Abstract
Campylobacter jejuni continues to be the leading cause of bacterial food-borne illness worldwide, so improvements to current methods used for bacterial detection and disease prevention are needed. We describe here the genome and proteome of C. jejuni bacteriophage NCTC 12673 and the exploitation of its receptor-binding protein for specific bacterial detection. Remarkably, the 135-kb Myoviridae genome of NCTC 12673 differs greatly from any other proteobacterial phage genome described (including C. jejuni phages CP220 and CPt10) and instead shows closest homology to the cyanobacterial T4-related myophages. The phage genome contains 172 putative open reading frames, including 12 homing endonucleases, no visible means of packaging, and a putative trans-splicing intein. The phage DNA appears to be strongly associated with a protein that interfered with PCR amplification and estimation of the phage genome mass by pulsed-field gel electrophoresis. Identification and analyses of the receptor-binding protein (Gp48) revealed features common to the Salmonella enterica P22 phage tailspike protein, including the ability to specifically recognize a host organism. Bacteriophage receptor-binding proteins may offer promising alternatives for use in pathogen detection platforms.
INTRODUCTION
Campylobacter belongs to the epsilon class of proteobacteria and is the leading cause of bacterial food-borne gastroenteritis worldwide (22). Campylobacter has also been associated with severe neurological disorders such as the Guillain-Barré and Miller-Fisher syndromes, as well as reactive arthritis and irritable bowel syndrome (32). Many Campylobacter phages have been characterized by their host-range characteristics, morphology, genome size, and susceptibility to restriction endonucleases (5, 7, 24). Only a few of the characterized Campylobacter phages are members of the B1 group of the family Siphoviridae (1), while most belong to the family Myoviridae possessing genomes which fall into three size classes: 110 to 150 kb (class III), 170 to 190 kb (class II), and 320 kb (class I) (24). One interesting characteristic of many of these phages is that their DNA appears to be resistant to cleavage by several common restriction endonucleases, thus making them “refractory to genomic analysis” (34). Two genomes of Campylobacter phages have been sequenced thus far: CPt10 and CP220 (34). Both appear to be closely related to each other and belong to the class II of Campylobacter Myoviridae phage genomes (34).
Bacteriophages possess enormous diagnostic and therapeutic potential, which provides promise for the development of a wide range of novel antimicrobials and diagnostic tools (14). Numerous phages have been isolated against Campylobacter for use in phage typing schemes (9, 10, 12, 18, 25), but it has only recently been shown that bacteriophages can effectively decrease Campylobacter contamination (4, 20, 36). For example, Campylobacter-specific phages were able to reduce the numbers of Campylobacter coli and Campylobacter jejuni in chickens when added directly to the chicken feed (6). It should be emphasized that bacteriophages specifically recognize their hosts through the use of receptor-binding proteins (RBPs) (15). We recently demonstrated that Salmonella enterica serovar Typhimurium Podoviridae phage P22 RBP can agglutinate and reduce the motility of host cells and act as a prophylactic agent reducing chicken colonization with S. Typhimurium (37). Immobilized RBP can also be exploited for diagnostic purposes and was shown to be more stable during storage than the whole phage (28). Use of RBPs instead of whole phages provides a novel family of antimicrobial and diagnostic platforms that can be easily engineered and produced in massive amounts using standard recombinant DNA technologies without the involvement of the whole phage/pathogen propagation system and the risk of pathogen gene transduction. We present data here on the genome and proteome of the C. jejuni lytic phage, NCTC 12673, which was originally isolated from poultry (10), as well as the identification and preliminary characterization of its RBP.
MATERIALS AND METHODS
Bacteriophage propagation.
Phage NCTC 12673 and the propagating C. jejuni strain NCTC 12661 were obtained from the Health Protection Agency National Collection of Type Cultures (NCTC; Salisbury, United Kingdom). Phages were propagated under microaerobic conditions (10% CO2, 5% O2, 85% N2) at 37°C either on plates or in liquid Mueller-Hinton medium (see File S1 in the supplemental material).
Isolation of phage DNA for sequencing.
Viral particles were eluted from the plates, incubated with RNase A and DNase I, and subjected to phenol-chloroform-isoamyl alcohol extraction (see File S1 in the supplemental material). DNA was precipitated with isopropanol and recovered by centrifugation. The pellet was air dried, washed twice with 70% ethanol, and resuspended in 10 mM Tris-HCl (pH 8.0). To further purify the DNA, 40 μg of RNase A/ml was added at room temperature for 30 min. The sample was then re-extracted using the phenol-chloroform-isopropanol extraction-precipitation procedure.
DNA sequencing and annotation.
The genome sequencing by Fidelity Systems was based upon the construction and sequencing of a mini-library and then primer walking using ThermoFidelase I and chemically modified primers (“fimers”) to inhibit unwanted reactions during cycle sequencing (8, 29). Coding regions were identified using Kodon (Applied Maths, Inc.) with homologues being screened using Batch BLAST (http://greengene.uml.edu/programs/NCBI_Blast.html) against the nonredundant database at the National Center for Biotechnology Information (NCBI). In questionable cases, PSI-BLAST (2) was also used. Bioinformatic analysis was performed as described previously (13, 16) (see also File S1 in the supplemental material).
TEM of phage particles.
Transmission electron microscopy (TEM) was performed with the phage suspension that was purified using a Sephacryl S-1000 SF (GE Healthcare) size exclusion column to get rid of bacterial proteins (data not shown). The samples were adsorbed onto copper-Formvar-carbon grids (Ted Pella, Inc.) for 10 min, negatively stained with 4% phosphotungstic acid (buffered to pH 6.8) for 2 min, air dried, and examined under a transmission electron microscope (Philips Morgagni 268; FEI Company). Images were captured with the charge-coupled device camera and controller (Gatan, Inc.) and processed using DigitalMicrograph (Gatan, Inc.). All steps were performed at room temperature.
Proteome analysis.
DNase and RNase were added to a final concentration of 1 μg/ml to the clarified phage lysate and incubated at room temperature for 30 min. The phages were precipitated using PEG 8000 (38) and then subjected to CsCl gradient purification and concentration (see File S1 in the supplemental material). Suspension of purified phage particles was subjected to SDS-PAGE. All visible bands were cut out and subjected to trypsin digestion. The digests were analyzed by liquid chromatography-mass spectrometry to acquire tandem mass spectra of the tryptic peptides (see File S1 in the supplemental material).
Cloning of genes 1 and 48 into the pGEX 6P-2 vector.
Phage DNA was subjected to a preamplification reaction with Phi29 polymerase (Fermentas) with subsequent PCR amplification (see File S1 in the supplemental material) of genes 1 and 48 by Vent DNA polymerase (NEB). The appropriate PCR product was ligated into a SmaI (Fermentas) cut pGEX 6P-2 vector (GE Healthcare) by overnight blunt ligation (done at room temperature) in the presence of SmaI (26). The correct orientation and product integrity were confirmed by sequencing of the insert performed by the Molecular Biology Service Unit, Department of Biological Sciences, University of Alberta.
Protein expression, purification, and immunoanalysis.
Gp1 and Gp48 proteins were expressed in Escherichia coli BL21 cells (Invitrogen) transformed with the pGEX 6P-2 plasmid containing either gene 1 or gene 48 (see File S1 in the supplemental material). These proteins were expressed and purified as soluble glutathione S-transferase (GST) fusions using glutathione-agarose affinity chromatography (Sigma-Aldrich) and ion-exchange chromatography using a MonoQ (GE Healthcare) column and the AKTA Explorer FPLC system (GE Healthcare). S. Typhimurium phage P22 tailspike protein (TSP) was expressed in E. coli BL21(DE3) (Invitrogen) cells using the P22 TSP gene cloned into the pET11a plasmid (Novagen). P22 TSP was purified by an immobilized metal affinity chromatography procedure using a HisTrap HP (GE Healthcare) column and AKTA Explorer FPLC system (GE Healthcare). All proteins were dialyzed against phosphate-buffered saline (PBS; 1.5 mM KH2PO4, 8 mM Na2HPO4, 137 mM NaCl [pH 7.2]), and the concentration was determined by measuring the absorbance at 260 and 280 nm. Purified proteins were subjected to standard immunoblot procedures (see File S1 in the supplemental material).
Agglutination of bacterial cells by GST-Gp48 and P22 TSP.
The microagglutination assays for C. jejuni and S. Typhimurium were performed as described previously (37) with minor modifications (see File S1 in the supplemental material). Agglutinated cells appeared as diffused aggregates contrary to the nonagglutinated cells, which formed compact sediments at the bottom of the well.
Analysis of the bacterial binding to the immobilized GST-Gp48.
The alcohol-washed fabricated gold substrates (28) were activated with dithiobis(succinimidyl propionate) (DTSP; Sigma-Aldrich) according to reference 3. The PBS-washed DTSP-covered substrates were incubated with a 5-μg/ml solution of GST-Gp48 in PBS for 1 h at room temperature, followed by similar incubation with a 1-mg/ml solution of bovine serum albumin to block the free substrate surface. The gold substrates were exposed to host (C. jejuni NCTC 11168H) or nonhost bacteria (E. coli K-12 and S. enterica subsp. enterica serovar Typhimurium ATCC 19585) at 109 CFU ml−1 in PBS for 30 min at room temperature. The substrates were washed, fixed, dried, and imaged using the scanning electron microscope (Hitachi S-4800) as described previously (28). The bacterial binding density was estimated from the images by using ImageJ software (National Institutes of Health).
Nucleotide sequence accession number.
The phage sequence was deposited with GenBank under accession number GU296433.
RESULTS AND DISCUSSION
General genome characterization.
At 26.2 mol% GC, the genome of the C. jejuni phage NCTC 12673 is significantly less than that of its host, C. jejuni NCTC 11168 (30.6 mol% GC), and C. jejuni RM1221 (30.3 mol% GC). An analysis of the base composition using a 50-bp window indicated regions of significantly higher GC-content at 19, 51, 61, 73, 108, 112, and 127 kb, suggesting that the genome arose through recombinational exchange of modules of different origins. A total of 172 ORFs were discovered in the 135-kb genome. In most cases, these were closely packed and were preceded by a sequence bearing strong similarity to the bacterial Shine-Dalgarno box (GGAGGT). The genetic map is presented in Fig. 1, with the specifics of each ORF and the putative protein products presented in Table S1 in the supplemental material. We were unable to define, in silico, the replication origin of this phage.
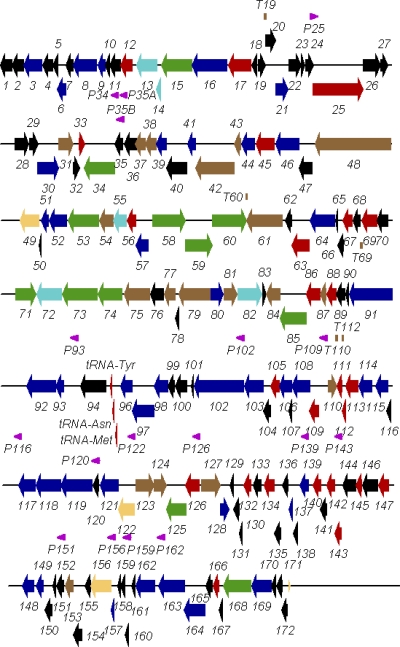
Fig. 1.
Genetic map of C. jejuni phage NCTC 12673. Color code: black, genes with no homologues; red, genes encoding proteins related to proteins of undefined function; dark blue, genes involved in nucleotide metabolism, replication, and recombination; beige, genes involved in transcription and transcript processing; light blue, gene fragments; green, hef homologues; brown, genes involved in morphogenesis. Putative promoters are illustrated as pink arrowheads with the name of the gene they transcribe. Rho-independent terminators are illustrated as vertical brown lines above the map line with the name of the gene that they follow. The map lines are 21 kb in length and overlap by 1 kb.
A full complement of genes exists for DNA replication, initiation, and elongation as well as dTTP biosynthesis (Fig. 1; see Table S1 in the supplemental material). Remarkably, the phage-specific DNA polymerase (Gp102) is homologous to cyanophages P-SSM4, Syn9, and S-PM2 replication proteins. Although phage NCTC 12673 does not encode its own RNA polymerase as do the T7-like phages, it has a number of genes that suggest complexity in its transcriptional mechanisms. Phage NCTC 12673 gp122 is homologous to the T4 late gene sigma factor gp55, showing, however, its closest homologues among the Myoviridae infecting cyanobacteria.
In addition to protein-encoding genes, NCTC 12673, as with many other phages with large genomes, possesses tRNA genes. The three tRNA encoding genes specify tRNA-Met(CAT), tRNA-Asn(GTT), and tRNA-Tyr(GTA). One can only assume that the presence of a phage-specific initiator tRNA aids in the initiation of translation of phage mRNAs (Fig. 1).
The phage NCTC 12673 genome also contains 12 copies of genes or pseudogenes homologous to homing endonuclease-like function (hef) nucleases. Interestingly, hef-like sequences were not detected in the Campylobacter genomes. The mean AT content of the hef elements is 71.67% ± 0.78%, and four DNA direct repeats were identified either within or adjacent to the hef homologues. A phylogenetic analysis of these Hef proteins indicates that considerable diversification has occurred (data not shown). Hef-encoded sequences show weak similarity to the Vsr (very short patch repair) endonuclease family (cd00221).
Remarkably, phage NCTC 12673 does not have any obvious means of DNA packaging. Its genome contains two fragments (gp55 and gp82) related to the large subunit of T4 terminase (gp17) and homology to the packaging protein from the Prochlorococcus myovirus P-SSM4. Also, the products of genes 55 and 82 show homology to the N and C termini of the large subunit terminase of vibriophage KVP40 (NP_899601.1), respectively. BLAST analysis against the intein database (InBase; NEB) resulted in the detection of a putative intein in Gp82. At the same time, the C terminus of Gp55 contains a CVxxD motif, which is typical for the intein N terminus (23). This suggested that an active form of the terminase might still result through intein splicing in trans.
Overall phylogeny of NCTC 12673 phage.
The C. jejuni myovirus NCTC 12673 contains 12 homing endonucleases, redundant and defective genes, a putative trans-splicing intein, and no visible means of packaging. Only two other genomes of Campylobacter phages, CPt10 and CP220 (34), have been sequenced thus far, and they bear very limited homology at the DNA level to NCTC 12673. Comparison at the protein level indicates that NCTC 12673 shares 71 (36.6%) and 72 (36.7%) homologues with Campylobacter phage CP220 and CPt10, respectively. Furthermore, the homologues are distributed over the length of the NCTC 12673 genome, suggesting that the relationships, though mosaic in nature, are not limited to the morphogenesis genes. It is of singular interest that a considerable number of the NCTC 12673 gene homologues lie within phages infecting members of the phylum Cyanobacteria rather than the class Epsilonproteobacteria. While these phages were considered the members of the T4 superfamily of viruses by virtue of containing 35 core genes, a recent reappraisal of these viruses resulted in them being excluded from the Teequatrovirinae (17). These results indicate that the T4-like phages are more diverse than previously determined.
DNA modification.
In silico analysis revealed the presence of numerous cleavage sites for different restriction endonucleases within the NCTC 12673 genome. In vitro experiments indicated that some of these enzymes were not effective in cleaving the DNA. Bsp143I (Sau3A) and EcoRI (224 and 16 sites, respectively) cleaved phage DNA effectively, whereas HaeIII and HindIII (22 and 38 sites, respectively) could not cleave it at all (data not shown). This would normally suggest that the DNA is protected through methylation either via host methyltransferases or the activity of the two putative phage encoded methylases. Gp97 is a protein of 392 amino acids containing COG2189 (adenine-specific DNA methylase, 71% aligned) and pfam01555 (N6/N4 DNA methylase, 68% coverage) motifs. It exhibits sequence similarity to much larger proteins described as type III restriction/modification enzymes. The 345-amino-acid Gp103 protein has no conserved motifs and exhibits low-level sequence similarity to smaller (i.e., 250 to 300 amino acids) bacterial and viral (Lactobacillus phage Lc-Nu, Pseudomonas phage B3, and Vibrio phage VP882) proteins identified as putative methylases. It should be noted that multiple restriction and modification systems are encoded within the C. jejuni genome (data not shown). In no case is the specificity of the cognate methylases known.
It was found that the phage DNA had tightly bound protein(s) which during phenol extraction went to the phenol phase, together with a significant amount (>80%) of the DNA. In addition, the protein(s) coeluted with DNA from the Qiagen genomic DNA purification kit columns and coprecipitated with phage DNA in 42% isopropanol. It did not appear that specific DNA sites were 100% occupied since sequencing from the GenomiPhi-amplified template allowed assembly of the entire genome (or, alternatively, φ29 DNA polymerase could displace some proteins bound to DNA). The results of the PCR amplification also suggest that small fractions of DNA contained protein-free sites. Similar problems were experienced with the C. coli phage in which bacteriophage DNA could not be accurately quantitated spectrophotometrically, nor could it be amplified with Taq polymerase or digested with most restriction endonucleases (C. M. Carvalho and J. Azeredo, unpublished data). All of these points strongly suggest that Campylobacter phage DNA has tightly bound protein(s) that impact its physicochemical properties, including estimated genome size by pulsed-field gel electrophoresis (170 kb for phage NCTC 12673 compared to the 135-kb genome; see Fig. S1 in the supplemental material).
Lysis.
None of the Campylobacter phage proteins exhibited homology to previously characterized lysins or holins. Gp11 (188 amino acids) is very similar to the predicted holins of T4-like myoviruses in size, number of predicted transmembrane domains, and possession of an extended C terminus. Upon iterative PSI-BLAST searches, Gp11 resembles bacterial predicted lytic murein transglycosylase possessing a weak Pfam soluble lytic transglycosylase domain. Experimental analysis is required to investigate whether this protein functions as a dual holin-lysin.
Morphogenesis.
Many of the structural proteins of this phage also displayed homology to existing phage proteins, especially those of the T4 supergroup of viruses (see Table S1 in the supplemental material). At the same time, no identifiable homologues of baseplate as well as Hoc or Soc head decoration proteins were revealed. Remarkably, three potential homologues of the T4 tail core protein Gp19 (Gp37, Gp38, and Gp84) were found. These three proteins differ in size and exhibit little overall sequence similarity to each other. In each case, the closest homologues are among cyanophage (S-PM2, P-SSM2, and S-RSM4) Gp19 homologues. The genome of NCTC 12673 also codes for Gp87, which possesses no homologues among viral proteins but contains a pfam00166 (chaperonin 10-kDa subunit) motif and sequence similarity to bacterial GroES proteins (E = 7 × 10−7; 40% identity). It may play a role in head morphogenesis, replacing host GroES similar to T4 Gp31.
Phage morphology.
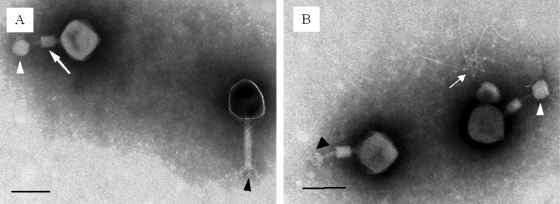
TEM analyses revealed that NCTC 12673 belongs to the Myoviridae family of viruses and possesses a long contracting tail with the length of ∼100 nm and a head of ca. 80 nm in diameter (Fig. 2A). In addition, this phage clearly shows evidence for a striated tail and a collar or neck whiskers. In its contracted state, nearly all of the observed phage particles were associated with spherical bodies, and in one case the tail plate appeared to be wrapped around this element (Fig. 2B). These spherical bodies (see also Fig. S2 in the supplemental material) are reminiscent of membrane vesicles originating from C. jejuni (19). It is possible to imagine active bacterial vesicle shedding as a primitive defense mechanism against phage predation. Alternatively, these vesicles are derived from lysed cells, and we are either observing the original bound phage or the resulting progeny attached to the vesicles. This observation requires further investigation.
Fig. 2.
Electron micrographs of NCTC 12673 bacteriophages. (A) Typical NCTC 12673 particles. Virions possess a long tail with fibers (black arrowhead). Virions where the tail is contracted (arrow) and attached to the spherical particles (white arrowhead) are also indicated. (B) Contracted tails (white and black arrowheads) showing altered tail plate morphology with potential collar (black arrowhead) and a star-shaped tail plate with associated tail fibers (arrow). Scale bars, 100 nm.
Proteome analysis.
Using CsCl-purified NCTC 12673 phage particles which were devoid of significant host protein contamination, we observed the following proteins (see Table S2 in the supplemental material): Gp1, Gp2, Gp69, Gp80, Gp90, Gp109, and Gp154 (all hypothetical proteins), Gp37 (tail tube protein), Gp75 (major coat protein), Gp79 (portal vertex protein), Gp81 (neck protein), and Gp123 (another major capsid protein homologue). In addition, Gp86 was identified among the structural proteins, although its annotation as a phosphatidylserine decarboxylase is perplexing since this enzyme is involved in the conversion of phosphatidylserine to phosphatidylethanolamine (through the removal of CO2), an enzymatic activity which might be associated with a temperate phage, which this is not, or a lipid-containing virus for which we have no evidence. Furthermore, the identification of a putative amidinotransferase (Gp45) is also puzzling. Typically, this enzyme transfers an amidino group from arginine to glycine in amino acid metabolism. It is intriguing that the C. jejuni flagella are heavily decorated with O-linked nonulosonic sugars that can be modified with both CO2 (35) and amidino (33) residues and that the NCTC 12673 phage may express surface enzymes that are capable of modifying these structures.
Identification of the RBP.
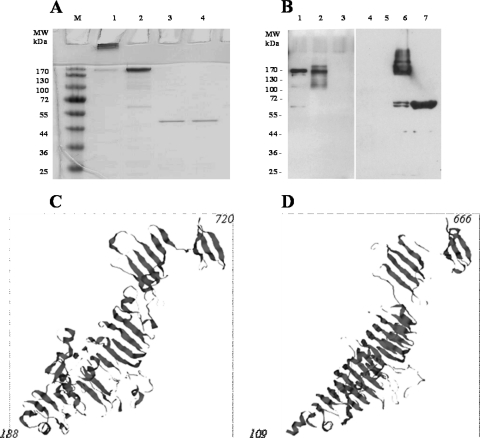
Identification of the phage NCTC 12673 RBP was among the main goals of this research. No gene with homology to any of the T4 tail fiber genes was identified in the genome of phage NCTC 12673. A further problem is that the “closest” relative of phage NCTC 12673 is the cyanobacterial phage S-PM2 which lamentably also lacks recognizable T4 fiber homologues. We used mass and synteny (gene position) and identified gene 48 as a potential gene for the fiber RBP (see File S1 in the supplemental material). Gp48 is also homologous to the C termini of CBJ93981.1 and CBJ94379.1, hypothetical proteins of Campylobacter phages CP220 and CPt10, respectively. Gene 48 was cloned into the pGEX 6P-2 plasmid and expressed as the soluble GST fusion protein with the total molecular mass of ∼180 kDa. Immobilized recombinant Gp48 was able to specifically recognize C. jejuni cells and thus was active as an RBP (Fig. 3A to D). It was also able to agglutinate C. jejuni suspensions similar to the phage P22 TSP agglutination of its host bacterium, S. Typhimurium (Fig. 3E and F). GST-Gp48 also formed SDS-resistant oligomers (Fig. 4A), a feature that is characteristic of a group of kinetically stable proteins. This group comprises several unrelated proteins including P22 TSP (21, 27). A similar experiment was performed with another NCTC 12673 virion protein— Gp1—that was also produced as a GST fusion. It can be seen that neither the GST tag nor Gp1 protein can form SDS-resistant oligomers (Fig. 4A). Thus, such behavior is specific to Gp48. Also, the GST-Gp48 antiserum was able to recognize the P22 TSP SDS-resistant trimers but not the unfolded monomers (Fig. 4B). This reactivity suggests that the P22 TSP and Gp48 may share a conformational epitope in spite of the lack of significant sequence homology between the two proteins. Interestingly, molecular modeling also revealed potential structural similarity between a portion of Gp48 and P22 TSP (Fig. 4C and D). The latter was shown to be a trimer with subunits having a characteristic beta-helix fold (30, 31). These findings reveal a novel RBP with potential application in the diagnosis and treatment of Campylobacter infections.
Fig. 3.
Cell binding properties of GST-Gp48. Scanning electron micrographs of C. jejuni 11168H cells (A and B), E. coli K-12 (C), or S. Typhimurium cells (D) captured on gold surfaces covered with GST-Gp48 (A, C, and D) or GST alone (B). (E) Agglutination of C. jejuni (ΔflaA) cells was observed upon incubation with the GST-Gp48 fusion protein. Wells 1 to 10 contained bacterial cells mixed with doubling dilutions of GST-Gp48. Well 11 contained no protein. Well 12 contained no cells. The minimal agglutinating concentration was determined to be 4 μg of GST-Gp48/ml. (F) A similar experiment was performed with S. Typhimurium cells using P22 TSP lacking endorhamnosidase activity. Well 6 contained no protein, and well 7 contained no cells. The minimal agglutinating concentration was determined to be 3 μg/ml for P22 TSP.
Fig. 4.
Experimental and model structural features of Gp48. (A) Electrophoretic behavior of the GST-Gp48 under SDS-PAGE conditions. Portions (5 μg) of protein per lane were loaded. M, molecular weight markers; lane 1, GST-Gp48 sample incubated in SDS-PAGE sample buffer (2% SDS) for 15 min at room temperature; lane 2, same as lane 1, but preheated at 95°C for 10 min; lanes 3 and 4, same as lanes 1 and 2, respectively, but experiments were performed with GST-Gp1. (B) Western blot analysis of Gp48 and wild-type P22 TSP with anti-Gp48 and anti-P22 antisera. Portions (10 μg) of protein per lane were loaded. For lanes 1 to 3, the membrane was probed with anti-GST-Gp48 antiserum; for lanes 4 to 7, the membrane was probed with anti-P22 TSP antiserum. Sample preparation was as described for panel A. Lane 1 (control), heated sample of GST-Gp48; lane 2, P22 TSP was not heated; lane 3, P22 TSP was heated; lane 4, GST-Gp48 was not heated; lane 5, GST-Gp48 was heated; lane 6, P22 TSP was not heated; lane 7, P22 TSP was heated. (C) Spatial model of Gp48. Gp48's three-dimensional structure was predicted using Phyre (11) software with a 95% probability score. (D) Structure of P22 TSP monomer (PDB ID 1TYV).
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the valuable assistance of Rakesh Bhatnagar, Randy Mandryk, and Arlene Oatway of the University of Alberta, Department of Biological Sciences Advanced Microscopy Facility. We also thank Brendan Wren for providing the C. jejuni flaA mutant. We especially thank Luc Tessier and Simon Foote for assistance with protein identification, as well as Jamshid Tanha for providing the anti-P22 TSP antiserum and P22 TSP expression plasmids. We are also grateful to J. Tanha and all of the Szymanski lab members—past and present—for helpful discussions.
Funding for these studies was provided to C.M.S. by Dow AgroSciences and the Alberta Innovates Centre for Carbohydrate Science. A.M.K. was supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada. C.M.S. holds an Alberta Innovates Scholar Award.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 30 September 2011.
REFERENCES
- 1. Ackermann H.-W., Abedon S. T. 2000. Bacteriophage names 2000: a compilation of known bacteriophages. Ohio State University, Mansfield, OH: http://mansfield.osu.edu/∼sabedon/names.htm [Google Scholar]
- 2. Altschul S. F., Koonin E. V. 1998. Iterated profile searches with PSI-BLAST: a tool for discovery in protein databases. Trends Biochem. Sci. 23: 444–447 [DOI] [PubMed] [Google Scholar]
- 3. Arya S. K., et al. 2011. Chemically immobilized T4-bacteriophage for specific Escherichia coli detection using surface plasmon resonance. Analyst 136: 486–492 [DOI] [PubMed] [Google Scholar]
- 4. Atterbury R. J., Connerton P. L., Dodd C. E., Rees C. E., Connerton I. F. 2003. Application of host-specific bacteriophages to the surface of chicken skin leads to a reduction in recovery of Campylobacter jejuni. Appl. Environ. Microbiol. 69: 6302–6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Atterbury R. J., Connerton P. L., Dodd C. E., Rees C. E., Connerton I. F. 2003. Isolation and characterization of Campylobacter bacteriophages from retail poultry. Appl. Environ. Microbiol. 69: 4511–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carvalho C. M., et al. 2010. The in vivo efficacy of two administration routes of a phage cocktail to reduce numbers of Campylobacter coli and Campylobacter jejuni in chickens. BMC Microbiol. 10: 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Connerton P. L., et al. 2004. Longitudinal study of Campylobacter jejuni bacteriophages and their hosts from broiler chickens. Appl. Environ. Microbiol. 70: 3877–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Forterre P. 2006. DNA topoisomerase V: a new fold of mysterious origin. Trends Biotechnol. 24: 245–247 [DOI] [PubMed] [Google Scholar]
- 9. Frost J. A., Kramer J. M., Gillanders S. A. 1999. Phage typing of Campylobacter jejuni and Campylobacter coli and its use as an adjunct to serotyping. Epidemiol. Infect. 123: 47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grajewski B. A., Kusek J. W., Gelfand H. M. 1985. Development of a bacteriophage typing system for Campylobacter jejuni and Campylobacter coli. J. Clin. Microbiol. 22: 13–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kelley L. A., Sternberg M. J. E. 2009. Protein structure prediction on the web: a case study using the Phyre server. Nat. Protoc. 4: 363–371 [DOI] [PubMed] [Google Scholar]
- 12. Khakhria R., Lior H. 1992. Extended phage-typing scheme for Campylobacter jejuni and Campylobacter coli. Epidemiol. Infect. 108: 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kropinski A. M., et al. 2009. In silico identification of genes in bacteriophage DNA. Methods Mol. Biol. 502: 57–89 [DOI] [PubMed] [Google Scholar]
- 14. Kutateladze M., Adamia R. 2010. Bacteriophages as potential new therapeutics to replace or supplement antibiotics. Trends Biotechnol. 28: 591–595 [DOI] [PubMed] [Google Scholar]
- 15. Kutter E., Sulakvelidze A. 2004. Bacteriophages: biology and applications. CRC Press, Inc., Boca Raton, FL [Google Scholar]
- 16. Lavigne R., Seto D., Mahadevan P., Ackermann H.-W., Kropinski A. M. 2008. Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tools. Res. Microbiol. 159: 406–414 [DOI] [PubMed] [Google Scholar]
- 17. Lavigne R., et al. 2009. Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiol. 9: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lin F. R. 1986. Studies on phage typing of Campylobacter jejuni/coli. Wei Sheng Wu Xue Bao Acta Microbiol. Sinica 26: 373–374 [PubMed] [Google Scholar]
- 19. Lindmark B., et al. 2009. Outer membrane vesicle-mediated release of cytolethal distending toxin (CDT) from Campylobacter jejuni. BMC Microbiol. 9: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Loc-Carrillo C., et al. 2005. Bacteriophage therapy to reduce Campylobacter jejuni colonization of broiler chickens. Appl. Environ. Microbiol. 71: 6554–6563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Manning M., Colon W. 2004. Structural basis of protein kinetic stability: resistance to sodium dodecyl sulfate suggests a central role for rigidity and a bias toward beta-sheet structure. Biochemistry 43: 11248–11254 [DOI] [PubMed] [Google Scholar]
- 22. Miller W. G., Mandrell R. E. 2005. Prevalence of Campylobacter in the food and water supply: incidence, outbreaks, isolation, and detection, p. 101–163 In Ketley J. M., Konkel M. E. (ed.), Campylobacter: molecular and cellular biology. Horizon Bioscience, Norfolk, United Kingdom [Google Scholar]
- 23. Perler F. B. 2002. InBase, the intein database. Nucleic Acids Res. 30: 383–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sails A. D., Wareing D. R., Bolton F. J., Fox A. J., Curry A. 1998. Characterisation of 16 Campylobacter jejuni and C. coli typing bacteriophages. J. Med. Microbiol. 47: 123–128 [DOI] [PubMed] [Google Scholar]
- 25. Salama S. M., Bolton F. J., Hutchinson D. N. 1990. Application of a new phagetyping scheme to campylobacters isolated during outbreaks. Epidemiol. Infect. 104: 405–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, NY [Google Scholar]
- 27. Sanchez-Ruiz J. M. 2010. Protein kinetic stability. Biophys. Chem. 148: 1–15 [DOI] [PubMed] [Google Scholar]
- 28. Singh A., et al. 2010. Bacteriophage tailspike proteins as molecular probes for sensitive and selective bacterial detection. Biosens. Bioelectron. 26: 131–138 [DOI] [PubMed] [Google Scholar]
- 29. Slesarev A. I., et al. 2002. The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens. Proc. Natl. Acad. Sci. U. S. A. 99: 4644–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Steinbacher S., et al. 1996. Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors. Proc. Natl. Acad. Sci. U. S. A. 93: 10584–10588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Steinbacher S., et al. 1994. Crystal structure of P22 tailspike protein: interdigitated subunits in a thermostable trimer. Science 265: 383–386 [DOI] [PubMed] [Google Scholar]
- 32. Ternhag A., Torner A., Svensson A., Ekdahl K., Giesecke J. 2008. Short- and long-term effects of bacterial gastrointestinal infections. Emerg. Infect. Dis. 14: 143–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thibault P., et al. 2001. Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J. Biol. Chem. 276: 34862–34870 [DOI] [PubMed] [Google Scholar]
- 34. Timms A. R., et al. 2010. Evidence for a lineage of virulent bacteriophages that target Campylobacter. BMC Genomics 11: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van Alphen L. B., et al. 2008. A functional Campylobacter jejuni maf4 gene results in novel glycoforms on flagellin and altered autoagglutination behavior. Microbiol. 154: 3385–3397 [DOI] [PubMed] [Google Scholar]
- 36. Wagenaar J. A., van Bergen M. A., Mueller M. A., Wassenaar T. M., Carlton R. M. 2005. Phage therapy reduces Campylobacter jejuni colonization in broilers. Vet. Microbiol. 109: 275–283 [DOI] [PubMed] [Google Scholar]
- 37. Waseh S., et al. 2010. Orally administered P22 phage tailspike protein reduces Salmonella colonization in chickens: prospects of a novel therapy against bacterial infections. PLoS One 5: e13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. 1970. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology 40: 734–744 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.