Abstract
We established a microarray for the simultaneous detection and identification of diverse putative pathogens often associated with fishery products by targeting specific genes of Listeria monocytogenes, Salmonella, Shigella, Staphylococcus aureus, Streptococcus pyogenes, Vibrio cholerae, Vibrio parahaemolyticus, Vibrio vulnificus, and Yersinia enterocolitica and the 16S-23S rRNA gene internal transcribed spacer (ITS) region of Proteus mirabilis and Proteus vulgaris. The microarray contained 26 specific probes and was tested against a total of 123 target bacterial strains that included 55 representative strains, 68 clinical isolates, and 45 strains of other bacterial species that belonged to 8 genera and 34 species, and it was shown to be specific and reproducible. A detection sensitivity of 10 ng DNA or 10 CFU/ml for pure cultures of each target organism demonstrated that the assay was highly sensitive and reproducible. Mock and real fishery product samples were tested by the microarray, and the accuracy was 100%. The DNA microarray method described in this communication is specific, sensitive, and reliable and has several advantages over traditional methods of bacterial culture and antiserum agglutination assays.
INTRODUCTION
Waterborne diseases such as cholera and typhoid in humans are caused by bacteria such as Vibrio, Salmonella, and Shigella spp. (13). In addition to these severe infectious pathogens, Listeria monocytogenes, Salmonella, Shigella, Streptococcus pyogenes, Staphylococcus aureus, Vibrio cholerae (non-O1), Vibrio parahaemolyticus, and Yersinia enterocolitica are also considered moderately hazardous (2). In China, many cases of Proteus mirabilis and Proteus vulgaris contamination of fishery products have been reported by the Entry-Exit Inspection and Quarantine Bureau. In this study, 11 groups of bacterial pathogens, namely, L. monocytogenes, Salmonella, Shigella, S. aureus, S. pyogenes, V. cholerae, V. parahaemolyticus, Vibrio vulnificus, Y. enterocolitica, P. mirabilis, and P. vulgaris (3), were targeted for development of a DNA microarray for detection.
The conventional culture-based methods used for microbial detection and identification are technically simple and inexpensive but laborious and time-consuming, as they take about 3 to 5 days to complete (4, 20). Molecular biology techniques based on microbial genotyping or DNA sequencing have emerged recently as common tools in biological research and pathogen detection. The reported cases include detection of Salmonella spp. by invA gene-based PCR techniques (17), detection of V. cholerae by PCR amplification of the tdh gene (24), and detection of L. monocytogenes by real-time PCR targeting the ssrA gene (22) and multiplex PCR to detect virulence-associated genes (prfA, plcA, hlyA, actA, and iap) (10). PCR and real-time PCR methods have gained significant popularity for use as molecular tools; however, multiplex PCR remains unstable, and the possibility of generating nonspecific products has hindered its wider application in diagnostics. Moreover, TaqMan probe-based real-time PCR is limited by its ability to detect only four of the commercially available fluorophores in a single reaction tube, and the use of SYBR green fluorescence followed by melting temperature determination has insufficient accuracy for detection of multiple target products (18). An oligonucleotide-based microarray assay is a more efficient approach for parallel analysis of a large number of specific sequences. In the microarray assay, the target molecule (DNA) to be analyzed is fluorescently labeled and then hybridized by base pair matching to its cognate recognition probe. Since the sequences of probes on the microarray are pathogen specific, the detection signals generated upon hybridization provide the basis for pathogen identification. Several studies have demonstrated the applicability of oligonucleotide arrays for detection of microbes in the environment (8, 14, 30, 31, 32).
In this work, the target genes for primer and probe design were hlyA for L. monocytogenes (27), invA for Salmonella (17), ipaH for Shigella (26), nuc for S. aureus (6), speB for S. pyogenes (5), rfbE for V. cholerae (15), toxR for V. parahaemolyticus (12), rpoS for V. vulnificus (11), ail for Y. enterocolitica (9), and the internal transcribed spacer (ITS) region for P. mirabilis and P. vulgaris (3).
MATERIALS AND METHODS
Bacterial strains.
The strains used in this study are listed in Table 1. Fifty-five representative strains and 68 isolates of target pathogens from the Tianjin Entry-Exit Inspection and Quarantine Bureau and nine hospitals in Tianjin, China, isolated from 2005 to 2007, were used. In addition, 45 strains of other bacterial species were used to validate the probe specificity of the microarray.
Table 1.
Bacterial strains used in this study
| Bacterial species | No. of strains from each source | Total no. of strains |
|---|---|---|
| Target bacterial species used to test the specificity of the probes | ||
| Listeria monocytogenes | 2,b 2,e 1k | 5 |
| Proteus mirabilis | 1,f 2,l 7,m 6n | 16 |
| Proteus vulgaris | 1,b 5,l 1,m 10n | 17 |
| Salmonella | 2,a 3,b 3,c 6,d 1,e 3,f 6g | 24 |
| Shigella | 1,a 2,b 1,d 3,g 3h | 10 |
| Staphylococcus aureus | 1,a 5,b 1,c 2,d 1i | 10 |
| Streptococcus pyogenes | 2,b 1e | 3 |
| Yersinia enterocolitica | 1,f 2,g 1,p 1q | 5 |
| Vibrio cholerae | 4r | 4 |
| Vibrio parahaemolyticus | 3,b 3,c 11,e 1,f 6j | 24 |
| Vibrio vulnificus | 1,f 3,j 1r | 5 |
| Other bacterial species used to test the specificity of the probes | ||
| Bacillus cereus | 2,d 1,e 3f | 6 |
| Bacillus subtilis | 1b | 1 |
| Bacillus thermodenitrificans | 1q | 1 |
| E. coli O157:H7 | 1t | 1 |
| Listeria innocua | 1m | 1 |
| Listeria welshimeri | 1m | 1 |
| Proteus myxofaciens | 1,c 1n | 2 |
| Proteus penneri | 1f | 1 |
| Staphylococcus caprae | 1b | 1 |
| Staphylococcus capitis | 1a | 1 |
| Staphylococcus epidermidis | 1,s 1d | 2 |
| Staphylococcus haemolyticus | 1,e 1k | 2 |
| Staphylococcus lentus | 1b | 1 |
| Staphylococcus saprophyticus | 1d | 1 |
| Staphylococcus sciuri | 1c | 1 |
| Staphylococcus simulans | 1,d 1,o 1m | 3 |
| Staphylococcus vitulinus | 1b | 1 |
| Staphylococcus warneri | 1c | 1 |
| Streptococcus agalactiae | 1a | 1 |
| Streptococcus bovis | 1c | 1 |
| Streptococcus canis | 1c | 1 |
| Streptococcus faecalis | 1d | 1 |
| Streptococcus faecium | 1b | 1 |
| Streptococcus lactis | 1d | 1 |
| Streptococcus mitis | 1d | 1 |
| Streptococcus porcinus | 1k | 1 |
| Streptococcus salivarius | 1d | 1 |
| Streptococcus suis | 1e | 1 |
| Vibrio hollisae | 1j | 1 |
| Vibrio fluvialis | 1,f 1r | 2 |
| Vibrio furnissii | 1k | 1 |
| Vibrio minicus | 1j | 1 |
| Vibrio alginolyticus | 1f | 1 |
| Yersinia rohdei | 1c | 1 |
| Bacterial species used to perform the double-blinded test (n = 20) | ||
| Bacillus cereus | 1f | 1 |
| Bacillus thermodenitrificans | 1q | 1 |
| E. coli O157:H7 | 1t | 1 |
| Listeria innocua | 1m | 1 |
| Listeria monocytogenes | 1,e 1k | 2 |
| Proteus mirabilis | 1,l 1m | 2 |
| Proteus vulgaris | 1l | 1 |
| Salmonella | 1g | 1 |
| Shigella | 1g | 1 |
| Staphylococcus haemolyticus | 1c | 1 |
| Group B Streptococcus type III | 1c | 1 |
| Streptococcus pyogenes | 1b | 1 |
| Streptococcus salivarius | 1d | 1 |
| Streptococcus sanguis | 1d | 1 |
| Vibrio parahaemolyticus | 1j | 1 |
| Vibrio vulnificus | 1j | 1 |
| Vibrio furnissii | 1j | 1 |
| Yersinia enterocolitica | 1p | 1 |
National Center for Veterinary Culture Collections (CVCC), Beijing, China.
National Center for Medical Culture Collections (CMCC), Beijing, China.
American Type Culture Collection (ATCC), Manassas, VA.
Institute of Microbiology, Chinese Academy of Sciences (AS), Beijing, China.
Academy of Military Medical Sciences (AMMS), Beijing, China.
Tianjin Entry-Exit Inspection and Quarantine Bureau, Tianjin, China.
School of Molecular and Microbial Biosciences, University of Sydney, Sydney, Australia.
Chinese Center for Disease Control and Prevention, Beijing, China.
Juntendo University, Tokyo, Japan.
Soochow University, Taiwan, China.
Agricultural Culture Collection of China (ACCC), Beijing, China.
Culture Collection of the University of Goteborg (CCUG), Gothenburg, Sweden.
Czech Culture Collection of Type Culture, Institute of Hygiene, Prague, Czech Republic.
Department of Immunobiology of Bacteria Institute of Microbiology and Immunology University of Lodz, Lodz, Poland.
Clinical isolate from Tianjin First Centre Hospital, Tianjin, China.
National Collection of Type Cultures (NCTC), Central Public Health Laboratory, London, United Kingdom.
German Collection of Microorganisms and Cell Cultures (DSMZ), Germany.
Shanghai Municipal Center for Disease Control and Prevention, Shanghai, China.
Tianjin Policy Hospital, Tianjin, China.
Robert Koch-Institut (RKI), Berlin, Germany.
Genomic DNA extraction.
All bacteria were cultured in 2YT medium (31), with the exception of L. monocytogenes, which was grown in tryptic soy broth-yeast extract (TSB-YE; Qingdao Hope Bio-Technology Co., Ltd., Shandong, China). Genomic DNA was extracted from 1.5 ml of overnight bacterial culture (approximately 108 CFU/ml) by using a DNA extraction kit (Tiangen, Beijing, China).
Two-step culture enrichment of diluted cultures and real samples.
Evaluation of the effectiveness of the diagnostic approach was carried out by comparing the accuracy of detection with diluted cultures as mock samples and real specimens. The diluted cultures were individual bacterial cultures that had been diluted serially to 1 to 10 CFU/ml, followed by a two-step culture process. (i) One hundred microliters of diluted culture, a swab of fish, and 250 ml of 2YT medium were mixed for activation of target bacteria at 37°C for 6 h, except for L. monocytogenes, which was mixed in TSB-YE. (ii) Ten milliliters of culture from step i was inoculated into 100 ml of alkaline peptone water (APW; Land Bridge Technology Co., Ltd., Beijing, China) for Vibrio spp., 100 ml of enterobacterium enrichment broth (EEB; Land Bridge Technology Co., Ltd., Beijing, China) for Gram-negative bacteria, 100 ml of culture meat infusion broth (MIB; Land Bridge Technology Co., Ltd., Beijing, China) for Gram-positive bacteria, or 100 ml of Half-Fraser medium (Qingdao Hope Bio-Technology Co., Ltd., Shandong, China) for L. monocytogenes. Cultures of these four groups were incubated overnight at 37°C with shaking.
Target genes and oligonucleotide primer design.
The target genes for primer and probe design were hlyA for L. monocytogenes, invA for Salmonella, ipaH for Shigella, nuc for S. aureus, speB for S. pyogenes, rfbE for V. cholerae, toxR for V. parahaemolyticus, rpoS for V. vulnificus, ail for Y. enterocolitica, and the ITS region for P. mirabilis and P. vulgaris. The primer pairs for ITS region (wl-5793 and wl-5794) and ipaH (wl-14621 and wl-14622) amplification were described in our previous study (28), and the other primers were designed using Primer Premier 5.0 software (Premier Boost International, CA). All primer sequences and concentrations used for the multiplex PCR are listed in Table 2.
Table 2.
Primers and their concentrations in multiplex PCR
| Primer | Target gene | Tm (°C) | Direction, sequence (5′-3′)a | Product size (bp) | GenBank accession no. | Primer concn (μM) in multiplex PCR | Primer concn (μM) for labeling |
|---|---|---|---|---|---|---|---|
| Group 1 | |||||||
| wl-5263 | nuc | 58 | F, 53-GAAAGGGCAATACGCAAAGA-72 | 481 | EF529608.1 | 0.1 | |
| wl-5262 | nuc | 65 | R, 533-AGCCAAGCCTTGACGAACTAAAGC-510 | EF529608.1 | 0.1 | 0.1 | |
| wl-5797 | speB | 49.2 | F, 283-CGCTATCACATTTATCCAA-301 | 688 | L26162.1 | 0.1 | |
| wl-5798 | speB | 49.9 | R, 970-AATACCAACATCAGCCATC-952 | L26162.1 | 0.1 | 0.1 | |
| Wl-5805 | invA | 49.7 | F, 536-CCTTTGACGGTGCGATG-552 | 1258 | U43272.1 | 0.2 | |
| Wl-5800 | invA | 51.9 | R, 1793-CCTTTA/GCGAATAACATCCT-1775 | U43272.1 | 0.2 | 0.2 | |
| Wl-37312 | rpoS | 50.4 | F, 331-CTGGCACTGCTTGATTTG-348 | 612 | NC_004459.2 | 0.26 | |
| Wl-37313 | rpoS | 52.4 | R, 943-TCAGAACTTCACGGAGGC-926 | AY187681.1 | 0.26 | 0.26 | |
| Wl-37317 | rfbE | 53.2 | F, 137-TAAAGCACGCCACAACAG-154 | 561 | DQ772987.1 | 0.26 | |
| Wl-37318 | rfbE | 53.1 | R, 697-CAGCACATAGATTCGTCATTC-677 | DQ772987.1 | 0.26 | 0.26 | |
| Wl-37382 | hlyA | 51.2 | F, 447-CAGGTGCTCTCGTGAAAG-464 | 598 | U25446.1 | 0.16 | |
| Wl-37383 | hlyA | 54.5 | R, 1044-TTCCCACTTACGGCAGC-1028 | U25446.1 | 0.16 | 0.16 | |
| Wl-5793 | 16S rRNA geneb | 55.4 | F, 1380-TGTACACACCGCCCGTC-1396 | 500–1,000 | AB553285.1 | 0.08 | |
| Wl-5794 | 23S rRNA geneb | 45.8 | R, 197-GGTACTTAGATGTTTCAGTTC-217 | AY987650.1 | 0.08 | 0.13 | |
| Group 2 | |||||||
| wl-5793 | 16S rRNA geneb | 55.4 | F, 1380-TGTACACACCGCCCGTC-1396 | 500–1,000 | AB553285.1 | 0.08 | |
| wl-5794 | 23S rRNA geneb | 45.8 | R, 197-GGTACTTAGATGTTTCAGTTC-217 | AY987650.1 | 0.08 | 0.1 | |
| wl-14621 | ipaH | 53.5 | F, 374-TTCCTTGACCGCCTTTC-390 | 731 | M76444.1 | 0.2 | |
| wl-14622 | ipaH | 53.3 | R, 1104-GCCAGTACCTCGTCAGTCA-1086 | M76444.1 | 0.2 | 0.16 | |
| wl-37330 | ail | 48.3 | F, 123-TGGGGATACATTGGATAA-140 | 384 | AM286415.1 | 0.3 | |
| wl-37329 | ail | 53.7 | R, 507-GGTGCCAACTTTTGTGCT-490 | M29945.1 | 0.3 | 0.43 | |
| Wl-5259 | toxR | 51 | F, 74-CCAAATAGTAATTCGCTCG-92 | 524 | AB029914.1 | 0.09 | |
| Wl-5260 | toxR | 47.9 | R, 597-CGTGATAATGATGGCTAAAC-578 | AB029914.1 | 0.09 | 0.2 |
F, forward primer; R, reverse primer.
For the ITS region, the forward and reverse primers target the conserved regions of the 16S rRNA and 23S rRNA genes, respectively.
Multiplex PCR and labeling of target genes.
Multiplex PCR was carried out in two groups: the first group consisted of L. monocytogenes, Salmonella, S. aureus, S. pyogenes, V. cholerae, and V. vulnificus, and the second group consisted of Shigella, V. parahaemolyticus, Y. enterocolitica, P. mirabilis, and P. vulgaris. The amplification was performed with 50 μl of a reaction mixture that consisted of 100 ng of DNA, 1× PCR buffer (50 mM KCl, 2.5 mM MgCl2, 10 mM Tris-HCl [pH 8.3]), a 100 μM concentration of each deoxynucleoside triphosphate (dNTP), 2.5 U Taq DNA polymerase (TaKaRa Biotechnology [Dalian] Co. Ltd., China), and each primer (concentrations are shown in Table 2). Reaction parameters were as follows: 94°C for 5 min; 35 cycles of 94°C for 30 s, 50°C for 1 min, and 72°C for 1 min; and a final extension at 72°C for 5 min. An aliquot of 2 μl of PCR product was run in an agarose gel to examine the amplified DNA (Fig. 1). To label PCR products, 10 μM Cy3-dUTP (Amersham Biosciences UK Ltd., Little Chalfont, England) and each reverse primer (concentrations are shown in Table 2) were included in a PCR mixture. Twelve microliters of amplification product generated from the above multiplex PCR was added as the template to 30 μl of PCR mixture. Thermal cycling conditions were the same as for the multiplex PCR. All labeled DNAs were stored at −20°C in the dark.
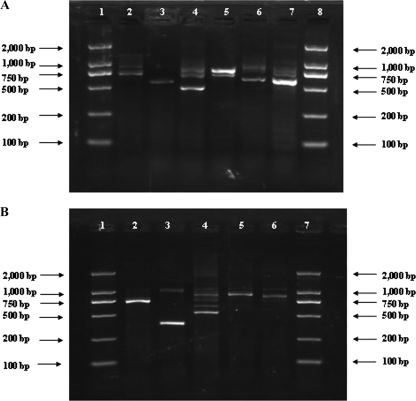
Fig. 1.
Agarose gel electrophoresis of multiplex PCR products. (A) Lanes: 1 and 8, molecular size standard (DL2000 marker); 2, Salmonella; 3, Listeria monocytogenes; 4, Staphylococcus aureus; 5, Streptococcus pyogenes; 6, Vibrio vulnificus; 7, Vibrio cholerae. (B) Lanes: 1 and 7, molecular size standard (DL2000 marker); 2, Shigella; 3, Yersinia enterocolitica; 4, Vibrio parahaemolyticus; 5, Proteus mirabilis; 6, Proteus vulgaris.
Oligonucleotide probe design.
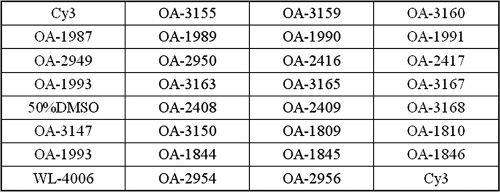
For each type of pathogen, one to four probes were designed by OligoArray 2.0, based on sequences in GenBank. One probe based on the ITS gene was designed as the positive control. A probe containing poly(T)40 was used as the negative control. A probe containing poly(T)40 labeled at the 3′ end with Cy3 was used as the positional reference and printing control. Each probe was 5′-amino modified and followed a spacer of poly(T)10-15 by a stretch of specific sequence (synthesized by AuGCT Biotechnology Corporation, Beijing, China). All of the oligonucleotide probes are listed in Table 3.
Table 3.
Oligonucleotide probes used in this study
| Probe | Target gene or virulence factor | Tm (°C)a | Sequence (5′-3′) | GenBank accession no. |
|---|---|---|---|---|
| OA-1809 | toxR | 74.6 | 115-AGTTGTACGATTAGGAAGCAACGAAAGCCGTATACTC-152 | AB029914.1 |
| OA-1810 | toxR | 74.9 | 177-AAGTTTTAACCCGTAACGAGCTTCACGAGTTTGTTT-212 | AB029914.1 |
| OA-1844 | nuc | 58.3 | 324-GATACACCTGAAACAAAGCATCC-346 | EF529608.1 |
| OA-1845 | nuc | 51.3 | 355-GTGTAGAGAAATATGGTCCTGA-376 | EF529608.1 |
| OA-1846 | nuc | 50.9 | 435-GACAAAGGTCAAAGAACTGAT-455 | EF529608.1 |
| OA-1987 | ipaH | 55.6 | 596-GATAATGATACCGGCGCTCTGCTCTCC-622 | M76444.1 |
| OA-1989 | ipaH | 50 | 702-AGATAGAAGTCTACCTGGCCTTCCAGACCA-731 | M76444.1 |
| OA-1990 | ipaH | 50 | 768-AGGAAATGCGTTTCTATGGCGTGTCG-793 | M76444.1 |
| OA-1991 | ipaH | 51.9 | 883-ACCATGGCATGCTGTACTGAAGCGTAC-909 | M76444.1 |
| OA-2408 | ITS | 62.2 | 209-GAATAACTAAGCTAATTCAAATGAGTTATCTTACT-243 | FJ518147.1 |
| OA-2409 | ITS | 68.2 | 481-CCACCCAGATAGTCTTTGAAAGAGACACTTT-511 | FJ518156.1 |
| OA-2416 | ITS | 70.9 | 405-AGCGCACAGTCAGCGCAACATACATTA-431 | FJ518156.1 |
| OA-2417 | ITS | 63.8 | 491-CCCAGACGTCATTAAGAAGAAACATCT-517 | FJ518583.1 |
| OA-2949 | invA | 73.2 | 1454-GCAACGTCAATGAATATTTCGGTATTCAGGAAAC-1487 | L26162.1 |
| OA-2950 | invA | 70.9 | 1741-GAATTACGAGCAGTAATGGTATCTGCTGAAGTTG-1774 | L26162.1 |
| OA-2954 | speB | 72.3 | 647-GGTAACCCTTACAACCTATTGACACCTGTTATTGAAA-683 | L26162.1 |
| OA-2956 | speB | 74.1 | 827-CCATATTTCAACCATCCTAAGAACTTGTTTGCAGC-861 | L26162.1 |
| OA-3147 | ail | 66.8 | 230-ATGATTTCTTCTATGGCAGTAATAAGTTTGGTC-262 | M29945.1 |
| OA-3150 | ail | 69.1 | 355-GGAAAGGTTAAGGCATCTGTATTTGATGAATC-386 | M29945.1 |
| OA-3155 | rfbE | 69 | 410-CTTTAAGAGATCTGTGTGATGAGCACGGC-438 | DQ772987.1 |
| OA-3159 | rfbE | 71.5 | 619-AACCAAGGGGTAGTGGCAGGGAAGC-643 | DQ772987.1 |
| OA-3160 | rfbE | 70.7 | 564-CATCACATCGGGCGAGGGTGGTAT-568 | DQ772987.1 |
| OA-3163 | rpoS | 71.7 | 563-ATGAGCCAACCGCTGAAGAGATCGC-587 | AY187681.1 |
| OA-3165 | rpoS | 70 | 597-GGATATTCCGGTTGACGATGTGAGCA-622 | AY187681.1 |
| OA-3167 | rpoS | 69.2 | 864-TGGGCAAGAGATTGGTTTAACTCGTGA-890 | AY187681.1 |
| OA-3168 | hlyA | 71.6 | 854-CCTACAAGACCTTCCAGATTTTTCGGCAA-882 | U25446.1 |
| OA-1993 | 16S rRNA gene | 71.9 | 1380-TTGTACACACCGCCCGTCACACCAT-1404b | X80725 |
| WL-4006 | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTc | |||
| Cy3 | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT-Cy3d |
Predicted using Primer Premier 5.0 software.
The 16S rRNA gene-based probe was used as the positive control.
The poly(T)40 probe was used as the negative control.
The poly(T)40 probe labeled with 3′-Cy3 was used as the positional reference and printing control.
DNA array preparation and hybridization.
The probes were dissolved in 50% dimethyl sulfoxide (DMSO) to a final concentration of 1 μg/μl and were printed onto aldehyde group-modified glass slides (CapitalBio Corporation, Beijing, China) by using a SpotArray 72 instrument (Perkin-Elmer Corporation, CA). Each probe was spotted in triplicate to eliminate irregular data due to physical defects in the glass slides. Printed slides were dried and stored at room temperature in the dark. Before use, the slides were scanned at 532 nm for spotting quality control. Each glass slide consisted of eight individual arrays framed with a 20-μl Geneframe (CapitalBio Corporation, Beijing, China), which constituted individual reaction chambers. A schematic diagram of the probe positions on the microarray is shown in Fig. 2.
Fig. 2.
Probe positions on the slide. OA-1993 is the positive-control probe based on the 16S rRNA gene. WL-4006 is the negative-control probe. Cy3 is the positional reference and printing control probe. The rest of the probes are specific probes for the target strains.
Hybridization was performed by the following procedure. All 30 μl of labeled PCR product was baked for about 2 h at 65°C until dry and then diluted in 20 μl of hybridization buffer (30% formamide, 0.5% SDS, 6× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 5× Denhardt's solution). The mixture was then applied to a hybridization chamber and incubated at 43°C for 12 h in a water bath. After incubation, the slide was washed sequentially in solution A (1× SSC, 0.2% SDS) for 3 min, solution B (0.2× SSC) for 3 min, and solution C (95% alcohol) for 1.5 min. The slide was dried under a gentle airstream before it was scanned.
Data acquisition and automated analysis.
The slide was scanned with laser beams at 532 nm by using a model 4100A biochip scanner (Axon Corporation, CA) with the following parameters: photomultiplier tube gain of 600 and pixel size of 5 μm. The images were saved as .tif files, and the signal intensities were saved as .gpr files. The signal-to-noise ratio was calculated for each spot by using Bactarray Analyzer 1.0, developed in-house, with the threshold set at 3.0. A detection result was recorded as positive only when all hybridization signals generated by the probes of the given target genes were above the signal-to-noise threshold.
RESULTS
Optimization of PCRs.
Multiplex PCR was used for amplification of the genes of pathogens, with the first group consisting of L. monocytogenes, Salmonella, S. aureus, S. pyogenes, V. cholerae, and V. vulnificus and the second group consisting of Shigella, V. parahaemolyticus, Y. enterocolitica, P. mirabilis, and P. vulgaris. Initially, a concentration of 0.2 μM was used for all primers. However, several pathogens failed to generate expected hybridization signals under these conditions. For those genes which failed to be amplified, the primer concentrations were adjusted upward, and for those genes which were readily amplified, the primer concentrations were adjusted downward to minimize interference. Consequently, primer concentrations in the range of 0.08 μM to 0.43 μM were tested to find the optimal amplification conditions (Table 2). With the optimized concentrations, the amplicons of the 11 groups of target pathogens were amplified, and the lengths of PCR products varied from 481 to 1,000 bp (Fig. 1).
Probe specificity.
The DNA microarray was tested using 81 representative strains, 87 environmental or clinical isolates of target pathogens, and 45 strains of other bacterial species. A total of 29 probes were used for the microarray, including 26 probes for specific genes of 11 groups of target pathogens, 1 for positive control, 1 for negative control, and 1 for a positional reference and printing control (Table 3). All of the strains belonging to the 11 target groups consistently hybridized to their corresponding probes, with 100% specificity, whereas their closely related strains failed to generate any positive signals. The hybridization results are shown in Fig. 3, panels 1 to 11.
Fig. 3.
Microarray differentiation of pathogens. (1) Listeria monocytogenes; (2) Salmonella; (3) Shigella; (4) Staphylococcus aureus; (5) Streptococcus pyogenes; (6) Vibrio cholerae; (7) Vibrio parahaemolyticus; (8) Vibrio vulnificus; (9) Yersinia enterocolitica; (10) Proteus mirabilis; (11) Proteus vulgaris; (12) Vibrio parahaemolyticus and Shigella; (13) Vibrio parahaemolyticus, Shigella, and Yersinia enterocolitica; (14) Salmonella, Vibrio cholerae, Vibrio vulnificus, and Streptococcus pyogenes.
To evaluate the reproducibility of the assay, 11 strains representing 11 groups of target pathogens were selected. The experiments were repeated three times, applying the same conditions for each strain. For such analysis, all of the target strains produced expected hybridization patterns, and the signal-to-noise ratio of each probe was above the threshold of 3.0.
Sensitivity of detection with genomic DNA.
Serial 10-fold dilutions of genomic DNAs of 11 target pathogens, ranging from 0.1 ng to 100 ng, were used as templates for multiplex PCR to test the sensitivity of the microarray assay. The positive signals generated were 0.1 ng DNA for V. parahaemolyticus; 1 ng DNA for L. monocytogenes, Salmonella, Shigella, S. aureus, S. pyogenes, and V. cholerae; and 10 ng DNA for V. vulnificus, Y. enterocolitica, P. mirabilis, and P. vulgaris. The sensitivity of detection with genomic DNA was set at 10 ng DNA.
Simultaneous detection of multiple pathogens.
Genomic DNAs from two pathogenic isolates of V. parahaemolyticus and Shigella, three pathogenic isolates of V. parahaemolyticus, Shigella, and Y. enterocolitica, and four pathogenic isolates of Salmonella, V. cholerae, V. vulnificus and S. pyogenes were mixed and used as templates to test the specificity of the microarray assay. The data demonstrated that the probes were able to be hybridized and also able to detect multiple pathogens in samples that contained multiple genomic profiles (Fig. 3, panels 12 to 14).
Double-blinded test.
A double-blinded test was performed in order to verify the reliability and specificity of the microarray. A total of 20 environmental and clinical isolates (Table 1) were selected to hybridize to the microarray without disclosure of their identity during testing, including isolates of Bacillus cereus (n = 1), Bacillus thermodenitrificans (n = 1), Escherichia coli O157:H7 (n = 1), Listeria innocua (n = 1), L. monocytogenes (n = 2), P. mirabilis (n = 2), P. vulgaris (n = 1), Salmonella (n = 1), Shigella (n = 1), Staphylococcus haemolyticus (n = 1), group B Streptococcus type III (n = 1), S. pyogenes (n = 1), Streptococcus salivarius (n = 1), Streptococcus sanguis (n = 1), V. parahaemolyticus (n = 1), V. vulnificus (n = 1), Vibrio furnissii (n = 1), and Y. enterocolitica (n = 1). The detection results were consistent with those obtained by conventional methods (data not shown).
Tests of mock samples.
Pure cultures of each of the 11 target bacterial pathogens, i.e., L. monocytogenes, Salmonella, Shigella, S. aureus, S. pyogenes, V. cholerae, V. parahaemolyticus, V. vulnificus, Y. enterocolitica, P. mirabilis, and P. vulgaris, were diluted from 101 to 106 CFU per ml, mixed with 250 ml of 2YT medium, and tested on the microarray. All of the targets were detected at levels as low as 10 CFU/ml (data not shown).
Tests of real samples and confirmation by sequencing.
A total of 20 batches of fish samples, including two catfishes, three loaches, seven croakers, and eight Chinese hooksnout carps, were collected from a local market and analyzed by the microarray. The hybridization profiles showed that one was contaminated by Shigella, two by S. aureus, one by P. mirabilis, three by P. vulgaris, and one by both P. mirabilis and P. vulgaris. These findings were confirmed consistently by nuc sequencing techniques for S. aureus, ipaH sequencing for Shigella, ureR sequencing for P. mirabilis, and ITS sequencing for P. vulgaris. In addition, 12 samples tested showed positive-control probe signals, which suggested that other bacteria besides the 11 pathogens studied were present. Overall, the microarray results produced 100% accuracy.
DISCUSSION
This is the first report of comprehensive detection and identification of 11 major groups of pathogens associated with fishery products. A two-step multiplex PCR was used for amplification and labeling: in the first step, the target genes were amplified by the forward and reverse primers, and in the second step, the single-stranded DNA was labeled by the reverse primers. The two-step PCR not only enhanced the amplification efficiency but also generated single-stranded PCR products for hybridization. The primer concentrations were optimized based on the intensities of hybridization signals, which were analyzed by interpretation software developed in-house. Initially, the same concentration (0.2 μM) was employed for all primers, but the signals were negative or weak for some of the target genes and were very strong for others. Therefore, different primer concentrations were tested to determine the best possible combination (Table 2). As little as 10 ng DNA could be detected by the microarray. Such a level of sensitivity permits the detection of bacteria after culture enrichment.
Previous studies have indicated that microarrays based on the 16S rRNA gene are not specific enough and cannot differentiate closely related species, such as Proteus mirabilis, Proteus penneri, and Proteus vulgaris, as they share high-level (up to 99%) homology in the 16S rRNA gene sequence. In comparison with the 16S rRNA gene, the sequence of the ITS region is considered to be under more evolutionary pressure and is therefore prone to more genetic variation. Sequence and length polymorphisms of ITS regions have been used increasingly as tools for bacterial species and/or subspecies identification (19, 21) and typing (16, 25), as well as for evolutionary studies (1, 7, 23, 29). Primers wl-5793 and wl-5794 were designed based on highly conserved regions of the 16S and 23S rRNA genes and were used successfully to amplify the ITS regions of all strains. Probe OA-1993 was used as the positive control, probes OA-2408 and OA-2409 were found to be specific for P. vulgaris, and probes OA-2416 and OA-2417 were found to be specific for P. mirabilis.
In the case of environmentally obtained samples, the bacterium number can be as low as a few CFU and could fall below the threshold of detection by immune-based or molecular assays. To overcome this problem, a two-step culture process was applied to enrich the target bacteria. In the first step, culture medium was used to enrich all bacteria that may have been present in the samples, and in the second step, a selective culture medium was employed. After the two-step culture, the bacterium content reached levels that were detectable by the microarray.
In conclusion, this study presents a new multiplex PCR-based microarray assay for the detection and identification of 11 groups of bacterial pathogens. The sensitivity and specificity of the described method make it suitable for applications in basic microbiological research, clinical diagnosis, food safety, and epidemiological surveillance.
ACKNOWLEDGMENTS
This study was supported by the National 863 Program (2006AA020703 and 2009AA06Z403), the National 973 Program of China (2009CB522603), and the Fundamental Research Funds for the Central Universities.
Footnotes
Published ahead of print on 30 September 2011.
REFERENCES
- 1. Anton A. I., Martinez-Murcia A. J., Rodriguez-Valera F. 1998. Sequence diversity in the 16S-23S intergenic spacer region (ISR) of the rRNA operons in representatives of the Escherichia coli ECOR collection. J. Mol. Evol. 47: 62–72 [DOI] [PubMed] [Google Scholar]
- 2. Brown A. C. 2008. Food safety, p. 53–55 In Understanding food—principles and preparation, 3rd ed. Thomson Learning Inc. Press, Belmont, CA [Google Scholar]
- 3. Cao B., et al. 2009. 16S-23S rDNA internal transcribed spacer regions in four Proteus species. J. Microbiol. Methods 77: 109–118 [DOI] [PubMed] [Google Scholar]
- 4. de Boer E., Beumer R. R. 1999. Methodology for detection and typing of foodborne microorganisms. Int. J. Food Microbiol. 50: 119–130 [DOI] [PubMed] [Google Scholar]
- 5. Dmitrieva N. F., et al. 2002. Frequency of genes speA, speB, and speC in Streptococcus pyogenes strains and the identification of the infective agent by polymerase chain reaction. Zh. Mikrobiol. Epidemiol. Immunobiol. 5: 3–6 [PubMed] [Google Scholar]
- 6. Elizaquivel P., Aznar R. 2008. A multiplex RTi-PCR reaction for simultaneous detection of Escherichia coli O157:H7, Salmonella spp. and Staphylococcus aureus on fresh, minimally processed vegetables. Food Microbiol. 25: 705–713 [DOI] [PubMed] [Google Scholar]
- 7. Gurtler V. 1999. The role of recombination and mutation in 16S-23S rDNA spacer rearrangements. Gene 238: 241–252 [DOI] [PubMed] [Google Scholar]
- 8. Han W., et al. 2007. DNA microarray-based identification of serogroups and virulence gene patterns of Escherichia coli isolates associated with porcine postweaning diarrhea and edema disease. Appl. Environ. Microbiol. 73: 4082–4088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang Y., et al. 2010. Possible use of ail and foxA polymorphisms for detecting pathogenic Yersinia enterocolitica. BMC Microbiol. 10: 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kaur S., Malik S. V., Vaidya V. M., Barbuddhe S. B. 2007. Listeria monocytogenes in spontaneous abortions in humans and its detection by multiplex PCR. J. Appl. Microbiol. 103: 1889–1896 [DOI] [PubMed] [Google Scholar]
- 11. Kim D. G., et al. 2008. Application of the rpoS gene for species-specific detection of Vibrio vulnificus by real-time PCR. J. Microbiol. Biotechnol. 18: 1841–1847 [DOI] [PubMed] [Google Scholar]
- 12. Kim Y. B., et al. 1999. Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J. Clin. Microbiol. 37: 1173–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leclerc H., Schwartzbrod L., Dei-Cas E. 2002. Microbial agents associated with waterborne diseases. Crit. Rev. Microbiol. 28: 371–409 [DOI] [PubMed] [Google Scholar]
- 14. Li Y., et al. 2006. Development of a serotype-specific DNA microarray for identification of some Shigella and pathogenic Escherichia coli strains. J. Clin. Microbiol. 44: 4376–4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu Y., et al. 2006. Electronic deoxyribonucleic acid (DNA) microarray detection of viable pathogenic Escherichia coli, Vibrio cholerae, and Salmonella typhi. Anal. Chim. Acta 578: 75–81 [DOI] [PubMed] [Google Scholar]
- 16. Maggi R. G., Chomel B., Hegarty B. C., Henn J., Breitschwerdt E. B. 2006. A Bartonella vinsonii berkhoffii typing scheme based upon 16S-23S ITS and Pap31 sequences from dog, coyote, gray fox, and human isolates. Mol. Cell. Probes 20: 128–134 [DOI] [PubMed] [Google Scholar]
- 17. Mainar-Jaime R. C., Atashparvar N., Chirino-Trejo M. 2008. Estimation of the diagnostic accuracy of the invA-gene-based PCR technique and a bacteriological culture for the detection of Salmonella spp. in caecal content from slaughtered pigs using Bayesian analysis. Zoonoses Public Health 55: 112–118 [DOI] [PubMed] [Google Scholar]
- 18. Maynard C., et al. 2005. Waterborne pathogen detection by use of oligonucleotide-based microarrays. Appl. Environ. Microbiol. 71: 8548–8557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mora D., Ricci G., Guglielmetti S., Daffonchio D., Fortina M. G. 2003. 16S-23S rRNA intergenic spacer region sequence variation in Streptococcus thermophilus and related dairy streptococci and development of a multiplex ITS-SSCP analysis for their identification. Microbiology 149: 807–813 [DOI] [PubMed] [Google Scholar]
- 20. Muytjens H. L., Roelofs-Willemse H., Jaspar G. H. 1988. Quality of powdered substitutes for breast milk with regard to members of the family Enterobacteriaceae. J. Clin. Microbiol. 26: 743–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nakanishi S., Kuwahara T., Nakayama H., Tanaka M., Ohnishi Y. 2005. Rapid species identification and partial strain differentiation of Clostridium butyricum by PCR using 16S-23S rDNA intergenic spacer regions. Microbiol. Immunol. 49: 613–621 [DOI] [PubMed] [Google Scholar]
- 22. O'Grady J., et al. 2009. Rapid detection of Listeria monocytogenes in food using culture enrichment combined with real-time PCR. Food Microbiol. 26: 4–7 [DOI] [PubMed] [Google Scholar]
- 23. Perez Luz S., Rodriguez-Valera F., Lan R., Reeves P. R. 1998. Variation of the ribosomal operon 16S-23S gene spacer region in representatives of Salmonella enterica subspecies. J. Bacteriol. 180: 2144–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Raghunath P., Acharya S., Bhanumathi A., Karunasagar I., Karunasagar I. 2008. Detection and molecular characterization of Vibrio parahaemolyticus isolated from seafood harvested along the southwest coast of India. Food Microbiol. 25: 824–830 [DOI] [PubMed] [Google Scholar]
- 25. Regassa L. B., et al. 2004. Differentiation of group VIII Spiroplasma strains with sequences of the 16S-23S rDNA intergenic spacer region. Can. J. Microbiol. 50: 1061–1067 [DOI] [PubMed] [Google Scholar]
- 26. Riyaz-Ul-Hassan S., Syed S., Johri S., Verma V., Qazi G. N. 2009. Application of a multiplex PCR assay for the detection of Shigella, Escherichia coli and Shiga toxin-producing Esch. coli in milk. J. Dairy Res. 76: 188–194 [DOI] [PubMed] [Google Scholar]
- 27. Rudi K., et al. 2003. Subtyping Listeria monocytogenes through the combined analyses of genotype and expression of the hlyA virulence determinant. J. Appl. Microbiol. 94: 720–732 [DOI] [PubMed] [Google Scholar]
- 28. Wang M., et al. 2009. Detection of Enterobacter sakazakii and other pathogens associated with infant formula powder by use of a DNA microarray. J. Clin. Microbiol. 47: 3178–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang M., et al. 2008. Analysis of the 16S-23S rRNA gene internal transcribed spacer region in Klebsiella species. J. Clin. Microbiol. 46: 3555–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang Q., et al. 2007. Development of a DNA microarray to identify the Streptococcus pneumoniae serotypes contained in the 23-valent pneumococcal polysaccharide vaccine and closely related serotypes. J. Microbiol. Methods 68: 128–136 [DOI] [PubMed] [Google Scholar]
- 31. Wen L., et al. 2006. Use of a serotype-specific DNA microarray for identification of group B Streptococcus (Streptococcus agalactiae). J. Clin. Microbiol. 44: 1447–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou G., et al. 2011. Development of a DNA microarray for detection and identification of Legionella pneumophila and ten other pathogens in drinking water. Int. J. Food Microbiol. 145: 293–300 [DOI] [PubMed] [Google Scholar]