Abstract
Gamma-butyrolactones (GBLs) produced by several Streptomyces species have been shown to serve as quorum-sensing signaling molecules for activating antibiotic production. The GBL system of Streptomyces chattanoogensis L10, a producer of antifungal agent natamycin, consists of three genes: scgA, scgX, and scgR. Both scgA and scgX contribute to GBL production, while scgR encodes a GBL receptor. ΔscgA and ΔscgX mutants of S. chattanoogensis behaved identically: they had a growth defect in submerged cultures and delayed or abolished the morphological differentiation and secondary metabolites production on solid medium. ScgR could bind to the promoter region of scgA and repress its transcription. Moreover, scgA seems also to be controlled by a GBL-mediated negative-feedback system. Hence, it is apparent that GBL biosynthesis is tightly controlled to ensure the correct timing for metabolic switch. An additional direct ScgR-target gene gbdA was identified by genomic SELEX and transcriptional analysis. Comparative proteomic analysis between L10 and its ΔscgA mutant revealed that the GBL system affects the expression of more than 50 proteins, including enzymes involved in carbon uptake system, primary metabolism, and stress response, we thus conclude that scgR-scgA-scgX constitute a novel GBL regulatory system involved in nutrient utilization, triggering adaptive responses, and finally dictating the switch from primary to secondary metabolism.
INTRODUCTION
Quorum sensing (QS) is a cell-cell communication process in which bacteria use the production and detection of extracellular chemicals called autoinducers to monitor cell population density and synchronize community behavior through regulation of their gene expression in response to changes in cell density. Acyl homoserine lactones (AHLs) are a major class of autoinducers used by Gram-negative proteobacteria for intraspecies quorum sensing (23) and has been studied intensively over the past decade. Streptomyces, probably as well as its related genera, use γ-butyrolactones (2,3-di-substituted-γ-butyrolactones) as autoinducers, the chemical structure of which is similar to that of AHLs except for the carbon side chain (31). In addition, other signal molecules, such as PI factor [2,3-diamino-2,3-bis(hydroxymethyl)-1,4-butanediol] and AHFCAs (2-alkyl-4-hydroxymethylfuran-3-carboxylic acids), have been described to play a similar role to that of GBLs in Streptomyces (6, 26).
Streptomycetes are Gram-positive soil bacteria that undergo a developmental program that leads to sporulating aerial hyphae. They are also characterized by a complex secondary metabolism, which makes them the largest antibiotic-producing genus, producing over two-thirds of the clinically used antibiotics of natural origin. The γ-butyrolactone (GBL) system of Streptomyces, typically consisting of a GBL synthase and a cognate receptor, is drawing more attention because of its close association with production of several antibiotics (31). Currently, there are four such systems with a known GBL and receptor, including the A-factor-ArpA system of Streptomyces griseus (12), the SCB1-ScbR system of S. coelicolor (32), the IM-2-FarA system of S. lavendulae (17), and the VB-BarA system of S. virginiae (19). Although they share high sequence similarity, the effects of GBL systems on secondary metabolism are different from species to species. Accumulated evidence revealed that the most extensively studied A-factor-ArpA system is atypical, in both signaling pathway and the location of GBL synthase and receptor genes. It can control both morphological differentiation and secondary metabolism, while most of other GBL systems can only regulate the production of secondary metabolite(s). All of the currently identified afsA-family genes (GBL synthases), apart from afsA itself, are located in the vicinity of or within antibiotic biosynthetic gene clusters and either adjacent or close to their cognate GBL receptor gene.
In contrast to the recent intensive studies of the GBL receptor controlling antibiotic production, little is known about the biosynthesis of the GBL itself and how the biosynthesis of GBL are regulated. Shikura et al. (30) identified a stereospecific reductase BarS1 participated in the later reduction step of virginiae butanolide (VB) biosynthesis, converting 1′-keto-type VB to VB. Recently, the biosynthetic pathway of A-factor was elucidated (14). By biochemical analysis, AfsA was proven to catalyze the condensation of dihydroxyacetone phosphate (DHAP) and a keto acid derivative, which is then followed by modifications with phosphatases and reductases, resulting in A factor formation. Very recently, ScbA, FarX, and BarX were also suggested to be involved in the biosynthesis of the corresponding GBLs, based on the observation that their mutants did not produce GBLs (13, 17, 19). However, the precise biochemical function and reaction mechanism are still obscure, and debates remain on the function of AfsA-family proteins, since some of them (e.g., ScbA and BarX) seem to have functions beyond those of an enzyme (15, 32).
Natamycin (NTM) is a polyene macrolide antifungal agent produced by several Streptomyces strains. As a high-efficiency natural food preservative, NTM has been widely used for the prevention of mold contamination of a variety of foods. Moreover, it is also used to treat fungal infections, such as fungal keratitis. The biosynthetic gene clusters of NTM have been cloned in S. natalensis and S. chattanoogensis (2, 9). Here, we reported the characterization of a GBL system of S. chattanoogensis, which consists of two GBL synthases ScgA and ScgX, a GBL receptor ScgR, and a yet-to-be-identified GBL molecule CHB. Our analysis shows that the GBL regulatory system of S. chattanoogensis (CHB system) is not restricted to secondary metabolism and developmental program but also participates in nutrient utilization and stress response. As far as we know, this is the first report showing the involvement of such signal molecule in the control of nutrient utilization, which will provide new insights into Streptomyces biology.
MATERIALS AND METHODS
Bacterial strains, plasmids, growth, and culture conditions.
Bacterial strains and plasmids used in the present study are listed in Table S1 in the supplemental material. S. chattanoogensis strains were normally maintained on YMG agar for spore preparation (9). The media (MSF, SMMS, and YEME) used for examination of morphological development and antibiotic production were prepared according to standard procedures (16). DNA manipulations in Escherichia coli and Streptomyces spp. were carried out according to standard procedures (16, 29). E. coli-S. chattanoogensis conjugation and flask fermentation of S. chattanoogensis strains were performed as described previously (9).
Cloning of GBL system genes in S. chattanoogensis.
All oligonucleotide primers used in the present study are presented in Table S2 in the supplemental material. The consensus degenerate primer pair HTH-F/HTH-R was selected based on the alignment of several previously characterized GBL receptors and used to amplify part of the GBL homologue in S. chattanoogensis. These primers were also used to screen a cosmid library of S. chattanoogensis L10 by PCR. PCR product used as a probe for Southern hybridization with restriction-endonuclease-digested cosmids was prepared by PCR in the presence of biotin-11-dUTP (Fermentas). The DNA fragments giving signals were recovered and cloned into pTA2 (Toyobo) for sequencing.
Mutagenesis and complementation of scgRAX genes.
(i) In-frame deletions of scgR and scgA were constructed by using a modified PCR targeting system (11), which was performed as follows. A 5.5-kb XbaI/HindIII-digested fragment from pMRD80 was cloned into pMRD400, resulting in pMRD401, which was then introduced into E. coli BW25113/pIJ790. The primer pair scgR-KO-F/scgR-KO-R was used to amplify the aac(3)IV cassette from pHY773, and the replacement of the part of scgR coding region (amino acids 30 to 220, containing the DNA-binding domain) by the amplified cassette was mediated by λ red recombination system. The targeted plasmid was then transformed into E. coli DH5α/BT340 in order to excise the aac(3)IV cassette as described previously (11). The resulting plasmid was designated pMRD105. After confirmation with restriction and PCR analysis, pMRD105 was introduced into E. coli ET12567/pUZ8002 for E. coli-Streptomyces conjugation. Exconjugants obtained after selection for thiostrepton were inoculated onto YMG plates for two rounds of nonselective growth before selection by replica plating for thiostrepton-sensitive colonies. Deletion of scgR was confirmed by PCR analysis. In-frame deletion of scgA was performed similarly to that of scgR, resulting in a mutant scgA allele, in which part of the scgA coding region (amino acids 40 to 277, containing the active residue) was deleted.
(ii) Cosmid SCH1F11 was used to construct the scgX-disrupted mutant by PCR-targeting system. After replacement of the whole open reading frame (ORF) with the aac(3)IV cassette, pMRD258 (targeted SCH1F11) was introduced into E. coli ET12567/pUZ8002, and this was followed by conjugation with S. chattanoogensis. Exconjugants were obtained after selection for apramycin. Replica plating was then used to search for the thiostrepton-sensitive and apramycin-resistant colonies. scgX-disrupted mutants were confirmed by PCR analysis.
For complementation, the primer pairs scgR-cpt-F/scgR-cpt-R, scgA-cpt-F/scgA-cpt-R, and scgX-cpt-F/scgX-cpt-R were used to amplify the DNA fragments containing promoter and coding regions of scgR, scgA, and scgX, respectively. The amplified PCR products were digested and ligated into the integrative plasmids pSET152 or pIJ8600 (16), resulting in pMRD157, pMRD184, and pMRD306. After transformation into E. coli ET12567/pUZ8002, these recombinant plasmids were introduced into the corresponding mutants for complementation.
RNA methods.
Isolation of total RNA and 5′RACE (5′ rapid amplification of cDNA ends) was performed as previously described (9). Two-step reverse transcription-PCR (RT-PCR) was performed, and cDNA was made from 2 μg of total RNA using a PrimeScript First Strand cDNA synthesis kit (TaKaRa) according to the manufacturer's guidelines. cDNA was amplified with the PCR conditions consisting of one cycle of denaturation at 94°C for 2 min, followed by different cycles of 94°C for 30 s, 55°C for 30 c, and 72°C for 60 s, with one extension cycle at 72°C for 5 min. Amplification and detection by quantitative RT-PCR were performed on Roche LightCycler 480 (Roche), and synthesized cDNA was amplified with the SYBR Premix Ex Taq (Takara) according to the manufacturer's instructions. The transcription of hrdB, which encodes the principal sigma factor of RNA polymerase, was used as the internal control. All values were normalized to the corresponding transcriptional level of hrdB. Reactions were performed in triplicate.
Introduction of ScgA mutants into Streptomyces.
pMRD402 was used to construct ScgA mutants. Site-directed mutagenesis was performed with a QuikChange site-directed mutagenesis kit (Stratagene) using the primers listed in Table S2 in the supplemental material. After confirmation by DNA sequencing, plasmids containing scgA mutants (including promoter and coding sequences) were transferred to S. chattanoogensis ZJUD7 by intergeneric conjugation. Complementary S. chattanoogensis mutants were verified by PCR with the primer pair scgA-cpt-F/scgA-cpt-R, which gave an amplified product corresponding to the inserted region. The amplified products were then cloned and sequenced again for confirmation.
Cloning, expression, and purification of recombinant ScgR.
The scgR coding region was amplified from SCH1F11 by using KOD DNA polymerase and the primer pair scgR-EX-F/scgR-EX-R (see Table S2 in the supplemental material). After digestion with NdeI and XhoI, the PCR products were ligated into the corresponding sites of pET22b, resulting in pMRD23. After confirmation by DNA sequencing, pMRD23 was introduced into E. coli Rosetta gami for recombinant protein expression. The culture was grown at 37°C to an optical density at 600 nm of 0.4 to 0.6. IPTG (isopropyl-β-d-thiogalactopyranoside) was then added to a final concentration of 0.25 mM. After 12 h of further incubation at 30°C, the cells were harvested by centrifugation and disrupted by sonication on ice, and the supernatant was recovered by centrifugation (13,000 × g for 20 min). His-tagged ScgR was separated using Ni-NTA His Bind Resin (Novagen) according to the manual of the supplier.
EMSAs.
For probe preparation, DNA fragments containing target regions were amplified by PCR. Afterward, the PCR products were cloned into pUC19 or pUC-mT (Sangon). Biotin-labeled probes for EMSA were obtained by PCR with 5′-biotin-labeled M13 universal primers, followed by gel recovery. About 5 ng of the probes were incubated with appropriate amounts of purified His-tagged ScgR at 30°C for 25 min in a buffer containing 2 μg of sheared salmon sperm DNA, 20 mM Tris-base (pH 7.5), 0.01% bovine serum albumin, and 5% glycerol in a total volume of 20 μl. Protein-bound DNA and free DNA were resolved on 6% acrylamide gels in 0.5× Tris-borate-EDTA buffer. Electrophoretic mobility shift assay (EMSA) gels were then electroblotted onto Biodyne B membrane (Pall). Labeled DNA was detected by using a chemiluminescent biotin-labeled nucleic acid detection kit (Beyotime, China) as described by the manufacturer.
Genomic SELEX (systematic evolution of ligands by exponential enrichment) search for ScgR target promoters.
For genomic library construction, genomic DNA digested with MboI was ligated to BamHI-cleaved and dephosphorylated pUC19 vector. The ligation products were then electrotransformed into E. coli DH5α. This primary library was estimated to contain 4 × 105 clones carrying recombinant vectors, which was sufficient for covering the genome of S. chattanoogensis L10. Plasmids isolated from primary library were used as a template for generating a collection of DNA fragments by PCR using M13 universal primers and KOD plus DNA polymerase (Toyobo); the PCR was performed according to the manufacturer's instructions. The resulting products were gel recovered and stored at −20°C for further use.
For binding experiments, 5-pmol DNA fragments were mixed with 10 pmol of His-tagged ScgR under the same conditions as described above for EMSA. After inoculation for 30 min, the mixture was applied onto a Ni column. Unbound DNA was washed with binding buffer containing 10 mM imidazole. DNA-ScgR complexes were then eluted with an elution buffer containing 400 mM imidazole. DNA fragments were recovered by phenol extraction and ethanol precipitation. This inoculation step was repeated several times when necessary. For sequencing, DNA fragments were cloned into pTA2 vector (Toyobo).
Extraction and treatments of ScgR ligand.
ScgR ligand was extracted either from solid or liquid cultures. For solid culture extraction, Five petri plates containing YMG agar medium were inoculated with the spores of S. chattanoogensis strain. After 3 days of growth at 26°C, the agar were cut into small pieces, combined, and extracted with ethyl acetate. For liquid culture extraction, 100 ml of 24- h-grown S. chattanoogensis cultures (grown in YEME medium) were extracted twice with 1 volume of ethyl acetate. The acid, alkali, heat, and proteinase K treatments were performed before the ethyl acetate extraction, as described elsewhere (33). All of the extract was dried under vacuum, and the residues were then redissolved in 50 μl of methanol and stored at −20°C for further use. In the EMSA, 1 μl of extract was added to the binding reaction mixture (20 μl), and 1 μl of pure methanol was used in a control experiment.
Quantification of NTM and extracellular pyruvate.
NTM production on the agar medium was detected by bioassays, which were conducted as previously described (9). For NTM determination in liquid culture, a 0.5-ml aliquot of the culture was extracted with 5 ml of methanol and further diluted with methanol if necessary. Quantification of NTM production was performed with an Agilent 1200 high-pressure liquid chromatography (HPLC) apparatus with a diode array UV detector set at 304 nm, fitted with a HC-C18 column (5 μm, 4.6 by 250 mm). Elution was performed with a mobile-phase mixture consisting of a linear gradient of water-methanol-acetonitrile (60:25:15 to 50:25:25, 0 to 5 min; 50:25:25, 5 to 20 min). The retention time for NTM was 12.7 min. Pure NTM (Sigma) was used as a control. Quantification of pyruvate was performed as described elsewhere (3).
Proteomic analysis.
For the preparation of cell extracts, S. chattanoogensis L10 and ZJUD7 were grown for 20 h in YEME medium. The mycelium was then collected and washed by centrifugation. Afterward, the mycelium was grounded in liquid nitrogen and suspended in buffer A consisting of 9 M urea, 4% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 1% dithiothreitol (DTT), and 1% IPG buffer (GE Healthcare). Proteins were precipitated by adding 10 volumes of acetone containing 0.07% DTT. After incubation at −20°C for 1 h, the precipitate was collected by centrifugation and allowed to dry under a vacuum. The final pellet was resuspended in an appropriate volume of buffer A, and samples were incubated for 1 h at room temperature. Insoluble material was removed by centrifugation. Protein concentration was determined by a Bradford assay (4). The protein samples were stored at −80°C until used.
For two-dimensional PAGE analysis, 1.0 mg of protein was diluted to 400 μl with buffer A containing a trace of bromophenol blue. Samples were applied to 24-cm IPG strips at pH 4 to 7 (GE Healthcare) and focused using the IPGphor isoelectric focusing systems (GE Healthcare). Second-dimension separation SDS-PAGE was performed using 12% acrylamide gels. Samples were run in triplicate. After staining with colloidal Coomassie G-250, the gels were scanned with an ImageScanner (GE Healthcare) and analyzed by using ImageMaster 5.0 software (GE Healthcare). Protein spots of interest were excised from gels, digested with trypsin, and identified by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry.
Nucleotide sequence accession number.
The nucleotide sequence of GBL cluster has been deposited in the NCBI database under accession number HQ241279.
RESULTS
Identification of a putative GBL cluster in S. chattanoogensis.
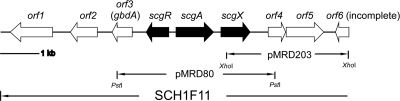
Since GBL synthases and receptors are usually located adjacently, degenerate PCR primers were designed according to the conserved regions of previously characterized GBL receptors. A 0.1-kb fragment amplified from the genomic DNA of S. chattanoogensis L10 was cloned and sequenced. Analysis of the sequence revealed that it displays a high degree of similarity to N-terminal region of GBL receptors, as expected. The same primer pair was then used to screen a cosmid library of S. chattanoogensis L10 (8, 14). Three cosmids (SCH1F11, SCH1C4, and SCH3F9) giving a positive signal were isolated. After a series of subcloning and walking experiments, the sequence of an 8.8-kb fragment in cosmid SCH1F11 was obtained (Fig. 1). Analysis of the this fragment identified eight complete ORFs. scgR was predicted to encode a 220-amino-acid protein, which showed sequence similarity to several GBL receptor homologs. Protein sequence alignments using ScgR and these GBL receptor homologs revealed the presence of a highly conserved N-terminal helix-tum-helix (HTH) DNA-binding domain, as well as a conserved tryptophan residue at the 121 position of ScgR, which was suggested to constitute a hydrophobic pocket for GBL binding. The translation start of the divergently transcribed scgA is separated from the translation start of scgR by 178 nucleotides. ScgA, consisting of 327 amino acids, shows homology to afsA-family proteins, such as AfsA (40% identity), which have recently been proven to be a key enzyme of A factor biosynthesis in S. griseus (14). The deduced amino acid sequence of the downstream scgX (separated by 153 nucleotides [nt]) showed homology with BarS1 (30% identity), which is responsible for the reduction of the C-6 position of VBs in S. virginiae. ScgX was predicted to belong to the short-chain dehydrogenase/reductases family as BarS1. Most members of this family are known to be NAD(P)-dependent oxidoreductases. A typical βαβ fold as an NAD(P)H-binding motif, GSSRGIG, can be found at the N-terminal region (13 to 19 amino acids) of ScgX, and the predicted active sites S147, Y160, and K164 are also present. Downstream of scgX, separated by 429 nt, are orf4 and orf5, which showed overlapping coding sequences. The deduced product of orf4 belongs to MarR family transcriptional regulator, while the predicted orf5 product shares homology with cytochrome P450 monooxygenase. orf1 and orf2 products were identified to be putative NADH ubiquinone oxidoreductase and serine/threonine protein kinase, respectively. The Orf3 product will be discussed later (see below). Overall, these data suggested that scgR, scgA, and scgX are likely to form a GBL-receptor system in S. chattanoogensis L10.
Fig. 1.
Gene organization in a 8.8-kb region containing scgR, scgA and scgX in S. chattanoogensis. The ORFs were deduced using Frameplot program. The horizontal lines below indicate the coverage of individual cosmid or plasmid clones.
Characterization of scgA, scgX, and scgR mutants.
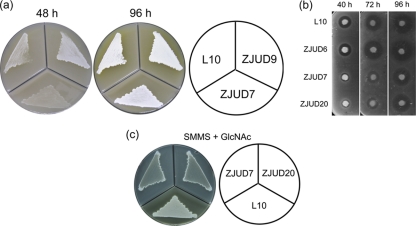
To assess the role of scgA, scgX, and scgR genes in S. chattanoogensis L10, gene deletion of each gene was carried out, resulting in strains ZJUD7 (ΔscgA), ZJUD20 (ΔscgX), and ZJUD6 (ΔscgR). These mutants were verified by PCR (see Fig. S1 in the supplemental material). To identify differences between the mutants and the parental strain, all strains were grown on YMG and MSF agar media. ZJUD7 and ZJUD20 were found to have identical phenotypes. Compared to L10, the morphological differentiation and yellow pigment (YP) production of those two mutants were delayed to various degrees depending on the medium tested. Since ZJUD7 and ZJUD20 behaved identically, only ZJUD7 grown on MSF agar plates was displayed in Fig. 2 a. To determine NTM production on MSF agar medium, bioassays were performed (Fig. 2b). L10 commenced NTM production after 40 h of growth, while its production in ZJUD7 exhibited a 24-h delay. Furthermore, the YP production of ZJUD7 delayed about 2 days. Unexpectedly, ZJUD6 (ΔscgR) showed growth and morphological characteristics identical to those of the parental strain when grown on these media (data not shown). To verify that the phenotypes were solely due to the inactivation of scgA and scgX, integrative vectors harboring scgA and scgX and their promoters were conjugated into the corresponding mutants. Introduction of recombinant plasmids fully restored phenotype to the parental strain level, underlining that the phenotype defects of mutant ZJUD7/ZJUD20 were indeed due to the inactivation of genes scgA and scgX.
Fig. 2.
Effect of GBL system genes on growth, morphological differentiation and natamycin production. (a) Mycelium formation and yellow pigment production on MSF medium. Only ZJUD7 is displayed, since ZJUD7 and ZJUD20 behaved identically. Aerial mycelium was readily detected for L10 after 48 h of growth, while the aerial mycelium was not observed for ZJUD7 until 72 h of growth. (b) Natamycin bioassay of different strains grown on MSF medium. (c) Comparison of morphological differentiation and yellow pigment production between L10, ZJUD7, and ZJUD20 grown on SMMS agar with different carbon sources.
ZJUD7 and ZJUD20 were also evaluated on SMMS agar with different carbon sources (0.4% [wt/vol]), including glucose, mannitol, and N-acetylglucosamine. When grown on the SMMS + glucose or SMMS + mannitol, ZJUD7 and ZJUD20 showed delayed morphological differentiation and YP production (data not shown). Interestingly, both development and YP production were totally blocked in ZJUD7 and ZJUD20 when grown on the GlcNAc-containing SMMS agar, on which L10 grew normally (Fig. 2c), suggesting that both ScgA and ScgX are also involved in nutrient utilization.
The characteristics of these mutants were also explored in more detail in YEME liquid medium (Fig. 3). Monitoring growth curves over time showed that both ΔscgA and ΔscgX mutants had a similar growth rate as L10 over the initial 20 h of incubation; however, they reached stationary phase earlier than the parental strain (Fig. 3a). Yellow pigment (YP) production was readily observed in L10 after 18 h of growth, while the mutants commenced YP production after 30 h (Fig. 3b). Compared to L10, NTM production of both mutants was reduced at the first 12 h after the initiation of NTM biosynthesis (Fig. 3c). However, an increased rate of NTM production was observed after 24 h, resulting in a similar yield of NTM as L10 at 36 h. Taken together, these results indicated that both ScgA and ScgX are involved in the nutrient utilization, growth control, morphological differentiation, and secondary metabolite production. The observation that ΔscgA and ΔscgX mutants showed identical phenotypes suggests that the growth deficiencies might be associated with an inactive GBL production.
Fig. 3.
(a) Growth curves of different strains grown in liquid YEME medium. (b) Yellow pigment production of different strains grown in liquid YEME medium at 24 and 36 h. (c) Natamycin production of different strains grown in liquid YEME medium.
Binding of ScgR to the promoter of scgA.
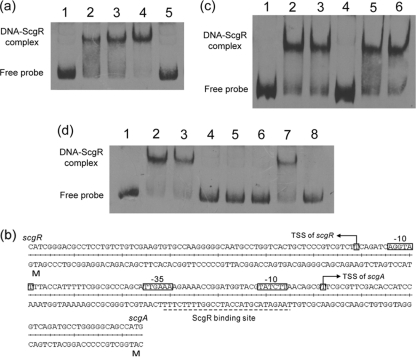
GBL receptor homologs share a highly conserved N-terminal HTH DNA-binding domain, which allow them to recognize similar DNA sequences (TNANAWACNNACYNNNCGGTTTKTTT) (10). Many GBL receptors, such as ScbR and FarA, were reported to bind to their own promoters and are thus subject to autorepression (18, 32). A 26-bp sequence (TAAGATACGTACCATCCGGTTTTCTT) with homology to the conserved binding motif of GBL receptor was indeed found in the intergenic region of scgA and scgR. To determine whether ScgR could bind to this region and whether this predicted cis-acting element is the only binding site in the intergenic region, purified rScgRs were used in EMSAs with 5′ biotin-labeled PCR products covering different regions. Strong shift signals were only observed when this putative binding motif was included (see Fig. S2 in the supplemental material). Competition with unlabeled probe DNA showed a release of the shift, verifying the specificity of the interaction (Fig. 4 a). The binding motif was designated GARE1 (gamma-butyrolactone autoregulator responsive element 1).
Fig. 4.
(a) Binding of rScgR to the scgA promoter region. Lane 1, free labeled probe; lanes 2 to 4, labeled probes with 25, 50, and 100 ng of rScgR; lane 5, probe with 100 ng of rScgR and 500× unlabeled probe. (b) Sequence of the scgRA promoter region. The transcriptional start sites of scgA and scgR were determined by 5′RACE. The binding site of ScgR covers the −10 and −35 regions of scgA promoter. (d) ScgR ligand properties. Lane 1, free probe; lane 2, probe + rScgR; lane 3, probe + rScgR + methanol; lane 4, probe + rScgR + extract from L10; lanes 5 to 7, probe + rScgR + extract exposed to various treatments (lane 5, heat; lane 6, acid; lane 7, alkali; lane 8, proteinase K). (c) Effect of ethyl acetate extract from different strains on the binding activity of rScgR. Lane 1, free probe; lane 2, probe + rScgR; lane 3, probe + rScgR + methanol; lane 4, probe + rScgR + extract from L10; lane 5, probe + rScgR + extract from ZJUD7; lane 6, probe + rScgR + extract from ZJUD20.
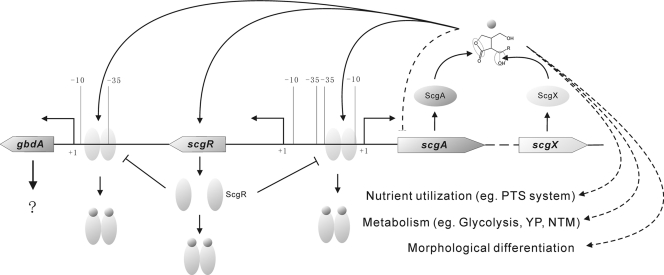
To assess whether ScgR is subject to autorepression like its counterparts, we first determined the transcriptional start sites (TSS) of scgR and scgA by 5′RACE. As shown in Fig. 4b, the TSS of scgR is located 65 nt upstream of the translation start codon, and the TSS of scgA was identified 39 nt upstream of the translation start codon. It is surprising to find that the ScgR-binding site GARE1 overlapped with the core promoter region of scgA rather than that of scgR. Combined with the in vivo analysis (see below), the results demonstrated that it is scgA rather than scgR that is transcriptionally repressed by ScgR.
Both ScgA and ScgX are involved in directing the biosynthesis of ScgR ligand.
Given that ScgR belongs to the GBL receptor family, the binding of ScgR to its target DNA might be relieved by γ-butyrolactone-like molecules. To establish whether similar signaling molecules were produced by S. chattanoogensis L10, GBL-containing extract was prepared as described in Materials and Methods. Gel retardation assays showed that formation of the shifted probe complex was significantly reduced in the presence of the GBL-containing fraction (Fig. 4c), indicating that the extract from L10 can prevent the binding of ScgR to its target DNA (containing GARE1). Since ScgA and ScgX exhibit homology to the enzymes involved in the biosynthesis of GBL molecules, the ethyl acetate extracts of ΔscgA and ΔscgX mutants were also tested for their ability to disrupt the ScgR-DNA interaction. As shown in Fig. 4c, neither of these extracts can prevent the formation of the shift complex. To further confirm that ScgR ligand is indeed a GBL molecule, similar EMSAs were carried out with ligand extracts exposed to different treatments (see Materials and Methods). The active compound was found to be resistant to heat, acidic, and proteinase K treatments but sensitive to alkali treatment (Fig. 4d). All of these properties are characteristics associated with GBLs (33). Taken together, these data suggested that both ScgA and ScgX are required for the biosynthesis of a cognate GBL of ScgR, which could inhibit its DNA-binding activity. We named the yet-to-be identified GBL CHB.
E90 of ScgA is essential for its enzymatic activity.
As shown above, ScgA belongs to the AfsA-family proteins. Similar to AfsA and ScbA (13, 14), structural modeling of ScgA also revealed an intramolecular dimer-like structure similar to dimeric FabZ/FabA (data not shown), which are important enzymes for fatty acid biosynthesis, catalyzing reactions of dehydration on 10-carbon thiol esters of the acyl carrier protein (20). Compared to dimeric FabZ, FabA homodimer has an additional isomerase activity. In dimeric FabZ, each of the two independent active sites is located in a deep narrow-shaped pocket formed at the dimer interface in which histidine from one subunit and glutamate from the other subunit are the active residues. Previous studies have shown that these residues involved in catalysis are also conserved in the AfsA repeats (Pfam03756) of AfsA homologues (13, 14). E78 and E240 of ScbA have been shown to be essential for SCB1 production, and these two residues are conserved in all AfsA-family proteins characterized to date (see Fig. S3 in the supplemental material). However, in ScgA, the conserved C-terminal glutamate residue (E240) is replaced by an aspartate residue (D253). Interestingly, a similar residue replacement was observed at the corresponding position of FabA, and this residue in FabA was suggested to be responsible for its ability to catalyze the isomerization reaction (20). Furthermore, a conserved arginine residue located in the C-terminal long nonpolar α-helix, which is required for the enzymatic activity of ScbA, was also replaced by a glutamine residue in ScgA (Q256).
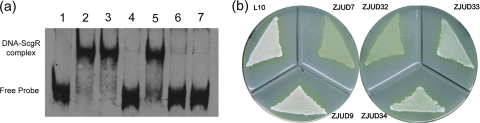
To establish whether these amino acid replacements in the C-terminal AfsA repeats have influence on the activity of ScgA and to confirm the role of ScgA in directing the biosynthesis of CHB, ScgA mutants containing point mutations were generated: ScgA[E90A], ScgA[D253A], and ScgA[Q256A]. These mutants were then introduced into the ZJUD7 (ΔscgA), resulting in strains ZJUD32, ZJUD33, and ZJUD34. The catalytic properties of mutated ScgA were evaluated by phenotype analysis and EMSA. Namely, the ability of ethyl acetate extracts to prevent the formation of DNA-ScgR complex was investigated. As shown in Fig. 5 a, only the ScgA[E90A] mutant ZJUD32 lost the ability to produce CHB, and this result was corroborated by the phenotype analysis (Fig. 5b), showing that only ZJUD32 has a phenotype identical to that of the ΔscgA mutant ZJUD7, whereas both ScgA[D253A] and ScgA[Q256A] retained the ability to produce CHB and successfully complemented strain ZJUD7, suggesting that D253 and Q256 are not involved in catalysis. These results support the idea that ScgA is involved in the catalytic pathway of CHB and that the phenotypes of ΔscgA mutant are associated with inactive production of CHB and also suggest that there might be different catalytic mechanisms among AfsA-family proteins. However, we cannot completely exclude the possibility that ScgA[E90A] is not stable.
Fig. 5.
(a) Effect of ethyl acetate extract from different strains on the binding activity of rScgR (lane 1, free probe; lane 2, probe + rScgR; lane 3, probe + rScgR + methanol; lane 4, probe + rScgR + extract from ZJUD9; lane 5, probe + rScgR + extract from ZJUD32; lane 6, probe + rScgR + extract from ZJUD33; lane 7, probe + rScgR + extract from ZJUD34). (b) Complementation of the scgA deletion mutant ZJUD7 by different mutated scgA (ZJUD9, ZJUD7 complemented with wild-type ScgA; ZJUD32, ZJUD7 complemented with ScgA[E90A]; ZJUD33, ZJUD7 complemented with ScgA[D253A]; ZJUD34, ZJUD7 complemented with ScgA[Q256A]).
Genome-wide selection of additional target promoters under the control of ScgR.
Thus far, signal transduction of almost all QS systems is mediated by the interaction of autoregulators with cognate receptors. Although no observable change was detected in ZJUD6 (ΔscgR), identification of additional target genes regulated by ScgR may provide insight into the CHB signaling cascade. For this, genomic SELEX was used to perform a genome-wide search for target promoters (see Materials and Methods). After six rounds of SELEX, discrete bands were identified, indicating that DNA fragments with high affinity to ScgR were enriched (data not shown). These DNA fragments were then gel-purified and cloned into pTA2 for sequencing. After sequencing of 20 independent clones, 19 of them could be classified into two sets. Group A contained 10 clones that had the same 177-bp MboI-digested region in the cloned fragments, while group B contained 9 clones that had the same 266-bp MboI-digested fragment. One clone was excluded by the EMSA since no shift band was observed. It is interesting to find that the 266-bp fragment contained the GARE1 box covering the promoter region of scgA, while the 177-bp fragment was located just downstream of scgR. To confirm ScgR binds to the 177-bp fragment identified by genomic SELEX, this fragment was biotin labeled and used in EMSAs with purified His-tagged ScgR, the results showed a clear gel shift (Fig. 6 a). Using the 177-bp sequence to search for the binding site of ScgR, no GARE1-like motif was found. However, a 26-bp imperfect inverted repeat sequence was observed in this region (Fig. 6b). To assess whether the 26-bp sequence represents the binding site of ScgR, purified rScgR was used in EMSAs such that the probes covered different fragments within this region (see Fig. S4 in the supplemental material). A strong shift signal was observed with probe containing this inverted repeat, indicating that it is indeed a binding site for ScgR, We thus designated the sequence as GARE2.
Fig. 6.
(a) Binding of rScgR to the upstream region of gbdA. Lane 1, free labeled probe; lanes 2 and 3, labeled probes with 25 and 50 ng of rScgR; lane 4, labeled probe with 50 ng of rScgR and 100× unlabeled probe. (b) Binding motifs of ScgR. GARE1 is located upstream of scgA; GARE2 is located upstream of gbdA. (c) Transcriptional comparison of scgA between S. chattanoogensis L10 and the mutant strains grown in YEME media by quantitative RT-PCR.
Since almost all of the isolated DNA fragments after six cycles could be bound by ScgR, to assess the possible presence of DNA fragments with lower affinity to ScgR, the DNA fragments from the fifth cycle were also cloned, and 20 recombinant clones were sequenced. Two clones were found to contain the DNA fragment carrying GARE1 and GARE2, respectively. Apart from these two fragments, no gel shift was observed for the other 18 SELEX-fragments in EMSA (data not shown), indicating that these DNA fragments represent nonspecific backgrounds in the genomic SELEX.
Both scgA and gbdA are transcriptionally repressed by ScgR, and scgA is also controlled by a CHB-mediated negative-feedback system.
The location of GARE2 and the direction of transcription of two neighboring genes suggested that the gene located downstream of scgR could be a potential ScgR-dependent gene, and we named this gene gbdA (gamma-butyrolactone dependent gene A). The TSS of gbdA was determined to be the T located 145 nt upstream of the translation start codon by 5′RACE. GARE2 is located between positions −13 and −38 relative to its TSS. To establish whether ScgR represses the transcription of gbdA and scgA in vivo and to analyze the transcriptional pattern of GBL system genes, semiquantitative RT-PCR and quantitative RT-PCR were performed to analyze the transcription of these genes in strains L10, ZJUD6, ZJUD7, and ZJUD20. Since these genes have identical transcriptional patterns in ZJUD7 and ZJUD20, only that of ZJUD7 was shown (see Fig. S5 in the supplemental material). Both scgR and scgX transcribed constitutively and showed no significant difference between mutant and parental strains (see Fig. S5 in the supplemental material), suggesting that scgR and scgX were not regulated by the GBL system. In contrast, scgA and gbdA showed growth-dependent expression profiles in L10. Both genes showed constitutive and dramatically increased expression in ZJUD6, supporting the conclusion that they were transcriptionally repressed by ScgR in vivo (Fig. 6c and see Fig. S5 in the supplemental material). No transcript of gbdA was detected in ZJUD7 and ZJUD20 at 24 h (see Fig. S5 in the supplemental material), indicating that ScgR might still bind to the promoter region of gbdA due to the absence of its ligand, another possibility is that scgA affects the gbdA expression independent of ScgR. Unexpectedly, unlike for gbdA, the transcription of scgA was not decreased but significantly increased in both ZJUD7 and ZJUD20 compared to the parental strain (Fig. 6c), suggesting that scgA might also be subject to a CHB-mediated negative-feedback system. A similar phenomenon was not observed when the transcriptional pattern of gbdA was analyzed, suggesting that CHB receptor ScgR may not participate in this CHB-mediated negative-feedback system. These results demonstrate that the expression of scgA was strictly controlled at the transcriptional level.
gbdA encodes a 182-amino-acid protein for which no homolog could be found in current protein databases. The sequence was predicted to contain a C-terminal leuA dimerization domain (Pfam 08502), identical to that of α-isopropylmalate synthase, which catalyzes the first committed step in the leucine biosynthetic pathway. To determine whether this gene mediates the CHB signaling cascade in S. chattanoogensis and accounts for the phenotype observed in CHB-deletion mutants, we constructed a gbdA-disrupted mutant ZJUD35, and a phenotypic analysis similar to those described for the above scg mutants were performed. No observable change was found (data not shown), suggesting that CHB systems may use other mechanisms to mediate its signaling pathways.
Pathway-specific regulators of NTM biosynthesis were not under the control of CHB regulatory system.
Production of NTM is absolutely dependent on pathway-specific positive regulators, In S. chattanoogensis, these activators are encoded by scnRI and scnRII (8). To determine whether the effect of CHB deletion on NTM production is mediated by these two regulator genes, RNA was isolated from liquid grown cultures of L10 and ZJUD7 (ΔscgA) at several time points corresponding to different growth phases. Measuring the transcriptional levels of scnRI and scnRII by quantitative RT-PCR showed no significant change between L10 and ZJUD7 over the time course (data not shown). Furthermore, semiquantitative RT-PCR revealed equal expression of all NTM cluster genes at 20 h (data not shown). Overall, these results suggest that the reduced production of NTM, at least in YEME medium, was not caused by a lower transcript abundance of NTM cluster in ZJUD7.
Proteomic analysis of the effects of CHB deletion.
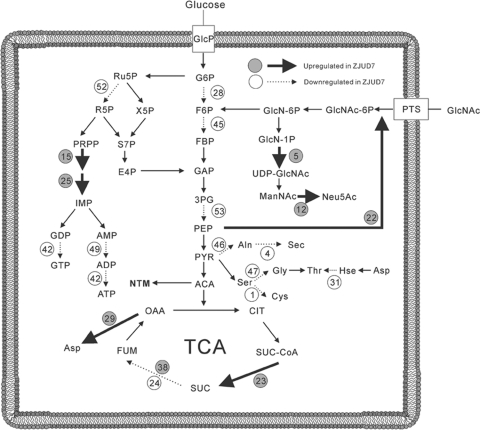
To examine the basis for growth deficiencies of CHB deletion mutants and to search for CHB regulon genes, comparative proteomic analysis between strains L10 and ZJUD7 (ΔscgA) was performed using two-dimensional gel electrophoresis in conjunction with MALDI-TOF mass spectrometry. Cells were harvested from culture grown in YEME liquid medium for 20 h, corresponding to the time just before ZJUD7 entered its transition phase. At that time point, S. chattanoogensis L10 had already produced yellow pigment and NTM, whereas strain ZJUD7 only produced a small amount of NTM. A total of 53 proteins that were over- or under-regulated in the mutant by >2-fold were identified (Table 1).
Table 1.
Differentially expressed spots of scgA mutant (ZJUD7) in two-dimensional gels
| Protein function | Spot no. | Characteristic(s) | Avg fold change(s) |
|---|---|---|---|
| Downregulated | |||
| Glycosis and TCA | 45 | 6-Phosphofructokinase | NDa |
| 53 | Phosphoglyceromutase | 0.014 | |
| 24 | Succinate dehydrogenase flavoprotein subunit, SDHA | 0.35 | |
| 28 | Glucose-6-phosphate isomerase | 0.43 | |
| Pantose phosphate pathway | 52 | Ribose-5-phosphate isomerase B | ND |
| Fatty acid biosynthesis and metabolism | 21 | Beta-ketoacyl-acyl carrier protein synthase III, KAS III | 0.47 |
| 8 | Acyl-coenzyme A dehydrogenase | 0.36 | |
| 27 | Short-chain dehydrogenase/reductase SDR | 0.42 | |
| Pyrimidine and energy metabolism | 42 | Nucleoside diphosphate kinase, Ndk | ND |
| 49 | Adenylate kinase | 0.019 | |
| 34 | F0F1 ATP synthase subunit delta | 0.46 | |
| Amino acid biosynthesis and metabolism | 46 | Putative l-alanine dehydrogenase | 0.019 |
| 4 | Putative selenocysteine lyase | 0.12 | |
| 31 | Threonine synthase | 0.40 | |
| 1, 3, 40 | Cystathionine beta-synthase | 0.23, 0.27, 0.39 | |
| 36 | Amidinotransferase | 0.16 | |
| 35 | Histidyl-tRNA synthetase | 0.37 | |
| 47 | Serine hydroxymethyltransferase | 0.035 | |
| Protein folding and degradation | 51 | Peptidyl-prolyl cis-trans isomerase | 0.017 |
| 19 | Molecular chaperone DnaK | 0.37 | |
| 16 | Chaperonin GroEL | 0.50 | |
| 48 | ATP-dependent Clp protease proteolytic subunit 1, ClpP | 0.20 | |
| Other | 50 | FeS assembly ATPase, SufC | ND |
| 44 | Elongation factor Tu, EF-Tu | 0.025 | |
| 2 | Subtilisin-like protease | 0.40 | |
| 9 | NADPH-dependent F420 reductase | 0.31 | |
| 10 | Thioredoxin,TrxA | 0.30 | |
| 13 | Siderophore-interacting protein | 0.42 | |
| 14 | GTP-binding protein TypA/BipA | 0.47 | |
| 18 | Putative O-methyltransferase | 0.46 | |
| 26 | Conserved hypothetical protein 1 | 0.36 | |
| 33 | Single-stranded DNA-binding protein | 0.28 | |
| 20 | Monooxygenase | 0.46 | |
| 39 | Putative cyclic nucleotide-binding domain-containing protein | 0.44 | |
| 32 | Aldo/keto reductase | 0.38 | |
| 41 | 1L-myo-inositol-1-phosphate synthase | 0.50 | |
| 30 | Cytochrome P450 monooxygenase | 0.22 | |
| Upregulated | |||
| Aminosugar metabolism | 5 | Bifunctional N-acetylglucosamine-1-phosphate uridyltransferase/glucosamine-1-phosphate acetyltransferase, GlmU | 4.74 |
| 12 | N-Acylneuraminate-9-phosphate synthase | 3.55 | |
| Glycosis and TCA | 23 | Succinyl-coenzyme A synthetase subunit alpha, SucD | 5.54 |
| 38 | Succinate dehydrogenase/fumarate reductase iron-sulfur subunit, SDHB | 4.34 | |
| Amino acid biosynthesis and metabolism | 29 | Aspartate aminotransferase | 3.07 |
| 15 | Phosphoribosyl isomerase A, HisA | 3.30 | |
| Purine metabolism | 25 | Bifunctional phosphoribosylaminoimidazolecarboxamide formyltransferase/IMP cyclohydrolase, PurH | 3.46 |
| Other | 43 | Elongation factor Ts, EF-Ts | 13.84 |
| 22 | Phosphoenolpyruvate-protein phosphotransferase, PstI | 6.088 | |
| 37 | hypothetical protein | 4.21 |
ND, not detected.
Based on their postulated function, these proteins could be assigned to different groups (Table. 1 and Fig. 7). Many proteins involved in the following processes were downregulated in ZJUD7: glycolysis (spots 28, 45, and 53), tricarboxylic acid (TCA) cycle (spot 24), fatty acid synthesis and metabolism (spots 8, 21, and 27), amino acid biosynthesis and metabolism (spots 1, 3, 4, 31, 36, 40, 46, and 47), protein synthesis (spot 50), pentose phosphate pathway (spot 52), nucleoside metabolism (spots 42, and 49), and energy metabolism (spot 34). These pathways are essential for active cell growth and primary metabolism. Among these downregulated proteins, 6-phosphofructokinase, phosphoglyceromutase, elongation factor Tu, ribose-5-phosphate isomerase B, l-alanine dehydrogenase, and serine hydroxymethyltransferase were hardly detectable in the ΔscgA mutant, which may hamper further growth. Coordinated changes in the expression of central metabolic enzymes were corroborated by growth kinetic curves, which displayed the presence of an early transition phase. These results demonstrate that the CHB-mediated regulatory system is involved in metabolism.
Fig. 7.
Different expressed proteins indicated in glycolysis, TCA cycle, pentose phosphate pathway, nucleotide metabolism, amino acid metabolism, and cell wall development.
It is also worth noting that many stress response-associated proteins were downregulated in ZJUD7, including molecular chaperone DnaK (spot 19), chaperonine GroEL (spot 16), ATP-dependent Clp protease proteolytic subunit 1 ClpP (spot 48), thioredoxin (spot 10), GTP-binding protein TyrA/BipA (spot 14), peptidyl-prolyl cis-trans isomerase (spot 51), and FeS assembly ATPase SufC (spot 50). Several of these proteins were identified as starvation-induced stress-response proteins (24), suggesting that CHB also has a role in activation of the stress response.
In view of the growth defect of ZJUD7, it is interesting to observe the increased expression level of several enzymes involved in amino acid biosynthesis (spots 15 and 29) and catabolism (spots 23 and 38). This might be caused by the cellular effort to overcome the metabolic imbalance in cellular pathways. It is also interesting to find that the PTS component PtsI and two enzymes (spots 5 and 12) involved in N-acetylglucosamine (GlcNAc) metabolism were upregulated, since the PTS system has been recently shown to play an important role in nutrient utilization and control of the onset of development and antibiotic production in Streptomyces (27). Taken together, these results clearly showed that the CHB regulon in S. chattanoogensis is not restricted to secondary metabolism and development, but is also involved in metabolism and stress response.
CHB system transcriptionally affects the phosphotransferase (PTS) system.
In bacteria, the central system for carbon uptake and signaling is the phosphotransferase system. Recently, the PTS system of Streptomyces has also been shown to play an important role in nutrient utilization and antibiotic production, especially those related to GlcNAc sensing, uptake, and metabolism (28). In S. chattanoogensis, ptsI and crr, encoding the PTS enzyme EI and EIIA, form an operon, as the situation in S. coelicolor. It was surprising to find that the expression of PtsI was significantly increased in the ΔscgA mutant. To assess whether the regulation was exerted at the transcriptional level, quantitative RT-PCR was performed (see Fig. S6a in the supplemental material). The result revealed the mRNA levels of the operon increased by 6.72-fold at 20 h in ZJUD7, which closely matched the data (6.088-fold) from the proteomic analysis, indicating that CHB system transcriptionally affected crr-ptsI operon. However, no significant change between L10 and ZJUD7 was observed at 12 h, suggesting that this regulation was not exerted at the initial rapid growth phase.
In the PTS system, PtsI is responsible for converting phosphoenolpyruvate to pyruvate, we thus determined the extracellular pyruvate concentrations in the culture of both L10 and ZJUD7 by HPLC. As shown in Fig. S6b in the supplemental material, no pyruvate was excreted from both strains at 6 h. Compared to L10, the extracellular pyruvate concentrations of ZJUD7 were significantly increased at 20 h and 24 h. No pyruvate was detected in the culture of both strains when they entered the stationary phase (36 h), indicating the reuse of accumulated pyruvate. The proteomics data were also verified by quantitative RT-PCR using another four genes (spots 5, 42, 43, and 48) (see Fig. S6c in the supplemental material), and the results showed a good correlation. These data confirmed that CHB system is involved in nutrient utilization and metabolism. Taken together, we propose a model for the CHB-mediated global regulatory system in S. chattanoogensis (Fig. 8).
Fig. 8.
Proposed model of CHB system in S. chattanoogensis. ScgR binds as a dimer to two targets sites, which are in front of scgA and gbdA promoters, respectively. CHB binds to ScgR and relieves the repression. Arrows denote activation and lines with a bar denote repression. Dash lines indicate that their precise mechanisms are under investigation.
DISCUSSION
In the present study, we cloned and characterized a GBL system in S. chattanoogensis. Compared to its counterparts, CHB system had several novel properties. First, the genes encoding the GBL system are clustered, but they are not located within or nearby a secondary metabolite gene cluster. Second, the CHB deletion mutants showed carbon source-dependent phenotypes and had growth deficiencies in submerged culture. Furthermore, proteomic and transcriptional analysis revealed that the CHB system negatively regulated the nutrient uptake system (PTS system), and many enzymes involved in primary metabolism were also under the control of the CHB system.
A common feature of nearly all of the QS systems is that signal transduction is mediated by the interaction of autoregulators with cognate receptors. We have made a genome-wide search for the ScgR binding sequence, and only two ScgR-dependent promoters (scgAp and gbdAp) were identified. The ΔgbdA mutant showed no observable change in phenotype, indicating that the only downstream gene we found (gbdA) cannot account for the phenotypes of CHB deletion mutants. It is possible that CHB regulation of gene expression is not mediated by a cognate receptor. Very recently, Chugani et al. (5) found that acyl-homoserine lactone (acyl-HSL) regulation in P. aeruginosa does not follow the classical QS tenet in that it is not mediated by a LuxR-type transcription factor: even after disrupting all LuxR-type receptors, there were at least 37 genes that still responded to acyl-HSL.
As shown above, the CHB regulatory system controlled growth, development, and antibiotic production. In Streptomyces, the onset of morphological differentiation and secondary metabolite production are controlled by various intra- and extracellular signals. Among the four known GBL-receptor systems described above, only A-factor-ArpA controls both morphological differentiation and secondary metabolite production. This could be attributed to the direct link between the A-factor receptor ArpA and AdpA, homologs of which have been proven to be pleiotropic regulators in several Streptomyces spp. (1, 9, 25). In S. coelicolor, AdpA-c appeared to be not associated with the SCB1 system. We found that ScgR cannot bind to the promoter of adpA-ch and that the transcriptional level of adpA-ch was not affected in ΔscgR or ΔscgA mutants (unpublished data). These results demonstrate that the growth arrest of CHB deletion mutants was not associated with AdpA-ch, indicating that there might be a novel mechanism mediating the signaling cascade of the CHB system.
Nutrient availability is pivotal for streptomycetes to choose between continuing vegetative growth or switching on the developmental program on solid medium. It also dictates the growth phase of Streptomyces in submerged cultures. Streptomycetes, as well as other bacteria, typically undergo multiphasic growth in liquid cultures, including exponential, transitional, and stationary phases. There are two distinct stages of exponential growth separated by a temporary growth called diauxic lag, which is an adaptive state and reflects depletion of a preferred nutritional source followed by activation of genes required for utilization of alternative substrates (24). Recently, these two stages of exponential growth were found to correspond to two different morphological phases: MI (the first compartmentalized mycelium) and MII (the second multinucleated mycelium) (21, 22). These phases were also characterized by distinct protein expression profiles. The switching on of secondary metabolism strongly correlated with hypha differentiation (MII). In view of the growth kinetics of ZJUD7 and ZJUD20 in YEME medium, as well as phenotype analysis, it is reasonable to believe that CHB deletion mutants of S. chattanoogensis were arrested at the transient diauxic lag between MI and MII phase and directly entered the stationary phase as a result of nutrient starvation. Proteomic analysis revealed CHB system directly or indirectly controlled the expression of more than 50 genes, among which a large number of metabolic enzymes involved in primary metabolism were dramatically decreased in ZJUD7 after the initial rapid growth. This should be responsible for the subsequent growth arrest. Coordinated downregulation of central metabolic enzymes is characteristic of the diauxic lag and transition phases (24). Expression of a variety of stress response proteins in L10 were higher than in ZJUD7, which is not surprising since it has been suggested that stress response and development are connected in Streptomyces (35). For instance, in S. coelicolor, the developmental regulator BldD, also directly controls a sigma factor gene, sigH, which is involved in the regulation of stress responses (7). These stress-related proteins were thought to be involved in adaptation to the stress generated by metabolic shifts. Several other genes that have been implicated in the activation of secondary metabolism and antibiotic production were also downregulated in ZJUD7, including ClpP (spot 48) and inositol-1-phosphate synthase (spot 41). These results, together with growth curves and phenotype analysis, strongly suggest that CHB deletion mutants failed to reorganize its metabolism for initiating the MII phase, which might be caused by the inability of activation of some global regulators or sigma factors. This idea is also supported by increased expression of aldo/keto reductase (spot 32), SufC (spot 50), and ssDNA-binding protein (spot 33) in L10, since the homologues of these proteins were also found upregulated at MII in S. coelicolor (22). Other supporting evidences are the profiles of pH level and extracellular pyruvate concentration of L10 and ZJUD7 (see Fig. S6b and S7 in the supplemental material). It has been demonstrated that the first round of growth is acidogenic and reinitiation of growth coincides with the consumption of organic acids in Streptomyces (34). When the pH level of L10 began to increase, the pH value of ZJUD7 remained at the same level (see Fig. S7 in the supplemental material). However, after a period of adaptation, an alternative mechanism seems to be able to bypass the CHB regulatory system and partially recover the phenotypes.
Compared to the production of the yellow pigment that was totally delayed or abolished in CHB deletion mutants, less NTM was produced in those mutants during the adaptation period. It is worth noting that the expression of the biosynthetic gene cluster of NTM is constitutive in S. chattanoogensis L10 at least when grown in YEME medium, and no significant change was observed at the transcriptional level of cluster genes between L10 and ZJUD7 at 20 h. These results suggest that reduced production of NTM in ZJUD7 might be at least in part attributable to scarce availability of its precursor acetyl-coenzyme A due to the coordinated downregulation of several enzymes involved in glycolysis. Overall, our results present a novel γ-butyrolactone regulatory system involved in nutrient utilization, triggering adaptive responses, and finally dictating the switch from primary to secondary metabolism.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Yun Song for critical reading and to Xinhang Jiang for technical assistance.
This study was supported by the National Natural Science Foundation of China (grant 31070040), the National Basic Research Program of China (973 Program, 2012CB721000), the National Key Project for New Drug Development, China (2011ZX09202-101-11), and the Fundamental Research Funds for the Central Universities (2010QNA6004).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 23 September 2011.
REFERENCES
- 1. Akanuma G., Hara H., Ohnishi Y., Horinouchi S. 2009. Dynamic changes in the extracellular proteome caused by absence of a pleiotropic regulator AdpA in Streptomyces griseus. Mol. Microbiol. 73: 898–912 [DOI] [PubMed] [Google Scholar]
- 2. Aparicio J. F., Fouces R., Mendes M. V., Olivera N., Martin J. F. 2000. A complex multienzyme system encoded by five polyketide synthase genes is involved in the biosynthesis of the 26-membered polyene macrolide pimaricin in Streptomyces natalensis. Chem. Biol. 7: 895–905 [DOI] [PubMed] [Google Scholar]
- 3. Borodina I., et al. 2008. Antibiotic overproduction in Streptomyces coelicolor A3(2) mediated by phosphofructokinase deletion. J. Biol. Chem. 283: 25186–25199 [DOI] [PubMed] [Google Scholar]
- 4. Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- 5. Chugani S., Greenberg E. P. 2010. LuxR homolog-independent gene regulation by acyl-homoserine lactones in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 107: 10673–10678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Corre C., Song L., O'Rourke S., Chater K. F., Challis G. L. 2008. 2-Alkyl-4-hydroxymethylfuran-3-carboxylic acids, antibiotic production inducers discovered by Streptomyces coelicolor genome mining. Proc. Natl. Acad. Sci. U. S. A. 105: 17510–17515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. den Hengst C. D., et al. 2010. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol. Microbiol. 78: 361–379 [DOI] [PubMed] [Google Scholar]
- 8. Du Y. L., et al. 2009. Identification of a novel Streptomyces chattanoogensis L10 and enhancing its natamycin production by overexpressing positive regulator ScnRII. J. Microbiol. 47: 506–513 [DOI] [PubMed] [Google Scholar]
- 9. Du Y. L., et al. 2011. The pleiotropic regulator AdpAch is required for natamycin biosynthesis and morphological differentiation in Streptomyces chattanoogensis. Microbiology 157: 1300–1311 [DOI] [PubMed] [Google Scholar]
- 10. Folcher M., et al. 2001. Pleiotropic functions of a Streptomyces pristinaespiralis autoregulator receptor in development, antibiotic biosynthesis, and expression of a superoxide dismutase. J. Biol. Chem. 276: 44297–44306 [DOI] [PubMed] [Google Scholar]
- 11. Gust B., Challis G. L., Fowler K., Kieser T., Chater K. F. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100: 1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Horinouchi S. 2002. A microbial hormone, A-factor, as a master switch for morphological differentiation and secondary metabolism in Streptomyces griseus. Front. Biosci. 7: d2045–2057 [DOI] [PubMed] [Google Scholar]
- 13. Hsiao N. H., et al. 2007. ScbA from Streptomyces coelicolor A3(2) has homology to fatty acid synthases and is able to synthesize gamma-butyrolactones. Microbiology 153: 1394–1404 [DOI] [PubMed] [Google Scholar]
- 14. Kato J. Y., Funa N., Watanabe H., Ohnishi Y., Horinouchi S. 2007. Biosynthesis of gamma-butyrolactone autoregulators that switch on secondary metabolism and morphological development in Streptomyces. Proc. Natl. Acad. Sci. U. S. A. 104: 2378–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kawachi R., et al. 2000. Identification of an AfsA homologue (BarX) from Streptomyces virginiae as a pleiotropic regulator controlling autoregulator biosynthesis, virginiamycin biosynthesis and virginiamycin M1 resistance. Mol. Microbiol. 36: 302–313 [DOI] [PubMed] [Google Scholar]
- 16. Kieser T., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, United Kingdom [Google Scholar]
- 17. Kitani S., Doi M., Shimizu T., Maeda A., Nihira T. 2010. Control of secondary metabolism by farX, which is involved in the gamma-butyrolactone biosynthesis of Streptomyces lavendulae FRI-5. Arch. Microbiol. 192: 211–220 [DOI] [PubMed] [Google Scholar]
- 18. Kitani S., Kinoshita H., Nihira T., Yamada Y. 1999. In vitro analysis of the butyrolactone autoregulator receptor protein (FarA) of Streptomyces lavendulae FRI-5 reveals that FarA acts as a DNA-binding transcriptional regulator that controls its own synthesis. J. Bacteriol. 181: 5081–5084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee Y. J., Kitani S., Nihira T. 2010. Null mutation analysis of an afsA-family gene, barX, that is involved in biosynthesis of the γ-butyrolactone autoregulator in Streptomyces virginiae. Microbiology 156: 206–210 [DOI] [PubMed] [Google Scholar]
- 20. Leesong M., Henderson B. S., Gillig J. R., Schwab J. M., Smith J. L. 1996. Structure of a dehydratase-isomerase from the bacterial pathway for biosynthesis of unsaturated fatty acids: two catalytic activities in one active site. Structure 4: 253–264 [DOI] [PubMed] [Google Scholar]
- 21. Manteca A., Alvarez R., Salazar N., Yague P., Sanchez J. 2008. Mycelium differentiation and antibiotic production in submerged cultures of Streptomyces coelicolor. Appl. Environ. Microbiol. 74: 3877–3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Manteca A., Jung H. R., Schwammle V., Jensen O. N., Sanchez J. 2010. Quantitative proteome analysis of Streptomyces coelicolor nonsporulating liquid cultures demonstrates a complex differentiation process comparable to that occurring in sporulating solid cultures. J. Proteome Res. 9: 4801–4811 [DOI] [PubMed] [Google Scholar]
- 23. Ng W. L., Bassler B. L. 2009. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 43: 197–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Novotna J., et al. 2003. Proteomic studies of diauxic lag in the differentiating prokaryote Streptomyces coelicolor reveal a regulatory network of stress-induced proteins and central metabolic enzymes. Mol. Microbiol. 48: 1289–1303 [DOI] [PubMed] [Google Scholar]
- 25. Pan Y., Liu G., Yang H., Tian Y., Tan H. 2009. The pleiotropic regulator AdpA-L directly controls the pathway-specific activator of nikkomycin biosynthesis in Streptomyces ansochromogenes. Mol. Microbiol. 72: 710–723 [DOI] [PubMed] [Google Scholar]
- 26. Recio E., Colinas A., Rumbero A., Aparicio J. F., Martin J. F. 2004. PI factor, a novel type quorum-sensing inducer elicits pimaricin production in Streptomyces natalensis. J. Biol. Chem. 279: 41586–41593 [DOI] [PubMed] [Google Scholar]
- 27. Rigali S., et al. 2006. The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Mol. Microbiol. 61: 1237–1251 [DOI] [PubMed] [Google Scholar]
- 28. Rigali S., et al. 2008. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 9: 670–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 30. Shikura N., Yamamura J., Nihira T. 2002. barS1, a gene for biosynthesis of a gamma-butyrolactone autoregulator, a microbial signaling molecule eliciting antibiotic production in Streptomyces species. J. Bacteriol. 184: 5151–5157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Takano E. 2006. Gamma-butyrolactones: Streptomyces signaling molecules regulating antibiotic production and differentiation. Curr. Opin. Microbiol. 9: 287–294 [DOI] [PubMed] [Google Scholar]
- 32. Takano E., Chakraburtty R., Nihira T., Yamada Y., Bibb M. J. 2001. A complex role for the gamma-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 41: 1015–1028 [DOI] [PubMed] [Google Scholar]
- 33. Takano E., et al. 2000. Purification and structural determination of SCB1, a gamma-butyrolactone that elicits antibiotic production in Streptomyces coelicolor A3(2). J. Biol. Chem. 275: 11010–11016 [DOI] [PubMed] [Google Scholar]
- 34. Viollier P. H., Minas W., Dale G. E., Folcher M., Thompson C. J. 2001. Role of acid metabolism in Streptomyces coelicolor morphological differentiation and antibiotic biosynthesis. J. Bacteriol. 183: 3184–3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vohradsky J., et al. 2000. Developmental control of stress stimulons in Streptomyces coelicolor revealed by statistical analyses of global gene expression patterns. J. Bacteriol. 182: 4979–4986 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.