than the other two complexes and contained the majority of food and environmental isolates (35/50), independent of their geographical origin (Fig. 1a). All remaining food and environmental isolates were closely linked to complex C (13/50), except one that was linked to complex A. According to Fisher's exact test, complex A isolates were significantly associated with clinical origin, while complex C isolates were significantly associated with food and the environment (P < 0.0001).
Fig. 1.
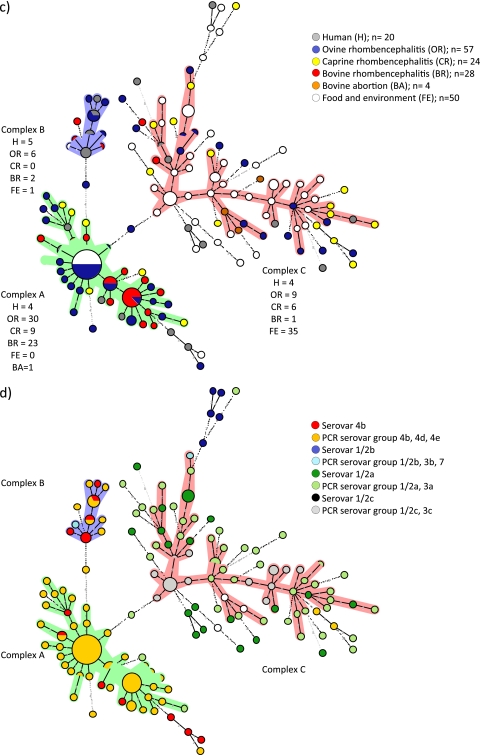
Minimum-spanning-tree analysis of 183 L. monocytogenes isolates based on eight genetic markers. Circles represent all isolates, and their size is proportional to the number of isolates. Colored zones surrounding the circles delineate the different clonal complexes. Clonal complexes were created based on the maximum neighbor distance of changes at two loci and the minimum size of five types. The length of the branches represents genetic distances (changes in loci are represented with numbers) between two neighboring types. The color codes indicate the clinical or nonclinical origin of the isolates (a), the specific origin of clinical isolates (b), the human and ruminant species (c), and the serovar or the PCR-based serovar group (d).
Ruminant rhombencephalitis isolates belong to the two related complexes A and B.
The majority of ruminant rhombencephalitis isolates (109 total), which were collected from a large geographical area and over a period of 14 years, belonged to the major complex A (62/109) and related minor complex B (8/109), whereas only 16 belonged to the large complex C (Fig. 1b). Hence, 93% of ruminant rhombencephalitis isolates were in complex A. This clustering was particularly striking in bovine rhombencephalitis isolates (total 27), of which all but 4 clustered in complex A (Fig. 1c). In addition to the bovine rhombencephalitis isolates, isolates of ovine rhombencephalitis (30) and caprine rhombencephalitis (9) were assigned to complex A. Notably, 29% (n = 32) of all ruminant rhombencephalitis isolates belonged to three identical MLVA-genotypes (Fig. 1b and c) within complex A. Only 5 of the 67 complex A isolates were of origins other than ruminant rhombencephalitis, including bovine placenta (n = 1), human neonatal infections (n = 2), and human CSF (n = 2) (Fig. 1b). Human clinical isolates from Switzerland were distributed nearly uniformly over the three complexes and their related SLV, independent of their origin (CSF, neonatal infection) (Fig. 1b). The minor clonal complex B (n = 14) contained mainly CNS-related isolates, including ovine rhombencephalitis (6/14), bovine rhombencephalitis (2/14), and human CSF isolates (3/14). The remaining isolates included two from human neonatal infections and one isolated from a cheese during a minor listeriosis outbreak (Fig. 1b and c).
The clinical isolates of complex C originated from ovine (nine), caprine (six) and bovine (one) rhombencephalitis, human CNS infection (one), human neonatal infections (three), and bovine abortion (two) (Fig. 1b and c).
Human clinical isolates were spread over the entire MST, and most had unique MLVA genotypes. However, three MLVA genotypes, two from complex B and one from complex A, were responsible for both ruminant rhombencephalitis and human infection (Fig. 1b).
Complex A and B isolates were significantly associated with ruminant rhombencephalitis, in contrast to complex C isolates, which were associated with food and the environment (Fisher's exact test, P < 0.0001 for complex A and P = 0.0086 for complex B).
Correlation of serovars with MLVA complexes and origin.
For 42 isolates, the serovar was known, as determined by serotyping. For the remaining isolates, the PCR serovar group type (1/2a-3a, 1/2b-3b-7, 1/2c-3c, and 4b-4d-4e) was identified according to the protocols of Doumith et al. (13) (see Table S1 in the supplemental material). The serovars and serovar group types correlated with MLVA clonal complexes (Fig. 1d). Complexes A and B contained exclusively isolates of serovar 4b and the serovar group type 4b-4d-4e, except two ruminant rhombencephalitis isolates, which were serovar group type 1/2a-3 and serovar group type 1/2b-3b-7 (Fig. 1d). In contrast, complex C comprised only isolates of serovar 1/2a and the serovar group types 1/2a-3a, and 1/2c-3c, except isolate L38/2007, which belongs to the PCR serovar group 1/2b-3b-7 (Fig. 1d). Of the isolates of serovar 1/2b and of the PCR serovar group 1/2b-3b, two were randomly spread over the MST, and most were allocated outside the MLVA complexes. Five closely related isolates of serovar 1/2b were linked to complex C, and one single isolate of this serovar was linked to complex B (Fig. 1d).
Ruminant rhombencephalitis isolates belonged predominantly to serovar groups 4b-4d-4e (75/109) and 1/2a-3a (31/109), with only two isolates affiliated with the serovar group 1/2b-3b-7. The vast majority of food and environmental isolates belonged to serovars 1/2a or serovar group 1/2a-3a (27/50) and serovar 1/2c or serovar group 1/2c-3c (16/50), respectively. Only three belonged to serovar 1/2b, and two isolates were serovar 4b.
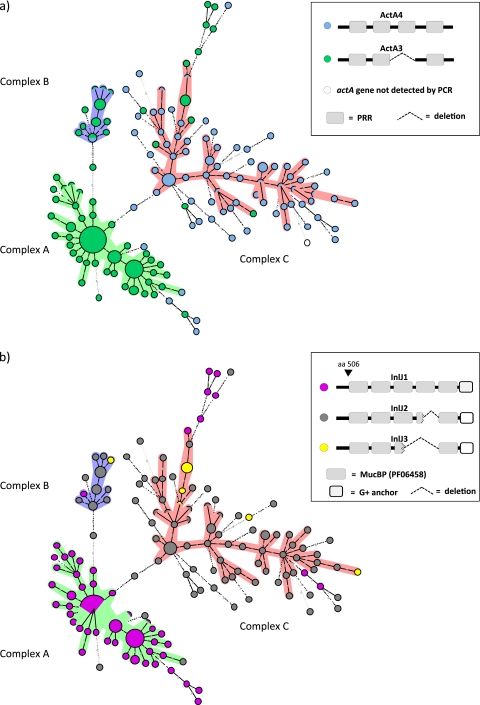
Allele profiles of the virulence-associated genes actA and inlJ correlate with MLVA complexes and origin of isolates.
PCR revealed major differences in the actA and inlJ genes of the three clonal complexes. Two different fragment sizes of actA and three fragments of inlJ were detected (Fig. 2a and b; also, see Table S1 in the supplemental material). For detailed characterization of the detected actA and inlJ variants, the entire actA from isolates L86/2007 and L11/2007 and inlJ from isolates L11/2007, L41/2007, and CHUV 206/2005 were sequenced.
Fig. 2.
Distribution of the actA (a) and inlJ (b) alleles in the minimum spanning tree. Characteristics of the minimum spanning tree are the same as in Fig. 1. (a) actA alleles are indicated with different colors. Schematic differences of the ActA profiles are shown in the inset. PRRs are depicted as gray boxes. (b) inlJ alleles are shown in different colors. The inset shows the schematic differences of the InlJ profiles. The mucin-binding protein (MucBP) domains are depicted as gray boxes. Broken boxes represent partial MucBP; dashed lines indicate deletions. “G+ anchor” indicates the LPXTG-motif cell wall anchor domain of Gram-positive organisms. The first amino acid of the first mucin-binding protein domain corresponds to position 506 for all three alleles.
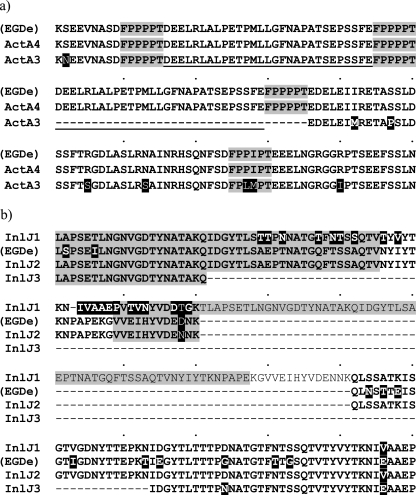
The two alleles of actA were designated actA3 (1,815 bp) and actA4 (1,920 bp) according to previous observations (61). DNA alignments showed that actA4 contains a segment of 105 bp that is absent in allele actA3. This segment in allele actA4 encodes a repeated 29-amino-acid stretch followed by a 6-amino-acid proline-rich region (PRR) (Fig. 3a).
Fig. 3.
(a) Alignment of the nucleotide sequence-derived amino acid sequences of the variable parts of ActA for the two different actA alleles. PRRs are marked in gray. The dashes indicate the 35 amino acids missing in ActA3, consisting of the 29 amino acids from the long repeat (underlined) and one 6-amino-acid-long PPR marked in gray. L86 (ActA4) and L11 (ActA3) predicted amino acid sequences are compared to ActA of the EGDe strain (accession number CAA42407). The amino acid variations in ActA3 are indicated by the black background. (b) Alignment of the nucleotide sequence-derived amino acid sequences of the variable part of InlJ for the three inlJ alleles. The predicted amino acid sequences of InlJ1, InlJ2, and InlJ3 are compared to InlJ of strain EGDe (accession number NP_466343). The dashes indicate the missing amino acids. The amino acid variations are indicated with a black background. Amino acids belonging to mucin-binding protein (MucBP) domains are marked in gray.
The three alleles of inlJ were designated inlJ1 (2,751 bp), inlJ2 (2,541 bp), and inlJ3 (2,343 bp). Partial DNA alignments of the three inlJ alleles and comparison with L. monocytogenes type strain EGDe are shown in Fig. 3b. Allele inlJ2 has the same length as inlJ in strain EGDe but codes for 13 different amino acids. Allele inlJ1 is significantly larger and inlJ3 is significantly shorter than inlJ in strain EGDe. These three alleles of inlJ differ mainly in the number of mucin-binding domains in their encoded proteins (MucBP; PF06458): InlJ1 has 5, InlJ2 has 4, and InlJ3 has 3 complete MucBP.
Differences in fragment sizes of the actA and inlJ genes correlate with the MLVA clonal complexes (Fig. 2). Generally, the shorter allele, actA3 (n = 98), is found mainly in the two clonal complexes that contain predominantly clinical isolates, including rhombencephalitis isolates, that is, A and B. Sixty-six of 67 isolates in complex A and 13/14 isolates in complex B contain actA3, while only 7 isolates of complex C (56) harbor this allele (Fig. 2a). In contrast, the larger allele, actA4 (n = 84), is mainly found in complex C (50 of 57), which contains predominantly isolates from food and the environment (Fig. 2a; also, see Table S1 in the supplemental material), and in SLVs linked to complex C (29/36). The remaining isolates harboring actA4 (n = 5) were in complex B and linked to complex A. Allele inlJ1 (n = 80) is mainly present in clonal complex A isolates and in its SLVs (5/7). Sixty-six of 67 complex A isolates and 5 of 7 isolates linked to complex A have inlJ1, whereas only one isolate of complex B and C each has this allele. Allele inlJ2 (n = 95) is predominantly clustered in clonal complex C, in which 50/57 isolates contain this allele, and is present in 28 of 36 SLVs linked to this cluster (Fig. 2b). Furthermore, 12 of the 14 complex B isolates contain inlJ2, while only one strain of complex A has this allele. Isolates with inlJ3 (n = 8) are primarily found in complex C (6).
With regard to the different complexes, 66 of 67 (98.5%) isolates of clonal complex A harbored an actA3-inlJ1 profile (Fig. 2a and b; also, see Table S1 in the supplemental material). In contrast, 48/58 (82.8%) isolates of clonal complex C had an actA4-inlJ2 profile, and 11/14 (78.6%) isolates of clonal complex B had an actA3-inlJ2 profile. These allelic profiles were significantly associated with complexes (Fisher's exact test, P < 0.0001).
There was also a significant association of the allele profile with the origin of the isolate. Most notably, all 27 bovine rhombencephalitis isolates contain actA3, while three of four isolates from bovine placenta have actA4 (Fig, 2A; also, see Table S1 in the supplemental material). Also, in sheep, the majority of rhombencephalitis isolates (42/57) carry allele actA3, whereas caprine rhombencephalitis isolates (16/24) predominantly carry allele actA4. With respect to inlJ, the majority of bovine rhombencephalitis isolates (23/27) carry allele inlJ1, while three of four isolates from bovine placenta have inlJ2. Sheep rhombencephalitis isolates (n = 57) carry all three alleles, inlJ1 (33/57), inlJ2 (21/57), and inlJ3 (3/57). Goat rhombencephalitis isolates (n = 24) also have inlJ1 (10/24), inlJ2 (13/24), and inlJ3 (1/24). In summary, the vast majority of bovine rhombencephalitis isolates harbor an actA3-inlJ1 profile, whereas in ovine and caprine rhombencephalitis, isolates are more heterogeneous. The actA3-inlJ1 profile was significantly associated with clinical origin and the actA4-inlJ2 profile with food and environmental origins (Fisher's exact test, P < 0.0001). More specifically, the actA3-inlJ1 profile was significantly associated with ruminant rhombencephalitis isolates compared to other clinical isolates (Fisher's exact test, P = 0.0122) or to food and environmental isolates (Fisher's exact test, P < 0.0001).
Major differences in inlF and inlG among MLVA complexes.
Major differences between MLVA complexes were observed in two additional virulence genes, inlF and inlG. The majority of isolates in all three clonal complexes and their corresponding SLVs carry the inlF gene. However, the partial predicted amino acid sequences of InlF of isolates from complex A and C share only 65% and 82% identity and similarity, respectively. The BLAST algorithm analysis of the predicted protein sequences revealed that isolates from complex A are 100% identical to InlF of the strain L. monocytogenes 4b F2365 (GenBank accession no. YP_013036.1), and those of complex C are 99% identical to the InlF of L. monocytogenes EGD-e (GenBank accession no. NP_463939.1). inlG was absent in all isolates of complex A and B, except for isolate L96/2007. In contrast, this internalin gene is found in the majority of complex C isolates and its linked SLVs (see Table S1 in the supplemental material).
The predicted proteins of the genes prfA, inlB, inlC, inlC2H, and inlA share at least 98% similarity between isolates of complexes A and C, and for the predicted protein sequences of inlE and inlD, the percent similarity was 90.3 and 93.7, respectively. Additionally, the BLAST algorithm analysis showed 99% identity between the predicted protein sequences of complex A and the protein sequence of L. monocytogenes 4b F2365 (GenBank accession no. NC_002973) and between complex C and the protein sequences of L. monocytogenes EGD-e (GenBank accession no. NC_003210). inlB of the L. monocytogenes strain F2365 is considered a pseudogene due to a premature stop codon. However, the partial predicted InlB sequences of complex A showed 99% identity to InlB of L monocytogenes 4b H7858 (GenBank accession no. ZP_00230293).
In vitro virulence does not differentiate between clinical isolates and isolates from food and the environment.
The plaque sizes of 99 isolates varied between 0 and 122% compared to that of the reference strain, L114. Small and large plaques were observed in both clinical and food/environmental isolates. Statistically significant differences in plaque size between L. monocytogenes isolates of the different complexes were not detected, as determined by MLVA or by actA and inlJ allele types, nor could the plaque sizes be correlated with the origins of the corresponding L. monocytogenes isolates.
DISCUSSION
Rhombencephalitis is the most common clinical phenotype of listeriosis in ruminants (8, 33, 41, 43). For that reason, the aims of this study included the investigation of the molecular epidemiology of ruminant rhombencephalitis L. monocytogenes isolates by MLVA and the analysis of their genetic relatedness to isolates originating from human clinical cases, food, and the environment.
Several molecular techniques have been evaluated for genotyping L. monocytogenes and for the investigation of the association between genotypes and virulence phenotypes (48). In general, these molecular approaches allow the classification of L. monocytogenes into three evolutionary genetic lineages (36, 61) that show high correlation with serotypes: lineage I comprises isolates of serotypes 1/2b, 3b, 4b, 4d, and 4e; lineage II comprises serotypes 1/2a, 1/2c, 3a, and 3c; and lineage III comprises serotypes 4a and 4c. In this study, MLVA typing of a collection of 183 isolates identified 143 MLVA types that clustered into two major complexes, A and C, a minor complex, B, and numerous single-locus variants (SLVs) linked to these complexes. The results clearly revealed that the MLVA genotyping assay grouped L. monocytogenes isolates according to their serotype and clinical origin (clinical versus food/environment). Complex A and the linked minor complex B are representative of serotype 4b and ruminant rhombencephalitis isolates, which were collected from all over Switzerland over a decade. Notably, 29% belong to only three major clonal variants of complex A, underlining the clonal nature of rhombencephalitis isolates. In contrast, non-disease-related isolates from food and environmental samples, irrespective of their geographical origin (Switzerland and Greece), are mostly of serotype 1/2a and are SLVs located either in complex C or linked to complex C. Hence, in accordance with previous studies in humans and sheep, the majority of CNS isolates were of serotype 4b, but a significant number were of serotype 1/2a (20, 34). Although only few isolates from abortions were included in the study, none of the MLVA types were detected in both abortion and rhombencephalitis, supporting the notion that clinical manifestations of listeriosis might be associated with L. monocytogenes subtypes (33, 34, 35, 48). The results of our study are in line with other genotyping studies, which have found differences in the distribution of clinical and nonclinical L. monocytogenes isolates between serotypic and genotypic subgroups (4, 29, 39, 61). In this regard, and considering the good correlation between the lineages and the serovars, the results of the PCR serovar group determination give clear evidence that complex A and B correlate with lineage I and complex C with lineage II, a model that is corroborated by the absence of inlG in complex A and B isolates. Hence, our results support the observation that lineage I (complex A and B) is highly clonal, whereas lineage II (complex C) is genetically more diverse (27). Interestingly, none of our ruminant isolates belong to lineage III, which is reported to account for up to 10% of animal listeriosis cases (27). In a previous study, lineage III isolates were observed by trend to be more prevalent in ruminant septicemia and neonatal infections than in encephalitis (48). Thus, the absence of isolates belonging to lineage III might be biased by the fact that our isolate collection comprised mainly rhombencephalitis isolates and very few ruminant isolates from other pathologies. In contrast to other studies (48, 61), in which differences in plaque size between lineages of L. monocytogenes were observed, our study did not reveal significant differences between MLVA complexes or clinical origins of isolates screening they were screened in vitro for plaque size.
Taken together, our MLVA data indicate that rhombencephalitis isolates of ruminants constitute a genetically homogeneous group that is distinct from food and environmental isolates. This observation might be linked to host tropism (ruminants), the ecological niche (the environment of ruminants) or to organotropism (neurovirulence) of the agent. Clustering of rhombencephalitis isolates is particularly evident in cattle, which is in accordance with previous studies that suggested an enhanced virulence and neurotropism of lineage I (complex A and B) strains in cattle (45, 48). There are indications that cattle are less susceptible to L. monocytogenes infection than small ruminants (8, 33, 42). Therefore, one may suspect an association between virulence and MLVA complexes, with complex A-containing L. monocytogenes isolates being particularly host-adapted and/or neurovirulent. In contrast, rhombencephalitis in small ruminants, given that they are apparently more susceptible to listeriosis, may be the result of infection with a wider spectrum of less neurovirulent isolates, including isolates from cluster C, which contains primarily food and environmental isolates. In agreement with this view, clinical isolates from human patients, who frequently suffer underlying immunity-compromising conditions, belong to all three complexes. However, genotype-related neurovirulence should be investigated further by subtyping more ruminant isolates from pathologies other than rhombencephalitis and by experimental infection studies assessing neurovirulence.
Interestingly, in addition to the different clustering of ruminant rhombencephalitis versus nonclinical isolates, none of the food isolates had a MLVA type identical to those of ruminant isolates, indicating that rhombencephalitis isolates generally do not contaminate food. However, food isolates related to human outbreaks were not included in this study. Furthermore, although few human isolates were included in this study, identical MLVA types were found in human clinical isolates and ruminant rhombencephalitis isolates, supporting the notion that ruminants might be a possible reservoir for human infection (4, 38, 44). Therefore, transmission of ruminant rhombencephalitis strains to humans, possibly via food, cannot be excluded.
As the pathogenicity of L. monocytogenes requires the activity of a particularly broad range of virulence factors that enable the pathogen to infect and replicate within the host (6, 32, 47, 52), the second aim of this study was to identify polymorphisms in virulence-associated genes, which are correlated with MLVA complexes and might be under selective pressure. The actA3 allele is mainly found in isolates of complexes A and B, which harbor most clinical isolates, whereas the actA4 allele is found in isolates of complex C. In particular, in our strain collection, actA3 is associated with rhombencephalitis. This observation is in agreement with other studies that detected two different actA allele types (actA3 and actA4) in clinically relevant versus environmental strains (61) and associated particular actA subtypes with CNS infection in cattle and humans (25, 48). The actA3 allele contains a deletion of one PRR-encoding region. Furthermore, the third PRR-encoding region of actA3 has a unique signature sequence, FPLMP (61). The PRR is involved in the stimulation of L. monocytogenes movement by providing profilin-actin complexes that fuel actin polymerization via vasodilator-stimulated phosphoprotein (VASP) (23). Although sequencing data indicate that actA3-harboring strains causing rhombencephalitis might be more efficient in intracellular movement and spread to the brain than isolates with other actA alleles, this is in contrast to our in vitro results and to in vitro studies showing a quantitative role for PRRs in the localization of VASP and profilin at the bacterium/actin tail interface and in the rate of actin-based motility (56).
Sabet and collaborators (51, 52) reported that inlJ is important for a fully virulent phenotype in mice. They demonstrated that inlJ is expressed only in vivo and that the virulence of inlJ deletion mutants is significantly attenuated in mice. Our study reveals that inlJ is characteristic of the species of L. monocytogenes, because we found this virulence gene in all 183 isolates. However, we identified, for the first time, three different alleles: inlJ1, inlJ2, and inlJ3. Our results suggest a correlation between the presence of the newly identified allele, inlJ1, and the clinical phenotype of rhombencephalitis. In contrast, isolates harboring inlJ2 or inlJ3 are mostly food and environmental isolates, which cluster in complex C or in isolates from clinical cases other than animal rhombencephalitis. These results indicate a possible role of inlJ1 in L. monocytogenes neurovirulence that leads to rhombencephalitis.
Together, the data show that isolates with the actA3 and inlJ1 alleles are significantly associated with rhombencephalitis in ruminants, whereas actA4 alleles, together with inlJ2 or inlJ3 alleles, are correlated with environmental or food origin. Therefore, it is tempting to speculate that actA and inlJ are implicated in L. monocytogenes host tropism and/or neurovirulence. Experimental-infection studies with L. monocytogenes deletion mutants are required to investigate this hypothesis. We did not observe significant differences between actA/inlJ allele types and origin with regard to in vitro cell-to-cell spread. This raises the question of the extent to which such plaque assays in bovine macrophages reflect the complex in vivo pathogenicity of L. monocytogenes isolates.
InlG was found solely in complex C, suggesting a role in survival outside the living host. inlF was amplified in isolates of all complexes. However, in agreement with the work of Nelson et al. (37), the InlF amino acid sequences of lineage I significantly differ from that of lineage II, suggesting that InlF might be specifically associated with either the host or the nonliving (environmental) habitat of L. monocytogenes. In contrast to actA, inlF, inlG, and inlJ, none of the other analyzed virulence genes showed modifications affecting amino acid sequences. This is also true for InlA, of which a truncated protein is known to occur in serotype 1/2a and 1/2c isolates (24). However, we sequenced only four selected isolates from each of the complexes A and C, and hence further polymorphisms might be detected with analysis of more isolates.
In summary, the results of this study showed that the MLVA method developed by Sperry et al. (57) has a high power of discrimination and allows tracking of L. monocytogenes strains from different origins, notably from bovine rhombencephalitis and from food and environmental isolates, supporting the view that L. monocytogenes strains are adapted to different hosts and ecological niches and differ in pathogenicity (4, 45, 48, 61). Moreover, MLVA clustering is strongly associated with particular alleles of three virulence genes, leading us to speculate that L. monocytogenes strains harboring the actA3 and inlJ1 alleles and a particular inlF genotype have increased virulence.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by the Swiss Federal Veterinary Office (grant no. 1.08.11).
We thank Jacques Bille (Centre Hospitalier Universitaire Vaudois CHUV, Lausanne, Switzerland) for kindly providing L. monocytogenes strains. We thank Edy Vilei for his work with the BioNumerics software and for his helpful comments.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 7 October 2011.
REFERENCES
- 1. Badaoui Najjar M., Chikindas M., Montville T. J. 2007. Changes in Listeria monocytogenes membrane fluidity in response to temperature stress. Appl. Environ. Microbiol. 73: 6429–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bartt R. 2000. Listeria and atypical presentations of Listeria in the central nervous system. Semin. Neurol. 20: 361–373 [DOI] [PubMed] [Google Scholar]
- 3. Bibb W. F., Schwartz B., Gellin B. G., Plikaytis B. D., Weaver R. E. 1989. Analysis of Listeria monocytogenes by multilocus enzyme electrophoresis and application of the method to epidemiologic investigations. Int. J. Food Microbiol. 8: 233–239 [DOI] [PubMed] [Google Scholar]
- 4. Boerlin P., Piffaretti J. C. 1991. Typing of human, animal, food, and environmental isolates of Listeria monocytogenes by multilocus enzyme electrophoresis. Appl. Environ. Microbiol. 57: 1624–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Borucki M. K., et al. 2004. Dairy farm reservoir of Listeria monocytogenes sporadic and epidemic strains. J. Food Prot. 67: 2496–2499 [DOI] [PubMed] [Google Scholar]
- 6. Braun L., Cossart P. 2000. Interactions between Listeria monocytogenes and host mammalian cells. Microbes Infect. 2: 803–811 [DOI] [PubMed] [Google Scholar]
- 7. Brosch R., Chen J., Luchansky J. B. 1994. Pulsed-field fingerprinting of listeriae: identification of genomic divisions for Listeria monocytogenes and their correlation with serovar. Appl. Environ. Microbiol. 60: 2584–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brugère-Picoux J. 2010. Ovine listeriosis. Small. Ruminant Res. 76: 12–20 [Google Scholar]
- 9. Büla C. J., Bille J., Glauser M. P. 1995. An epidemic of food-borne listeriosis in western Switzerland: description of 57 cases involving adults. Clin. Infect. Dis. 20: 66–72 [DOI] [PubMed] [Google Scholar]
- 10. Cabanes D., Dehoux P., Dussurget O., Frangeul L., Cossart P. 2002. Surface proteins and the pathogenic potential of Listeria monocytogenes. Trends Microbiol. 10: 238–245 [DOI] [PubMed] [Google Scholar]
- 11. Camejo A., et al. 2011. The arsenal of virulence factors deployed by Listeria monocytogenes to promote its cell infection cycle. Virulence 2 [DOI] [PubMed] [Google Scholar]
- 12. Cossart P., Pizarro-Cerda J., Lecuit M. 2003. Invasion of mammalian cells by Listeria monocytogenes: functional mimicry to subvert cellular functions. Trends Cell Biol. 13: 23–31 [DOI] [PubMed] [Google Scholar]
- 13. Doumith M., Buchrieser C., Glaser P., Jacquet C., Martin P. 2004. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 42: 3819–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Esteban J. I., Oporto B., Aduriz G., Juste R. A., Hurtado A. 2009. Faecal shedding and strain diversity of Listeria monocytogenes in healthy ruminants and swine in Northern Spain. BMC Vet. Res. 5: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Farber J. M., Peterkin P. I. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55: 476–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Filioussis G., Johansson A., Frey J., Perreten V. 2010. Prevalence, genetic diversity and antimicrobial susceptibility of Listeria monocytogenes isolated from open-air food markets in Greece. Food Control. 20: 314–317 [Google Scholar]
- 17. Gaillard J. L., Berche P., Mounier J., Richard S., Sansonetti P. 1987. In vitro model of penetration and intracellular growth of Listeria monocytogenes in the human enterocyte-like cell line Caco-2. Infect. Immun. 55: 2822–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Glaser P., et al. 2001. Comparative genomics of Listeria species. Science 294: 849–852 [DOI] [PubMed] [Google Scholar]
- 19. Gouin E., et al. 2010. The Listeria monocytogenes InlC protein interferes with innate immune responses by targeting the IκB kinase subunit IKKα. Proc. Natl. Acad. Sci. U. S. A. 107: 17333–17338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goulet V., Hedberg C., Le Monnier A., De Valk H. 2008. Increasing incidence of listeriosis in France and other European countries. Emerg. Infect. Dis. 14: 734–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hain T., et al. 2007. Pathogenomics of Listeria spp. Int. J. Med. Microbiol. 297: 541–557 [DOI] [PubMed] [Google Scholar]
- 22. Hunter P. R., Gaston M. A. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26: 2465–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ireton K., Cossart P. 1997. Host-pathogen interactions during entry and actin-based movement of Listeria monocytogenes. Annu. Rev. Genet. 31: 113–138 [DOI] [PubMed] [Google Scholar]
- 24. Jacquet C., et al. 2004. A molecular marker for evaluating the pathogenic potential of foodborne Listeria monocytogenes. J. Infect. Dis. 189: 2094–2100 [DOI] [PubMed] [Google Scholar]
- 25. Jacquet C., Gouin E., Jeannel D., Cossart P., Rocourt J. 2002. Expression of ActA, Ami, InlB, and listeriolysin O in Listeria monocytogenes of human and food origin. Appl. Environ. Microbiol. 68: 616–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jaradat Z. W., Schutze G. E., Bhunia A. K. 2002. Genetic homogeneity among Listeria monocytogenes strains from infected patients and meat products from two geographic locations determined by phenotyping, ribotyping and PCR analysis of virulence genes. Int. J. Food Microbiol. 76: 1–10 [DOI] [PubMed] [Google Scholar]
- 27. Jeffers G. T., et al. 2001. Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology 147: 1095–1104 [DOI] [PubMed] [Google Scholar]
- 28. Jensen N. E., Aarestrup F. M., Jensen J., Wegener H. C. 1996. Listeria monocytogenes in bovine mastitis. Possible implication for human health. Int. J. Food Microbiol. 32: 209–216 [DOI] [PubMed] [Google Scholar]
- 29. Jersek B., et al. 1999. Typing of Listeria monocytogenes strains by repetitive element sequence-based PCR. J. Clin. Microbiol. 37: 103–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kathariou S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 65: 1811–1829 [DOI] [PubMed] [Google Scholar]
- 31. Kirchner M., Higgins D. E. 2008. Inhibition of ROCK activity allows InlF-mediated invasion and increased virulence of Listeria monocytogenes. Mol. Microbiol. 68: 749–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lecuit M., et al. 2001. A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science 292: 1722–1725 [DOI] [PubMed] [Google Scholar]
- 33. Low J. C., Donachie W. 1997. A review of Listeria monocytogenes and listeriosis. Vet. J. 153: 9–29 [DOI] [PubMed] [Google Scholar]
- 34. Low J. C., Wright F., McLauchlin J., Donachie W. 1993. Serotyping and distribution of Listeria isolates from cases of ovine listeriosis. Vet. Rec. 133: 165–166 [DOI] [PubMed] [Google Scholar]
- 35. McLauchlin J. 1990. Distribution of serovars of Listeria monocytogenes isolated from different categories of patients with listeriosis. Eur. J. Clin. Microbiol. Infect. Dis. 9: 210–213 [DOI] [PubMed] [Google Scholar]
- 36. Nadon C. A., Woodward D. L., Young C., Rodgers F. G., Wiedmann M. 2001. Correlations between molecular subtyping and serotyping of Listeria monocytogenes. J. Clin. Microbiol. 39: 2704–2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nelson K. E., et al. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32: 2386–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nightingale K. K., et al. 2004. Ecology and transmission of Listeria monocytogenes infecting ruminants and in the farm environment. Appl. Environ. Microbiol. 70: 4458–4467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nightingale K. K., Windham K., Wiedmann M. 2005. Evolution and molecular phylogeny of Listeria monocytogenes isolated from human and animal listeriosis cases and foods. J. Bacteriol. 187: 5537–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nishibori T., et al. 1995. Correlation between the presence of virulence-associated genes as determined by PCR and actual virulence to mice in various strains of Listeria Spp. Microbiol. Immunol. 39: 343–349 [DOI] [PubMed] [Google Scholar]
- 41. Oevermann A., et al. 2008. Neuropathological survey of fallen stock: active surveillance reveals high prevalence of encephalitic listeriosis in small ruminants. Vet. Microbiol. 130: 320–329 [DOI] [PubMed] [Google Scholar]
- 42. Oevermann A., et al. 2010. Neuropathogenesis of naturally occurring encephalitis caused by Listeria monocytogenes in ruminants. Brain Pathol. 20: 378–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Oevermann A., Zurbriggen A., Vandevelde M. 2010. Rhombencephalitis caused by Listeria monocytogenes in humans and ruminants: a zoonosis on the rise? Interdiscip. Perspect. Infect. Dis. 2010: 632513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Okwumabua O., et al. 2005. Characterization of Listeria monocytogenes isolates from food animal clinical cases: PFGE pattern similarity to strains from human listeriosis cases. FEMS Microbiol. Lett. 249: 275–281 [DOI] [PubMed] [Google Scholar]
- 45. Orsi R. H., Bakker H. C., Wiedmann M. 2011. Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 301: 79–96 [DOI] [PubMed] [Google Scholar]
- 46. Piffaretti J. C., et al. 1989. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc. Natl. Acad. Sci. U. S. A. 86: 3818–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pizarro-Cerda J., Bonazzi M., Cossart P. 2010. Clathrin-mediated endocytosis: what works for small, also works for big. Bioessays 32: 496–504 [DOI] [PubMed] [Google Scholar]
- 48. Pohl M. A., Wiedmann M., Nightingale K. K. 2006. Associations among Listeria monocytogenes genotypes and distinct clinical manifestations of listeriosis in cattle. Am. J. Vet. Res. 67: 616–626 [DOI] [PubMed] [Google Scholar]
- 49. Ragon M., et al. 2008. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 4: e1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rodríguez-Lázaro D., et al. 2004. Quantitative detection of Listeria monocytogenes and Listeria innocua by real-time PCR: assessment of hly, iap, and lin02483 targets and AmpliFluor technology. Appl. Environ. Microbiol. 70: 1366–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sabet C., Lecuit M., Cabanes D., Cossart P., Bierne H. 2005. LPXTG protein InlJ, a newly identified internalin involved in Listeria monocytogenes virulence. Infect. Immun. 73: 6912–6922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sabet C., et al. 2008. The Listeria monocytogenes virulence factor InlJ is specifically expressed in vivo and behaves as an adhesin. Infect. Immun. 76: 1368–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Seeliger H. P. R., Höhne K. 1979. Serotyping of Listeria monocytogenes and related species, p. 31–49 In Bergan T., Norris J. R. (ed.), Methods in microbiology, vol. 13 Elsevier Ltd., London, United Kingdom [Google Scholar]
- 54. Severino P., et al. 2007. Comparative transcriptome analysis of Listeria monocytogenes strains of the two major lineages reveals differences in virulence, cell wall, and stress response. Appl. Environ. Microbiol. 73: 6078–6088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Siegman-Igra Y., et al. 2002. Listeria monocytogenes infection in Israel and review of cases worldwide. Emerg. Infect. Dis. 8: 305–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Smith G. A., Theriot J. A., Portnoy D. A. 1996. The tandem repeat domain in the Listeria monocytogenes ActA protein controls the rate of actin-based motility, the percentage of moving bacteria, and the localization of vasodilator-stimulated phosphoprotein and profilin. J. Cell Biol. 135: 647–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sperry K. E., Kathariou S., Edwards J. S., Wolf L. A. 2008. Multiple-locus variable-number tandem-repeat analysis as a tool for subtyping Listeria monocytogenes strains. J. Clin. Microbiol. 46: 1435–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stabel J. R., Stabel T. J. 1995. Immortalization and characterization of bovine peritoneal macrophages transfected with SV40 plasmid DNA. Vet. Immunol. Immunopathol. 45: 211–220 [DOI] [PubMed] [Google Scholar]
- 59. Sun A. N., Camilli A., Portnoy D. A. 1990. Isolation of Listeria monocytogenes small-plaque mutants defective for intracellular growth and cell-to-cell spread. Infect. Immun. 58: 3770–3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vazquez-Boland J. A., et al. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14: 584–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wiedmann M., et al. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65: 2707–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zundel E., Bernard S. 2006. Listeria monocytogenes translocates throughout the digestive tract in asymptomatic sheep. J. Med. Microbiol. 55: 1717–1723 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.