Abstract
The cell wall of the fruiting body of the mushroom Lentinula edodes is degraded after harvesting by enzymes such as β-1,3-glucanase. In this study, a novel endo-type β-1,3-glucanase, GLU1, was purified from L. edodes fruiting bodies after harvesting. The gene encoding it, glu1, was isolated by rapid amplification of cDNA ends (RACE)-PCR using primers designed from the N-terminal amino acid sequence of GLU1. The putative amino acid sequence of the mature protein contained 247 amino acid residues with a molecular mass of 26 kDa and a pI of 3.87, and recombinant GLU1 expressed in Pichia pastoris exhibited β-1,3-glucanase activity. GLU1 catalyzed depolymerization of glucans composed of β-1,3-linked main chains, and reaction product analysis by thin-layer chromatography (TLC) clearly indicated that the enzyme had an endolytic mode. However, the amino acid sequence of GLU1 showed no significant similarity to known glycoside hydrolases. GLU1 has similarity to several hypothetical proteins in fungi, and GLU1 and highly similar proteins should be classified as a novel glycoside hydrolase family (GH128).
INTRODUCTION
The cell walls of filamentous fungi have been investigated in several species of basidiomycota, including Schizophyllum commune (26), Agaricus bisporus (15), Coprinopsis cinerea (1), and Lentinula edodes (24a). These reports have indicated that the major components of the cell wall are chitin and β-1,3-glucan with β-1,6-linked branches. Several β-1,3-glucans from basidiomycetous mushrooms display antitumor activity; examples include lentinan from L. edodes (2) and schizophyllan from S. commune (17). Although lentinan can be purified from fresh shiitake mushrooms (L. edodes), it is degraded during storage as a result of β-1,3-glucanase activity (13, 14). This suggests that β-1,3-glucanases are involved in cell wall lysis in the process of fruiting body senescence after harvesting.
During the filamentous fungal life cycle, cell walls are synthesized, reoriented, and lysed (27, 16). Cell wall lysis and changes in the constituent polysaccharides are essential for fruiting body development in basidiomycota (5, 6, 7). Autolysis of the pileus of fruiting bodies in C. cinerea also occurs following basidiospore formation (11). These observations suggest that cell wall-lytic enzymes, such as β-1,3-glucanase, are very important to the morphology of basidiomycetous fungi. There have been few reports on purified glucanases in basidiomycetes, such as endo-β-1,3-glucanase in A. bisporus (3) and β-1,3(4)-glucanase in Phanerochaete chrysosporium (8); however, the physiological and biological functions of endo-β-1,3-glucanase in basidiomycetous fungi are unclear.
Previously, we reported that two exo-β-1,3-glucanase-encoding genes (exg1 and exg2) are involved in morphogenesis of L. edodes (21, 22) and that the enzyme encoded by exg2 is also involved in postharvest cell wall degradation (22). One endo-β-1,3-glucanase, TLG1, which is a homolog of thaumatin-like protein, was purified from L. edodes fruiting bodies after harvest (4). The gene, tlg1, encoding the enzyme was isolated and characterized (23). TLG1 is involved in cell wall degradation after harvest (23). Sakamoto et al. (23, 24) also pointed out that endoglucanase(s) other than TLG1 are expressed in the L. edodes fruiting body. β-1,3-Glucanases are classified into several families based on their amino acid sequence similarities in the Carbohydrate-Active enZYmes (CAZy) server database (http://www.cazy.org/Glycoside-Hydrolases.html). Major β-1,3-glucanases are classified into glycoside hydrolase (GH) families 5, 16, 17, 55, 64, and 81. Of the β-1,3-glucanases from L. edodes, EXG1 is classified into GH5 and EXG2 is classified into GH55. However, endoglucanase TLG1 has not been classified into any GH family.
In this study, we purified and characterized a novel endo-β-1,3-glucanase, GLU1, from the fruiting body of L. edodes. We also cloned a gene, glu1, encoding GLU1, and based on its amino acid sequence, proposed that GLU1 belongs to a new GH family.
MATERIALS AND METHODS
Strain and culture conditions.
The commercially cultivated L. edodes dikaryotic strain H600 (22) was used in all experiments. For RNA and protein extraction, harvested mature fruiting bodies were transferred immediately to a desiccator at 25°C (21) through day 4. Following postharvest preservation, all samples were separated into pileus, gills (lamellae), and stipe and then frozen immediately in liquid nitrogen. All samples frozen in liquid nitrogen were stored at −80°C.
Measurement of glucanase activity.
β-1,3-Glucanase activity was measured using 0.5% laminarin (Sigma-Aldrich, Inc., St. Louis, MO) in 50 mM sodium acetate buffer (pH 4.2) at 37°C for 30 min. Reducing sugars liberated from the substrate were analyzed according to the Somogyi-Nelson method (25). One unit (U) of enzyme activity was defined as the amount of enzyme that produces 1 μmol reducing sugar per minute under the above-described conditions. To determine the kinetic properties of GLU1, the reactions were performed with 0.25 to 10 mg/ml of laminarin. To examine substrate specificity, 1% of various glucans (laminarin, lentinan [13], pachyman, CM-pachyman, curdlan, CM-curdlan, lichenan, barley glucan [Megazyme International Ireland Ltd., Ireland], and pustulan [Calbiochem, CA]) were incubated with GLU1. The effects of pH and temperature on enzyme function were analyzed as described previously (10). Assays to assess the effect of pH on activity and stability were carried out using the following buffers: 50 mM citric acid-sodium phosphate at pH 3 to 5, 50 mM sodium acetate at pH 4.0 to 6.0, 50 mM sodium phosphate at pH 6.0 to 8.0, and 50 mM Tris-HCl at pH 8.0 to 9.0. The effect of temperature on the activity was determined at temperatures in the range of 10 to 80°C. To analyze thermostability, aliquots of the enzyme were treated in 50 mM sodium phosphate buffer (pH 7.0) at 10 to 80°C for 30 min.
Purification of β-1,3-glucanase.
Proteins were extracted from 20 g of fruiting body 4 days after harvest. Samples were crushed in liquid nitrogen, suspended in 50 ml of 10 mM sodium phosphate buffer (pH 7.0), and incubated with rotation for 15 min at room temperature. Ammonium sulfate was added until the concentration reached 30% saturation. The resulting precipitate was removed by centrifugation at 12,000 × g for 20 min. The supernatant was applied to a HiPrep phenyl column (GE Healthcare, United Kingdom) that had been equilibrated with 10 mM sodium phosphate buffer (PB) (pH 7.0) containing ammonium sulfate at a concentration of 30% saturation. The column was washed with the same buffer, and proteins were eluted using a linear concentration gradient (30 to 0%) of saturated ammonium sulfate (160 ml) in PB at a flow rate of 4 ml/min. During the purification steps, β-1,3-glucanase activities were assessed using AZCL-pachyman (Megazyme) as a substrate following the method of Sakamoto et al. (23). Fractions containing glucanase activity were collected and concentrated using an Amicon Ultra 5,000 NMWL (nominal molecular weight limit) filter (Millipore, Bedford, MA). The concentrated sample was dialyzed against PB and then applied to a HiLoad Sepharose Q anion exchange column (GE Healthcare). Absorbed proteins were eluted using a linear concentration gradient of NaCl (0 to 0.5 M) at a flow rate of 3 ml/min. Fractions containing β-1,3-glucanase activity were collected and concentrated by ultrafiltration through an Amicon Ultra 10,000 NMWL filter (Millipore). Concentrated proteins were then applied to a Superdex 75 10/30 gel filtration column (GE Healthcare) that had been equilibrated in 10 mM sodium phosphate buffer (pH 6.5), and proteins were eluted with the same buffer at a flow rate of 0.25 ml/min. Purified GLU1 was analyzed using SDS-PAGE, and the N-terminal amino acid sequence of GLU1 was analyzed as described by Sakamoto et al. (21).
Cloning and sequencing of the glu1 gene.
cDNA was synthesized from total RNA extracted from the gills of fruiting bodies preserved at day 4 postharvest (day 4 RNA) using the SMART RACE cDNA amplification kit (BD Biosciences, CA), according to the manufacturer's protocol. 3′ Rapid amplification of cDNA ends (RACE) was performed using degenerate primers (glu-1U, 5′-YTN GCN TGG CCN TGG TAY AA-3′, and glu-2U, 5′-TGG CCN TGG TAY AAY WSN CC-3′) designed using the N-terminal amino acid sequence of GLU1, as described by Sakamoto et al. (21). cDNA for the 5′ RACE template was synthesized from day 4 RNA using a GeneRacer kit (Invitrogen, CA), and PCR was performed as described previously (22) using glu1-specific (glu1-182L-RACE, 5′-CAT CCT TGT TGG GCT TGA CGG GTA G-3′, and glu1-461L-RACE, 5′-GAC GGA AAT TGG TTG TGG GCT TGT T-3′) and GeneRacer primers (Invitrogen).
Heterologous expression of glu1.
Vector pCIZα-A in an EasySelect Pichia expression kit (Invitrogen) was used for transformation and expression of recombinant GLU1. A DNA fragment that encodes mature GLU1 was amplified using a gene-specific primer pair (glu1-Nt-pCIZ, 5′-CCG GAA TTC GGA AAA CGA GGA TTG GCT TG-3′, and glu1-TGAL-pCIZ, 3′-GTA CATC AAG TTAC CAA CAG TCT AGA CTA G-5′) and was introduced into the EcoRI and XbaI cleavage sites of the vector pCIZα-A (pPCIZ-glu1). The plasmid pPCIZ-glu1 was linearized with BPU-1102I (TaKaRa Bio, Inc., Japan) and transformed into Pichia pastoris strain KM18. The transformant was cultivated in growth medium and then in induction medium as described by Kawai et al. (8). Recombinant protein was purified in steps similar to those described above for the purification of native GLU1 from L. edodes fruiting body.
Western blot analysis.
Rabbit anti-GLU1 was prepared (custom service of TaKaRa Bio, Inc.) using three peptides (SSPVAQLATRQAQQGC, NEPDINGITPAEAASC, and KKALPAVTSSTTSGQGC) identified as potential GLU1 epitopes using Epitope Adviser 2.1 software (FQS, Japan). Protein samples were prepared from the gills of fruiting bodies by crushing them in liquid nitrogen, followed by suspension in extraction buffer (200 mM sodium acetate, pH 4.2). Protein concentrations were determined using the Bio-Rad protein assay kit (Bio-Rad Laboratories, CA) with a bovine serum albumin (BSA) standard. Samples were separated by electrophoresis and transferred to a polyvinylidene difluoride (PVDF) membrane following Sakamoto et al. (22). Western blot analysis was carried out as described previously (22); rabbit anti-GLU1 was used as the first antibody, followed by horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (GE Healthcare) as the secondary antibody. Hybridization was visualized using an enhanced chemiluminescence (ECL) detection kit (GE Healthcare) following the manufacturer's instructions.
TLC analysis of reaction products.
To analyze the mode of action of GLU1, laminarin (1%, wt/vol) was incubated with GLU1 (54 nM) in 50 mM sodium acetate (pH 4.2) at 30°C for 24 h, and the reaction mixture (4 μl) was applied to a thin-layer chromatography (TLC) plate (silica gel grade 60 F254; Merck Co., Germany). The reaction products were developed with ethyl acetate (EtOAc)-CH3COOH-water (3:2:1, by volume) and detected using 5% (vol/vol) sulfuric acid and 0.5% (vol/vol) thymol in ethanol.
Nucleotide sequence accession number.
The glu1 gene sequence was submitted to DDBJ under accession number AB571655.
RESULTS AND DISCUSSION
Purification of GLU1 and cloning of glu1.
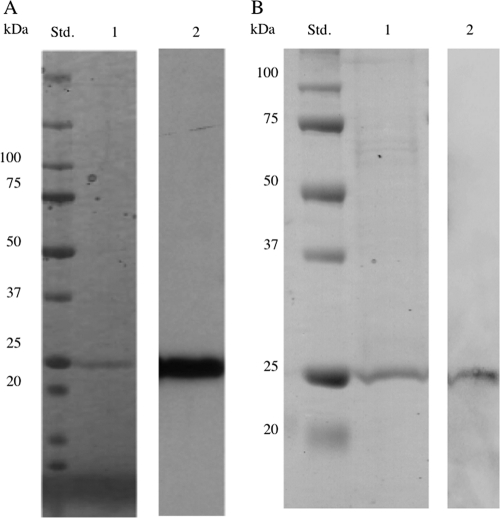
GLU1 was purified from fruiting bodies 4 days after harvest by hydrophobic chromatography and continuous ion-exchange chromatography while monitoring its activity based on degradation of AZCL-pachyman. An unknown peak of β-1,3-glucanase activity was clearly separated from β-1,3-glucanases EXG2 (22) and TLG1 (23) by ion-exchange chromatography. After gel filtration, a peak of activity corresponding to a single major band (Fig. 1A, lane 1) was observed by SDS-PAGE and Coomassie brilliant blue staining. The molecular mass of the purified glucanase (GLU1) was determined to be 25 kDa from SDS-PAGE (Fig. 1A). The N-terminal amino acid sequence of GLU1 was GKRGLAWPWYNSPLD.
Fig. 1.
(A) SDS-PAGE and Western blot analysis of purified GLU1. Lanes: Std., molecular mass standards; 1, SDS-PAGE and Coomassie brilliant blue staining of purified GLU1; 2, Western blot analysis of GLU1 using anti-GLU1 antipeptide serum. (B) SDS-PAGE and Western blot analysis of purified recombinant GLU1. Lanes: Std., molecular mass standards; 1, SDS-PAGE and Coomassie staining of purified recombinant GLU1; 2, Western blot analysis of purified recombinant GLU1 using anti-GLU1.
The glu1 gene was cloned using the N-terminal amino acid sequence of GLU1. The fragment amplified by 3′ RACE using degenerate primers was subcloned and sequenced, and the putative N-terminal amino acid sequence was identical to that of the GLU1 N terminus. Following 5′ RACE, the gene encoding GLU1 was cloned and sequenced. The nucleotide sequence of glu1 was deposited in the DDBJ/EMBL/GenBank databases under accession number AB571655. Using the PSORT II program (http://psort.ims.u-tokyo.ac.jp/form2.html), the mature protein was predicted to be an extracellular or cell wall protein, with a potential cleavage site between residues 20 and 21 (see Fig. S1 in the supplemental material). Thus, the putative mature protein is comprised of 247 amino acid residues with a molecular mass of 26 kDa and a pI of 3.87. The predicted N-terminal amino acid sequence was identical to that of GLU1 (see Fig. S1). GLU1 antipeptide serum (anti-GLU1) was prepared from epitopes predicted from the deduced amino acid sequence of glu1. The anti-GLU1 cross-reacted with purified GLU1 (Fig. 1A, lane 2). The recombinant GLU1 was expressed in P. pastoris, and purified protein was detected as a single band by SDS-PAGE with Coomassie staining (Fig. 1B). The purified protein immunoreacted with anti-GLU1 (Fig. 1B), showing that it was recombinant GLU1. When recombinant GLU1 was incubated with laminarin, its hydrolyzing activity was as good as that of the wild-type enzyme (data not shown). These data collectively indicated that glu1 encodes active β-1,3-glucanase.
The deduced amino acid sequence of the glu1 product was analyzed using the BLASTP algorithm; one significantly similar hypothetical protein (<e−100) was found in the S. commune genome (XP_003029493), and several less-similar hypothetical proteins (>e−20) were found in several fungal genomes. However, the deduced amino acid sequence of the glu1 did not have any similarities to known GH families in CAZy.
Characterization of enzymatic properties of GLU1.
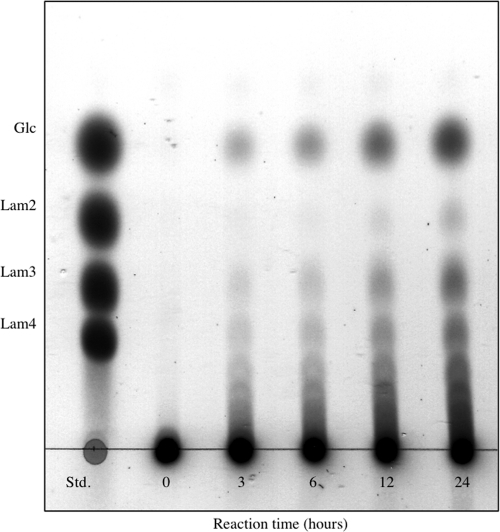
To study its mode of action, GLU1 was incubated with laminarin and the enzymatic reaction products were analyzed by TLC (Fig. 2). The reaction led to the formation of glucose and a series of laminarioligosaccharides, indicating that GLU1 is an endo-type β-1,3-glucanase. However, oligosaccharides formed during the reaction accumulated without further degradation even after incubation for 24 h. These results suggested a low hydrolyzing activity of GLU1 toward laminarioligosaccharides.
Fig. 2.
TLC pattern of the hydrolytic products from laminarin by GLU1. Laminarin (1%, wt/vol) was incubated with wild-type GLU1 (54 nM) in 50 mM sodium acetate buffer (pH 4.2) at 30°C. Standards (Std.) were glucose (Glc), laminaribiose (Lam2), laminaritriose (Lam3), and laminaritetraose (Lam4).
Effects of pH and temperature on GLU1 function were analyzed. The optimal conditions for degradation of laminarin by the enzyme were pH 4 at 50°C. GLU1 was stable over a broad pH range, from 3 to 9, when kept at 4°C for 20 h. The enzyme was completely inactivated after incubation at 60°C for 30 min.
The substrate specificity of purified GLU1 was examined using various polysaccharides (Table 1). Both soluble and insoluble glucans composed of β-1,3-linked main chains were readily hydrolyzed by GLU1, and laminarin was the most favorable substrate for the enzyme, with a specific activity of 1.85 U/mg (Km, 1.67 ± 0.18 [mean ± standard deviation], and kcat, 0.84 ± 0.051 s−1). Lentinan, a β-1,3/1,6-glucan from L. edodes (β-1,3:1,6 linkage ratio of 3:2) (18, 12), was also degraded by GLU1, but the activity for lentinan was less than the activity for laminarin (a ratio of 7:1) (20), suggesting that β-1,6 linkages prevent hydrolysis of β-1,3 linkages by GLU1. The enzyme did not react with β-1,3/1,4-glucans, such as barley glucan or lichenan, or with β-1,6 glucans, such as pustulan. Thus, the hydrolytic activity of GLU1 was confirmed to be strictly limited to the β-1,3 glycoside linkage in β-1,3-glucans and β-1,3/1,6-glucans. Endo-β-1,3-glucanases are divided into two classes, EC 3.2.1.6 [endo-1,3(4)-β-glucanase] and EC 3.2.1.39 (endo-1,3-β-glucanase), based mainly on substrate specificity. Because GLU1 did not degrade β-1,3 linkages within β-1,3/1,4-glucans, such as barley glucan, the enzyme was categorized into EC 3.2.1.39.
Table 1.
Substrate specificity of wild-type GLU1a
| Substrate | Glycoside linkage(s) | Activity (U/mg) | Relative activity (%)c |
|---|---|---|---|
| Laminarin | β-1,3/1,6 | 1.85 | 100.0 |
| Lentinan | β-1,3/1,6 | 0.081 | 4.4 |
| Pachyman | β-1,3 | 0.28 | 14.9 |
| Curdlan | β-1,3 | 0.17 | 9.1 |
| CM-pachyman | β-1,3 | 0.17 | 9.1 |
| CM-curdlan | β-1,3 | 0.27 | 13.7 |
| Lichenan | β-1,3/1,4 | NDb | ND |
| Barley glucan | β-1,3/1,4 | ND | ND |
| Pustulan | β-1,6 | ND | ND |
Assays were performed using a reaction mixture containing 50 mM sodium acetate buffer (pH 4.2) and 1% (wt/vol) substrate.
ND, not detectable.
Relative activity when the activity using laminarin as a substrate is calculated as 100%.
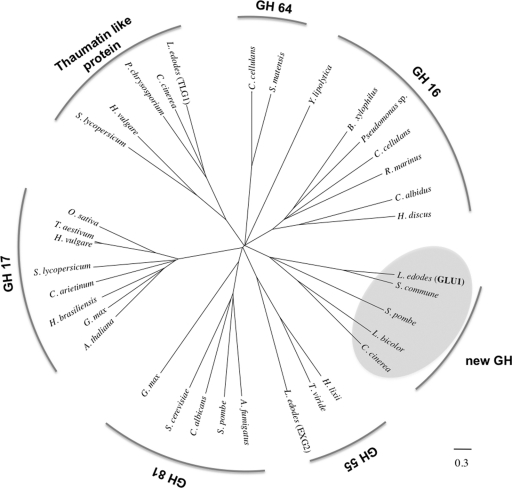
GLU1 belongs to a new GH family.
A phylogenetic tree was drawn based on several sequences: GLU1 and proteins with homology to GLU1, thaumatin-like proteins with β-1,3-glucanase activity, and other EC 3.2.1.39 glucanases on the CAZy server (http://www.cazy.org) (Fig. 3). As shown in Fig. 3, GLU1 does not belong to existing GH families containing EC 3.2.1.39 glucanases. Moreover, amino acid sequence analysis of GLU1 revealed no significant homology with any previously described functional proteins, indicating that this enzyme should be classified in a new family, GH128. The amino acid sequence of GLU1 has significant similarity (e−108) to that of a hypothetical protein (XP_003029493) in the S. commune genome and has low similarity (e−15 to e−12) to hypothetical proteins in fungal genomes, including Schizosaccharomyces pombe NP_592836, C. cinerea XP_001829384, and Laccaria bicolor XP_001873506. These hypothetical proteins have two conserved glutamic acids in their putative amino acid sequences (E103 and E195 in GLU1) (see Fig. S1 in the supplemental material). The deduced sequence of GLU1 was also used to predict its three-dimensional structure using the Phyre server (http://www.sbg.bio.ic.ac.uk/∼phyre/) (9). The result indicated that the three-dimensional structure of GLU1 matches some (β/α)8 TIM barrel structures, such as GH39 β-xylosidase (PDB code, Q9ZFM2; E-value, 0.00012) and GH5 β-mannanase (PDB code, Q4W8M3; E-value, 0.011). Therefore, GLU1 would be categorized in the GH-A clan by the CAZy server, and two conserved glutamic acids in GLU1 (see Fig. S1) are expected to perform as the catalytic residues. These results suggest that GLU1 and similar proteins belong to a new GH family, GH128. This is the first known record of enzyme activity in GH128, and further enzymatic characterization of GLU1 will be helpful for understanding characteristic properties of the GH128 family.
Fig. 3.
Phylogenetic relationship of L. edodes glucanases (GLU1, TLG1, and EXG2), thaumatin-like proteins, and EC 3.2.1.39 β-1,3-glucanases on the CAZy server. Sequences of GLU1, TLG1, EXG2, fungal proteins having homology with GLU1 (XP_003029493 from Schizophyllum commune, NP_592836 from Schizosaccharomyces pombe, XP_001873506 from Laccaria bicolor, and XP_001829384 from Coprinopsis cinerea, highlighted by gray circles), thaumatin-like proteins (CAA50059 from Solanum lycopersicum, AAK55325 from Hordeum vulgare, genome database of Phanerochaete chrysosporium, and XP_001837765 from Coprinopsis cinerea), GH family 16 members (BAE02683 from Bursaphelenchus xylophilus, AAZ04385 from Chlamys albidus, BAH84971 from Haliotis discus, XP_500934 from Yarrowia lipolytica, AAC44371 from Cellulosimicrobium cellulans, BAC16331 from Pseudomonas sp., and AAC69707 from Rhodothermus marinus), GH family 17 members (AAA32756 from Arabidopsis thaliana, CAA10167 from Cicer arietinum, AAA33946 from Glycine max, CAB38443 from Hevea brasiliensis, AAA32958 from Hordeum vulgare, AAD10382 from Oryza sativa, AAA03617 from Solanum lycopersicum, and CAA77085 from Triticum aestivum), GH family 55 members (CAA58889 from Hypocrea lixii and BAD67019 from Trichoderma viride), GH family 64 members (AAA25520 from Cellulosimicrobium cellulans and BAA34349 from Streptomyces matensis), and GH family 81 members (AF121133 from Aspergillus fumigatus, CAB62580 from Candida albicans, BAA11407 from Glycine max, AAB82378 from Saccharomyces cerevisiae, and CAA91245 from Schizosaccharomyces pombe) were aligned using MAFFT (version 6; http://align.bmr.kyushu-u.ac.jp/mafft/online/server/) with the E-INS-I algorithm. A phylogenetic tree was inferred from the alignments with the minimum linkage method and drawn by using FigTree software (version 1.1.2; http://tree.bio.ed.ac.uk/software/figtree/).
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by a Grant-in-Aid for Scientific Research (no. 21880050) to N.K. from the Japan Society for the Promotion of Science (JSPS).
We are grateful to Bernard Henrissat of CNRS and University of Aix-Marseille I and II in Marseille, France, for his help with sequence analysis of GLU1. We also thank Kiyohiko Igarashi of Tokyo University, Daniel Cullen of the USDA, and Toshitsugu Sato of Kitami Institute of Technology, who gave us helpful suggestions.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 30 September 2011.
REFERENCES
- 1. Bottom C. B., Sierhr D. J. 1979. Structure of an alkali-soluble polysaccharide from the hyphal wall of the basidiomycete Coprinus macrorhizus var. microsporus. Carbohydr. Res. 77: 169–181 [Google Scholar]
- 2. Chihara G., Maeda Y., Hamuro J., Sasaki T., Fukuoka F. 1969. Inhibition of mouse sarcoma 180 by polysaccharides from Lentinus edodes (Berk.) sing. Nature 222: 687–688 [DOI] [PubMed] [Google Scholar]
- 3. Galán B., Mendoza C. G., Calonje M., Novaes-Ledieu M. 1999. Production, purification, and properties of an endo-1,3-β-glucanase from the basidiomycete Agaricus bisporus. Curr. Microbiol. 38: 190–193 [DOI] [PubMed] [Google Scholar]
- 4. Grenier J., Potvin C., Trudel J., Asselin A. 1999. Some thaumatin-like proteins hydrolyse polymeric β-1,3-glucans. Plant J. 19: 473–480 [DOI] [PubMed] [Google Scholar]
- 5. Kamada T., Takemaru T. 1977. Stipe elongation during basidiocarp maturation in Coprinus macrorhizus: mechanical properties of stipe cell wall. Plant Cell Physiol. 18: 831–840 [Google Scholar]
- 6. Kamada T., Takemaru T. 1977. Stipe elongation during basidiocarp maturation in Coprinus macrorhizus: changes in polysaccharide composition of stipe cell wall during elongation. Plant Cell Physiol. 18: 1291–1300 [Google Scholar]
- 7. Kamada T., Fujii T., Takemaru T. 1980. Stipe elongation during basidiocarp maturation in Coprinus macrorhizus: changes in activity of cell wall lytic enzymes. Trans. Mycol. Soc. Jpn. 21: 359–367 [Google Scholar]
- 8. Kawai R., Igarashi K., Yoshida M., Kitaoka M., Samejima M. 2006. Hydrolysis of β-1,3/1,6-glucan by glycoside hydrolase family 16 endo-1,3(4)-β-glucanase from the basidiomycete Phanerochaete chrysosporium. Appl. Microbiol. Biotech. 71: 898–906 [DOI] [PubMed] [Google Scholar]
- 9. Kelley L. A., Sternberg M. J. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4: 363–371 [DOI] [PubMed] [Google Scholar]
- 10. Konno N., Sakamoto Y. 2011. An endo-β-1,6-glucanase involved in Lentinula edodes fruiting body autolysis. Appl. Microbiol. Biotechnol. 91: 1365–1373 [DOI] [PubMed] [Google Scholar]
- 11. Kües U. 2000. Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol. Mol. Biol. Rev. 64: 316–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Martin K., McDougall B. M., Mcllroy S., Chen J., Seviour R. J. 2007. Biochemistry and molecular biology of exocellular fungal β-(1,3)- and β-(1,6)-glucanase. FEMS Microbiol. Rev. 31: 168–192 [DOI] [PubMed] [Google Scholar]
- 13. Minato K., Mizuno M., Terai H., Tsuchida H. 1999. Autolysis of lentinan, an antitumor polysaccharide, during storage of Lentinus edodes, Shiitake mushroom. J. Agric. Food Chem. 47: 1530–1532 [DOI] [PubMed] [Google Scholar]
- 14. Minato K., Kakawami S., Nomura K., Tsuchida H., Mizuno M. 2004. An exo β-1,3 glucanase synthesized de novo degrades lentinan during storage of Lentinula edodes and diminishes immunomodulating activity of the mushroom. Carbohydr. Polym. 56: 279–286 [Google Scholar]
- 15. Mol P. C., Wessels J. G. H. 1990. Differences in wall structure between substrate hyphae and hyphae of fruit body stipes in Agarucus bisporus. Mycol. Res. 94: 472–479 [Google Scholar]
- 16. Moore D. 1998. Fungal morphogenesis. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 17. Morikawa K., Takeda R., Yamazaki M., Mizuno D. 1985. Induction of tumoricidal activity of polymorphonuclear leukocytes by a linear β-1,3-D-glucan and other immunomodulators in murine cells. Cancer Res. 45: 1496–1501 [PubMed] [Google Scholar]
- 18. Read S. M., Currie G., Bacic A. 1996. Analysis of the structural heterogeneity of laminarin by electrospray-ionisation-mass spectrometry. Carbohydr. Res. 281: 187–201 [DOI] [PubMed] [Google Scholar]
- 19. Reference deleted.
- 20. Saito H., Tabeta R., Sasaki T., Yoshioka Y. 1986. A high-resolution solid-state 13C NMR study of (1→3)-β-d-glucans from various sources. Conformation characterization as viewed from the conformation-dependent 13C chemical shifts and its consequence to gelation property. Bull. Chem. Soc. Jpn. 59: 2093–2101 [Google Scholar]
- 21. Sakamoto Y., Irie T., Sato T. 2005. Isolation and characterization of a fruiting body-specific exo-β-1,3-glucanase-encoding gene, exg1, from Lentinula edodes. Curr. Genet. 47: 244–252 [DOI] [PubMed] [Google Scholar]
- 22. Sakamoto Y., et al. 2005. Characterization of the Lentinula edodes exg2 gene encoding a lentinan-degrading exo-β-1,3-glucanase. Curr. Genet. 48: 195–203 [DOI] [PubMed] [Google Scholar]
- 23. Sakamoto Y., et al. 2006. Lentinula edodes tlg1 encodes a thaumatin-like protein that is involved in lentinan degradation and fruiting body senescence. Plant Physiol. 141: 793–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sakamoto Y., Nakade K., Sato T. 2009. Characterization of the post-harvest changes in gene transcription in the gill of the Lentinula edodes fruiting body. Curr. Genet. 55: 409–423 [DOI] [PubMed] [Google Scholar]
- 24a. Shida M., Ushioda Y., Nakajima T., Matsuda K. 1981. Structure of the alkali-insoluble skeletal glucan of Lentinus edodes. J. Biochem. 90: 1093–1100 [DOI] [PubMed] [Google Scholar]
- 25. Somogyi M. 1952. Notes on sugar determination. J. Biol. Chem. 195: 19–23 [PubMed] [Google Scholar]
- 26. Wessels J. G. H., Kreger D. R., Marchant R., Regensburg B. A., De Vries O. M. 1972. Chemical and morphological characterization of the hyphal wall surface of the basidiomycete Schizophyllum commune. Biochim. Biophys. Acta 273: 346–358 [DOI] [PubMed] [Google Scholar]
- 27. Wessels J. G. H. 1993. Fruiting in the higher fungi. Adv. Microb. Phys. 34: 147–202 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.