Abstract
We detected and identified genotypes of human-pathogenic microsporidia in fecal samples from 51 asymptomatic captive-bred pet parrots in South Korea. Microsporidia were identified in 8 samples (15.7%); 7 parrots tested positive for Encephalitozoon hellem, and 1 parrot tested positive for both E. hellem and Encephalitozoon cuniculi. In genotypic identifications, E. hellem was present in genotypes 1A and 2B and E. cuniculi was present in genotype II. Pet parrots might be a source of human microsporidian infection.
TEXT
Microsporidia are obligate, intracellular, eukaryotic, single-celled fungi that infect a wide variety of vertebrate and invertebrate hosts (5). Microsporidiosis is increasingly recognized as an opportunistic disease in immunocompromised and immunocompetent individuals (5). To date, among the eight microsporidian species identified in humans, Enterocytozoon bineusi, Encephalitozoon intestinalis, Encephalitozoon hellem, and Encephalitozoon cuniculi are newly emerging zoonotic pathogens. E. bineusi and E. intestinalis induce chronic diarrhea in immunocompromised patients and induce acute self-limiting diarrhea in immunocompetent persons. E. hellem and E. cuniculi have been less commonly diagnosed and cause ocular or disseminated infections in immunocompromised patients (15).
All microsporidian species that infect humans have also been detected in various bird species. Microsporidiosis in birds was first described in masked lovebirds (Agapornis personta) in 1975 (13). After identification of microsporidia in birds, birds were considered the primary hosts of E. hellem, and droppings of asymptomatic pet birds were considered a potential environmental source of E. hellem infection in humans (2). In captive birds, microsporidiosis caused by E. hellem was first reported in budgerigars (Melopsittacus undulatus) in 1997 (3). Since then, numerous cases have been reported in a variety of pet psittacine birds (4, 16, 17). The less common microsporidian species, E. bineusi and E. cuniculi, have also been detected in psittacine birds, but E. intestinalis has been detected only in a few birds (11, 14). Encephalotozoon infections in birds typically present nonspecific signs, such as depression, decreased appetite, weight loss, and diarrhea, and unusually show ocular signs, such as conjunctivitis and keratitis. However, many infected birds present as an unexpected death without clinical signs (20). The purpose of this study was to evaluate the prevalence of microsporidia in fecal samples of asymptomatic parrots kept in private households in South Korea.
Sample collection.
A total of 51 fresh fecal samples were collected from parrots which were bred in captivity and kept in private households. All birds surveyed appeared healthy at the time of sampling. Samples were collected by placing paper sheets under the cages of birds. Fresh droppings of birds were placed in sterile tubes and frozen immediately at −70°C until analysis.
PCR and genotyping.
To extract DNA from fecal samples, the samples were resuspended in phosphate-buffered saline (PBS) (pH 7.0) and then mixed thoroughly by vortexing. The supernatants were used to extract nucleic acid. DNAzol (Invitrogen, Karlsruhe, Germany) was used according to the manufacturer's instructions, and the nucleic acid was eluted with 50 μl of Tris-EDTA (TE) buffer. For identification of microsporidian species in fecal samples, primer sets were used as previously published (12). Nested PCR was carried out using the Maxime PCR premix kit (Intron Biotechnology Inc., Seongnam, South Korea). Each product was examined via electrophoresis in 2% agarose gels stained with ethidium bromide. The PCR products were sequenced with ABI PRISM BigDye terminator v3.0 cycle sequencing kit (Applied Biosystems, Foster City, CA). Each PCR product was genetically identified through DNA homology analyses of a nucleotide sequence database using the BLAST algorithms, and subspecies and genotypes were identified.
Detection and genotype identification of Encephalitozoon spp.
No clinical signs of microsporidiosis were observed in any bird examined in this study. Eight samples of a total of 51 (15.7%) tested positive. E. hellem was detected in 7 birds (13.7%). Coinfection with E. cuniculi and E. hellem was detected in 1 bird (2.0%). No E. intestinalis and E. bineusi were detected. To identify genotypes of microsporidian species, sequences of the positive samples were compared with the reference genotype from GenBank. E. hellem was present in two different genotypes, genotypes 1A and 2B, and E. cuniculi was present in one genotype, genotype II (Table 1). The close relatedness between the isolates was also reflected in the phylogram (Fig. 1). The sequence homology was about 97 to 99% compared with reference strain of E. hellem genotype 1A (GenBank accession number AF338367.1). However, the nucleotide sequence homology of genotype 2B was comparatively low to the reference genotypes from GenBank, 90% with reference strain of E. hellem genotype 2B (GenBank accession number AF338368.1). This was suspected to be due to the low quality of the template DNA. In the case of E. cuniculi genotype II, the sequence homology was about 95% (GenBank accession number EU001241.1). In the study population, 6 new psittacine bird hosts were found: African gray parrot (Psittacus erithacus), blue-streaked lory (Eos reticulata), chestnut-fronted macaw (Ara severus), eclectus (Eclectus roratus), green-cheeked parakeet (Pyrrhura molinae), red-shouldered macaw (Diopsittaca nobilis), and rose-ringed parakeet (Psittacula krameri).
Table 1.
Differentiation and genotype identification of isolated microsporidian species in fecal samples from pet parrots
| Species and genotype | No. of positive samples (n = 51) | Prevalence (%) |
|---|---|---|
| E. cuniculi genotype II | 1a | 2.0a |
| E. hellem genotype 1A | 7 | 13.7 |
| E. hellem genotype 2B | 1a | 2.0a |
| Total | 8 | 15.7 |
One bird was found to be coinfected with E. cuniculi and E. hellem.
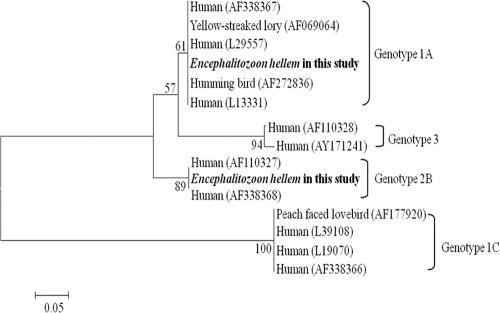
Fig. 1.
Phylogenetic tree for nucleotide sequences of Microsporidium hellem from pet parrots examined in the present study compared to other genotypes based on internal transcribed spacer (ITS) ribosomal DNA (rDNA) sequence. GenBank accession numbers are shown in parentheses. The evolutionary relationships of these genes were generated by the neighbor-joining (NJ) method with a bootstrap value of 1,000 using MEGA4 software. The numbers at the nodes represent the percentage with a bootstrap value of 1,000. Bar, 0.05 substitution per site.
Discussion.
Because microsporidia are important opportunistic pathogens in humans, more research must be done to diagnose microsporidiosis and to determine the potential host range. Diagnostic procedures such as specific stains are not readily available in diagnostic facilities due to their low detection limit (7, 11). Molecular methods have been used to diagnose microsporidiosis, and these methods are more sensitive in the diagnosis of spore shedding in naturally infected asymptomatic birds (6, 18). Currently, molecular methods are used to identify potentially human-pathogenic microsporidian genotypes in birds, since they are a potential environmental source of microsporidian infection (2, 8, 10, 11, 14).
Using a molecular method, we detected microsporidia in 15.7% of fecal samples from pet parrots in South Korea, which is a relatively low prevalence. Recently reported prevalences of microsporidia were 11%, 28.9%, and 40.1% in various birds in The Netherlands, Portugal, and the Czech Republic, respectively (1, 11, 14). The most prevalent microsporidian species in this study was E. hellem (13.7%). E. bieneusi and E. intestinalis were not detected. This was an expected result, because birds were considered to be a primary host of E. hellem (15). Barton et al. also suggested that the most frequent microsporidia in birds is E. hellem (2). However, recent studies reported that E. hellem was the least frequently identified microsporidia and that E. bieneusi was assumed to be the most frequent microsporidia in birds (1, 11, 14).
By sequencing the positive products using the internal transcribed spacer (ITS) gene, two genotypes from E. hellem, genotype 1A and genotype 2B, were identified. Genotype 1A (9.8%) was more common than genotype 2B (2.0%). Genotype 1 is the most commonly identified subtype in humans and birds (19). Although genotype 1A was previously reported in birds (11), genotype 2B has so far not been identified in birds. Genotype 2B has been reported from a patient in Switzerland and is thought to be a genotype derived from humans (21). According to previous studies analyzing E. hellem genotypes, isolates from humans show regional differences, and isolates from birds have not been subjected to these types of genetic analysis. These results suggest the existence of extensive genetic diversity in E. hellem isolates between humans and birds worldwide.
Rabbits and rodents are the main hosts of E. cuniculi, but two recent reports suggest that birds may be a new host of E. cuniculi (1, 11). In this study, one E. cuniculi genotype, genotype II (2.0%), was identified from one bird. Each E. cuniculi genotype has a specific host preference and geographical distribution. Genotype II is classified as a mouse strain and has been identified only in Europe. Genotype I was identified in rabbits in America, Australia, and Europe, and genotype III infects dogs from America, South Africa, and Europe (15). E. cuniculi genotype II isolation from birds in Korea supports the expansion of its host diversity and geographical distribution. Genotypes I and III have been reported in humans, but genotype II has not been reported. However, E. cuniculi infection may occur in imported animals from other countries and may subclinically infect animals (9). Potential spreading of genotypes not previously identified in humans should be considered.
The results of the present study demonstrated that microsporidia are widespread in asymptomatic psittacine birds kept in private households. The relatively high prevalence in pet parrots suggests that pet parrots might be a potential source of human infection and environmental contamination in South Korea. Moreover, genotypic analysis of microsporidia implies the existence of extensive genetic diversity between humans and birds and the expansion of host diversity and geographical distribution.
Nucleotide sequence accession number.
The unique E. hellem ITS sequence obtained in this study has been deposited in GenBank under accession number JF836368.
Footnotes
Published ahead of print on 30 September 2011.
REFERENCES
- 1. Bart A., Wentink-Bonnema E. M., Heddema E. R., Buijs J., van Gool T. 2008. Frequent occurrence of human-associated microsporidia in fecal droppings of urban pigeons in Amsterdam, The Netherlands. Appl. Environ. Microbiol. 74: 7056–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barton C. E., Phalen D. N., Snowden K. F. 2003. Prevalence of microsporidian spores shed by asymptomatic lovebirds: evidence for a potential emerging zoonosis. J. Avian Med. Surg. 17: 197–202 [Google Scholar]
- 3. Black S. S., Steinohrt L. A., Bertucci D. C., Rogers L. B., Didier E. S. 1997. Encephalitozoon hellem in budgerigars (Melopsittacus undulatus). Vet. Pathol. 34: 189–198 [DOI] [PubMed] [Google Scholar]
- 4. Canny C. J., Ward D. A., Patton S., Orosz S. E. 1999. Microsporidial keratoconjunctivitis in a double yellow-headed Amazon parrot (Amazona ochrocephala oratrix). J. Avian Med. Surg. 13: 279–286 [Google Scholar]
- 5. Didier E. S. 2005. Microsporidiosis: an emerging and opportunistic infection in humans and animals. Acta Trop. 94: 61–76 [DOI] [PubMed] [Google Scholar]
- 6. Fedorko D., Hijazi Y. 1996. Application of molecular techniques to the diagnosis of microsporidial infection. Emerg. Infect. Dis. 2: 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Garcia L. 2002. Laboratory identification of the microsporidia. J. Clin. Microbiol. 40: 1892–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Graczyk T. K., et al. 2007. Urban feral pigeons (Columba livia) as a source for air- and waterborne contamination with Enterocytozoon bineusi spores. Appl. Environ. Microbiol. 73: 4357–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guscetti F., Mathis A., Hatt J. M., Deplazed P. 2003. Overt fatal and chronic subclinical Encephalitozoon cuniculi microsporidiosis in a colony of captive emperor tamarins (Saguinus imperator). J. Med. Primatol. 32: 111–119 [DOI] [PubMed] [Google Scholar]
- 10. Haro M., et al. 2005. First detection and genotyping of human-associated microsporidia in pigeons from urban parks. Appl. Environ. Microbiol. 71: 3153–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kasickova D., Sak B., Kvac M., Ditrich O. 2007. Detection of Encephalitozoon cuniculi in a new host—cockatiel (Nymphicus hollandicus) using molecular methods. Parasitol. Res. 101: 1685–1688 [DOI] [PubMed] [Google Scholar]
- 12. Katzwinkel-Wladarsch S., Lieb M., Heise W., Loster T., Rinder H. 1996. Direct amplification and species determination of microsporidian DNA from stool specimens. Trop. Med. Int. Health 1: 373–378 [DOI] [PubMed] [Google Scholar]
- 13. Kemp R. L., Kluge J. P. 1975. Encephalitozoon sp. in the blue-masked lovebirds (Agapornis personata): first confirmed report of microsporidian infection in birds. J. Protozool. 22: 489–491 [DOI] [PubMed] [Google Scholar]
- 14. Lobo M., et al. 2006. Identification of potentially human-pathogenic Enteroxytozoon bieneusi genotypes in various birds. Appl. Environ. Microbiol. 72: 7380–7382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mathis A., Weber R., Deplazes P. 2005. Zoonotic potential of the microsporidia. Clin. Microbiol. 18: 423–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Phalen D. N., Logan K. S., Snowden K. F. 2006. Encephalitozoon hellem infection as the cause of a unilateral chronic keratoconjunctivitis in an umbrella cockatoo (Cacatua alba). Vet. Ophthalmol. 9: 59–63 [DOI] [PubMed] [Google Scholar]
- 17. Pulparampil N., Graham D., Phalen D., Snowden K. 1998. Encephalitozoon hellem infection in two eclectus parrots (Eclectus roratus): identification from archival tissues. J. Eukaryot. Microbiol. 45: 651–655 [DOI] [PubMed] [Google Scholar]
- 18. Sak B., Kasickova D., Kvac M., Kvetonova D., Ditrich O. 2010. Microsporidia in exotic birds: intermittent spore excretion of Encephalitozoon spp. in naturally infected budgerigars (Melopsittacus undulatus). Vet. Parasitol. 168: 196–200 [DOI] [PubMed] [Google Scholar]
- 19. Snowden K., Logan K., Phalen D. N. 2000. Isolation and characterization of an avian isolate of Encephalitozoon hellem. Parasitology 121: 9–14 [DOI] [PubMed] [Google Scholar]
- 20. Snowden K., Phalen D. N. 2004. Encephalitozoon infection in birds. Semin. Avian Exotic Pet Med. 13: 94–99 [Google Scholar]
- 21. Xiao L., et al. 2001. Genotyping Encephalitozoon hellem isolates by analysis of the polar tube protein gene. J. Clin. Microbiol. 39: 2191–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]