Abstract
We describe a simple method for stabilizing and extracting high-quality prokaryotic RNA from meat. Heat and salt stress of Escherichia coli and Salmonella spp. in minced meat reproducibly induced dnaK and otsB expression, respectively, as observed by quantitative reverse transcription-PCR (>5-fold relative changes). Thus, the method is applicable in studies of bacterial gene expression in a meat matrix.
TEXT
A key component in understanding the ability of pathogens to survive in food products is to determine their response to food-related stresses. In recent years, advances in molecular techniques have enabled global studies of gene expression, using tools such as quantitative reverse transcription-PCR (qRT-PCR) and DNA microarrays. While these techniques have been employed in numerous studies examining the behavior of food-borne pathogens in broth or food-like broth models, only a few studies have investigated quantitative gene expression of pathogens in complex food matrices like dairy products (3, 7, 8, 11) or meat (4, 5, 9). Such investigations have been impeded by the complexity of food matrices, by inherent PCR-inhibiting compounds, and by low levels of target molecules (1). Diversity in foods necessitates methods applicable for different categories of food products. We wished to establish a method to investigate gene expression of bacteria directly from a complex food matrix that is often associated with transmission of pathogenic bacteria. As a model system we chose to study Salmonella spp. and verocytotoxic Escherichia coli in artificially contaminated minced meat.
Minced beef (8 to 15% fat) obtained in local Danish supermarkets was inoculated with a mix of strains of either Escherichia coli or Salmonella enterica. The E. coli mix consisted of E. coli serotype O26:H- (D3410), serotype O111:H- (D3411) obtained from Statens Serum Institut (SSI; Copenhagen, Denmark), and serotype O157:H7 from Danish beef meat (D3423) (2). The Salmonella mix consisted of S. enterica serovar Dublin (D3414), S. enterica serovar Typhimurium DT193 (D3415), and S. enterica serovar Derby (D3420) obtained from SSI. From −80°C frozen stocks, all strains were cultured at 37°C for 24 h on Luria-Bertani agar (LB; Oxoid, Greve, Denmark). A single colony from each strain was cultured individually and diluted 1:10 in prewarmed LB, and the E. coli mixed culture and the Salmonella mixed culture were prepared by pooling equal volumes of the three E. coli or the three Salmonella strain cultures. The mixed cultures were incubated until an absorbance at 600 nm (A600) of 0.5 was reached, corresponding to approximately 5 ×108 CFU/ml as measured by plate counts. Samples of 5.0 g minced beef meat were weighed in stomacher filter bags and were stored at refrigeration temperature (5°C) or on ice during processing. All experiments with meat samples were performed in two technical replicates and were repeated in three biological replications. Ten-milliliter mixed cultures, or dilutions thereof, were pelleted by centrifugation (5,000 × g for 10 min) and resuspended in 0.5 ml LB to be used as inocula. Meat samples were inoculated and manually massaged for about a minute to distribute inocula. For heat stress conditions, meat samples were incubated at 45°C in a water bath for 30 min immediately after inoculation, and untreated controls were left at room temperature. For salt stress conditions, 1 ml of 17.5% NaCl solution was added to meat samples (5.0 g) to obtain 5% NaCl (in water phase) and were massaged manually before inoculation. Meat samples were stabilized with 10 ml RNAlater tissue collection medium (Ambion, Nærum, Denmark) and were manually massaged and left for 10 min in order to generate meat juice. The salt stress samples were stabilized with RNAlater 0 and 60 min after inoculation.
Meat juice (∼9 ml) was collected and transferred to centrifuge tubes. Bacteria in the suspension (approximately 1 × 108 CFU per sample, determined by total plate counts) were harvested by centrifugation (5,000 × g for 10 min), and RNA was extracted from the pellets. Noninoculated meat samples, determined to contain a negligible level of bacteria (<5 × 104 CFU/g), served as controls to examine the presence of RNA from the natural microflora as well as eukaryotic RNA from the meat and were processed as described above.
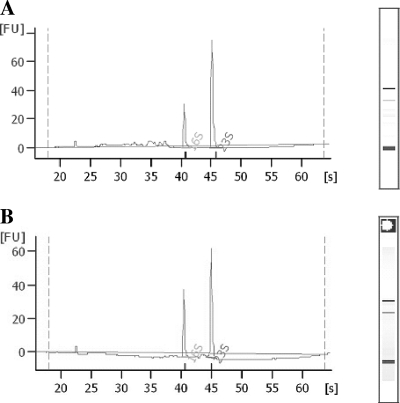
RNA extractions were performed with the RNeasy kit (Qiagen, Copenhagen, Denmark). Pellets from meat samples and broth cultures were treated with 0.2 ml Tris-EDTA buffer (Tris-EDTA, 10:1; pH 8) containing 3 mg lysozyme and 0.004 mg proteinase K and were vortexed for 10 s every second minute during a 10-min incubation period at room temperature. Cells were disrupted using Fastprep (MP Biomedicals, Illkirch, France) as recommended by the manufacturer, followed by a wash step with buffer RLT containing 2-mercaptoethanol (14.3 M; 10 μl per ml of buffer RLT). Samples were centrifuged at 14,000 × g for 2 min, supernatants were transferred to new tubes, and 500 μl of 96% ethanol was added. RNA extractions were continued according to Qiagen recommendations, including an on-column DNase (Qiagen) treatment for 15 min. Purified RNA was quantified using a Nanodrop ND-1000 apparatus (ThermoScientific, Wilmington, DE), and the RNA quality was verified based on an optical density at 260 nm (OD260)/OD280 absorption ratio of >1.95 and an OD230/OD260 absorption ratio of >2.0, and the integrity was further assessed either electrophoretically following ethidium bromide staining or with a 2100 Bioanalyzer (Agilent, Palo Alto, CA), resulting in clear patterns with prominent 16S and 23S ribosomal bands and RNA integrity number values of >7 (Fig. 1).
Fig. 1.
Electropherograms (created using an Agilent 2100 Bioanalyzer) showing integrity of RNA samples extracted from an E. coli mixed culture inoculated into minced beef (A) and an E. coli mixed culture in LB broth (B). FU, fluorescence units.
cDNA was synthesized using a SuperScript VILO cDNA synthesis kit (Invitrogen,Tåstrup, Denmark) with random hexamer primers and following the manufacturer's instruction, using 500 ng total RNA per sample (Invitrogen). Real-time PCR was completed on an ABI 7900HT Fast PCR system with SYBR green (Molecular Probes) as the fluorescent reporter. Primers (Table 1) were designed with Primer-BLAST (NCBI, Bethesda, MD). Amplification was carried out in triplicate in a 20-μl final volume containing 2.0 μl cDNA (100 ng), 10 μl Express SYBR greenER qPCR supermix with premixed ROX reference dye (Invitrogen), and each primer at a concentration of 0.2 μM. The cycling conditions were 95°C for 2 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. A subsequent melting curve determination between 60 and 95°C at a transition rate of 0.1°C/s confirmed the specificity of the PCR product. Each run included a negative control and a cDNA reaction without reverse transcriptase to rule out DNA contamination. Spectrophotometric measurements as well as quantitative PCR results showed none or very low contamination by RNA from eukaryotic tissue cells or background flora in noninoculated meat samples.
Table 1.
Primer sequences of stress genes and reference genes used in this study
| Gene (genus or species) | Primer orientation and sequence (5′-3′) |
|---|---|
| dnaK (E. coli) | Forward: CACCACGCCTTCTATCAT |
| Reverse: GCCTTTAACTTCGACCCA | |
| otsB (E. coli) | Forward: AGCAGGGAAAGTGTGTTGTC |
| Reverse: ATCATCGCCCAGAAATACGG | |
| gapA (E. coli) | Forward: CATCATCCCGTCCTCTACC |
| Reverse: CGCCATACCAGTCAGTTT | |
| dnaK (Salmonella) | Forward: CGATTATGGATGGAACGCAGG |
| Reverse: GGCTGACCAACCAGAGTT | |
| otsB (Salmonella) | Forward: GGTAGTCCGTGAGGTAGAGG |
| Reverse: GGAGCCTGACGGTAGTGC | |
| rpoD (Salmonella) | Forward: CTGAAAATACCACCAGCACC |
| Reverse: CGGGTCAACAGTTCAACAGTG |
To investigate potential bias due to inhibiting components in the meat, extractions of RNA from bacteria inoculated in meat and LB broth culture were concomitantly conducted using the same protocol. Threshold cycle (CT) values of the investigated genes were consistently found at expected levels in the control samples over the range of experimental setups (data not shown). Amplification efficiencies for the primers (Table 1) recognizing dnaK (heat stress), otsB (salt stress), and rpoD and gapA (reference genes) were assessed based on the slopes of standard curves. The efficiencies were >90%, and the correlation coefficients were above 0.99 after optimization.
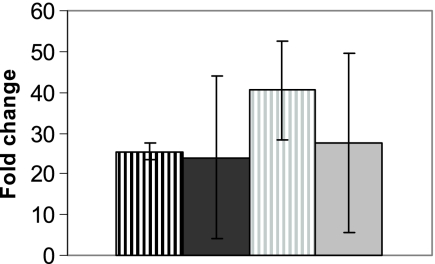
Measured mRNA levels of the target gene were normalized to the mRNA levels of the reference genes (rpoD and gapA), which were chosen for transcript stability under the given experimental conditions (data not shown). Normalized values were used to calculate ratios of expression levels (relative fold changes) in treated samples versus untreated control samples (Fig. 2) based on the 2−ΔΔCT method (6).
Fig. 2.
Relative expression of dnaK after heat treatment (45°C, 30 min) of samples versus untreated samples and normalized to reference genes gapA (E. coli [dark gray]) and rpoD (Salmonella [light gray]). RNA was extracted from inoculated meat samples (vertically striped bars) and LB broth cultures (solid bars). Error bars show variations between two biological replicates, each measured in technical triplicate.
Using our RNA extraction method, it was possible to measure expression of bacterial stress genes in a food matrix by applying salt and heat stress. Following heat stress, an induction of dnaK expression was observed compared to untreated controls. Relative changes in expression of dnaK ranging from 24- to 41-fold were calculated for E. coli and Salmonella cultures (Fig. 2), although with some variation between biological replicates. Likewise, E. coli and Salmonella expression of otsB was increased >30- and 5-fold, respectively, after salt stress (data not shown). The variation in fold induction may have been related to small variations in timing of the sampling, which can influence the induction level (10). The extraction protocol was applied to meat samples inoculated with target bacteria levels ranging from 106 to 5 × 109 per gram of meat, which all yielded sufficient RNA (>50 ng/μl) and qPCR detection of reference genes (CT values of <30).
The current study demonstrates that it is possible to extract bacterial RNA suitable for gene expression analyses directly from minced meat. The initial stabilization of the RNA without lengthy procedures or toxic chemicals as well as the easy and fast extraction procedure renders the method highly suitable for the study of gene expression related to changes in a minced meat environment.
Acknowledgments
The work was supported by the Danish Research Council for Strategic Research.
Footnotes
Published ahead of print on 7 October 2011.
REFERENCES
- 1. Brehm-Stecher B., Young C., Jaykus L. A., Tortorello M. L. 2009. Sample preparation: the forgotten beginning. J. Food Prot. 72: 1774–1789 [DOI] [PubMed] [Google Scholar]
- 2. Breum S. Ï., Boel J. 2010. Prevalence of Escherichia coli O157 and verocytotoxin producing E. coli (VTEC) on Danish beef carcasses. Int. J. Food Microbiol. 141: 90–96 [DOI] [PubMed] [Google Scholar]
- 3. Calles-Enriquez M., Ladero V., Fernandez M., Martin M. C., Alvarez M. A. 2010. Extraction of RNA from fermented milk products for in situ gene expression analysis. Anal. Biochem. 400: 307–309 [DOI] [PubMed] [Google Scholar]
- 4. de Wet S. C., Denman S. E., Sly L., McSweeney C. S. 2008. An improved method for RNA extraction from carcass samples for detection of viable Escherichia coli O157:H7 by reverse-transcriptase polymerase chain reaction. Lett. Appl. Microbiol. 47: 399–404 [DOI] [PubMed] [Google Scholar]
- 5. Fratamico P. M., Wang S., Yan X., Zhang W., Li Y. 2011. Differential gene expression of E. coli O157:H7 in ground beef extract compared to tryptic soy broth. J. Food Sci. 76: M79–M87 [DOI] [PubMed] [Google Scholar]
- 6. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- 7. Makhzami S., et al. 2008. In situ gene expression in cheese matrices: application to a set of enterococcal genes. J. Microbiol. Methods 75: 485–490 [DOI] [PubMed] [Google Scholar]
- 8. Monnet C., Ulve V. M., Sarthou A. S., Irlinger F. 2008. Extraction of RNA from cheese without prior separation of microbial cells. Appl. Environ. Microbiol. 74: 5724–5730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Olesen I., Thorsen L., Jespersen L. 2010. Relative transcription of Listeria monocytogenes virulence genes in liver pâtés with varying NaCl content. Int. J. Food Microbiol. 141: S60–S68 [DOI] [PubMed] [Google Scholar]
- 10. Rosen R., Ron E. Z. 2002. Proteome analysis in the study of the bacterial heat-shock response. Mass Spectrom. Rev. 21: 244–265 [DOI] [PubMed] [Google Scholar]
- 11. Ulve V. M., et al. 2008. RNA extraction from cheese for analysis of in situ gene expression of Lactococcus lactis. J. Appl. Microbiol. 105: 1327–1333 [DOI] [PubMed] [Google Scholar]