Abstract
Arbuscular mycorrhizal fungi (AMF) play important roles as plant protection agents, reducing or suppressing nematode colonization. However, it has never been investigated whether the galls produced in roots by nematode infection are colonized by AMF. This study tested whether galls produced by Meloidogyne incognita infection in Prunus persica roots are colonized by AMF. We also determined the changes in AMF composition and biodiversity mediated by infection with this root-knot nematode. DNA from galls and roots of plants infected by M. incognita and from roots of noninfected plants was extracted, amplified, cloned, and sequenced using AMF-specific primers. Phylogenetic analysis using the small-subunit (SSU) ribosomal DNA (rDNA) data set revealed 22 different AMF sequence types (17 Glomus sequence types, 3 Paraglomus sequence types, 1 Scutellospora sequence type, and 1 Acaulospora sequence type). The highest AMF diversity was found in uninfected roots, followed by infected roots and galls. This study indicates that the galls produced in P. persica roots due to infection with M. incognita were colonized extensively by a community of AMF, belonging to the families Paraglomeraceae and Glomeraceae, that was different from the community detected in roots. Although the function of the AMF in the galls is still unknown, we hypothesize that they act as protection agents against opportunistic pathogens.
INTRODUCTION
Arbuscular mycorrhizal fungi (AMF) are obligate, symbiotic fungi which form mutualistic associations with the roots of 80% of terrestrial plant species. Apart from improving plant nutrition, AMF play important roles in the reduction of pathogen infections (7, 25). The protective effect of AMF against a broad range of soilborne fungi and bacteria (31, 35), as well as against root-feeding nematodes (14, 17), has been well documented. In some crops, it has been shown that mycorrhizal associations have a suppressive effect on endoparasitic nematodes (12, 32, 39), so AMF could be considered biological control agents (24).
A previous molecular study using PCR-denaturing gradient gel electrophoresis (DGGE) indicated that the presence of nematodes alters the composition of AMF communities inside Ammophila arenaria roots (27). However, when this methodology is used without subsequent cloning procedures, it is not possible to determine the identities of the AMF, since this technique is based on the observation of the composition of the fungal or bacterial communities using DGGE profiles (3).
Earlier reports showed that AMF can colonize nitrogen-fixing legume root nodules (29) and senescent nodules after nitrogen fixation has ceased (30). Also, mycorrhizal nodules belonging to the genus Glomus have been found in the root systems of angiosperms, such as Gymnostoma deplancheanum and Gymnostoma nodiflorum (13). In spite of the ecological and economic relevance of the interactions between AMF and root-knot nematodes, it has never been investigated whether the galls produced in roots by nematode infection are colonized by AMF.
Meloidogyne incognita is the most widespread root-knot nematode and probably the most serious plant-parasitic nematode pest of tropical and subtropical regions throughout the world (41). M. incognita has been found to be associated with Prunus persica (L.) Batsch in Venezuela, where it causes severe decreases in the productivity of this important fruit crop (9, 10).
In the present study, we intended to elucidate whether galls produced by M. incognita infection in P. persica roots are colonized by AMF and to elucidate the changes in AMF composition and biodiversity mediated by infection with this root-knot nematode.
MATERIALS AND METHODS
Study site and sampling.
The study was conducted in a commercial orchard located in Colonia Tovar, Aragua State, in the northern part of Venezuela (10°29′N, 67°07′W; 1,790 m above sea level). Its climate is temperate, with a mean annual temperature of 16.8°C and an average annual rainfall of 1,271 mm (mostly concentrated in a rainy season between June and October). The soil in the experimental area was a sandy loam inceptisol in the USDA soil classification system (33). The soil characteristics were as follows: a pH of 5.18, 5.75% clay, 40.5% silt, 53.75% sand, a cation-exchange capacity of 6.46 cmol kg of soil−1, total N of 2.7 g kg−1, available P of 32 μg g−1, 5.9% organic matter, and a bulk density of 1.29 g cm−3.
The plants used in this survey were 13-year-old peach trees (Prunus persica [L.] Batsch cv. Criollo Amarillo). The experimental sampling was a randomized factorial design with six replication blocks (100 m2 each) in an experimental area of approximately 1,800 m2. Each block consisted of 20 trees, some of them naturally infected by the root-knot nematode. The sampling was conducted during fruiting. Six infected plants and six uninfected plants, one of each in each of the six replication blocks, were sampled. The roots were sampled using three soil cores from three points in a single tree in each block. Roots and galls of M. incognita-infected plants and roots of uninfected plants were selected.
The root samples (secondary and tertiary) were washed with distilled water, and the nematode galls were separated and thoroughly surface washed. The material was frozen until processing.
Nematode identification.
Meloidogyne incognita worms were isolated from the infected roots, and their identity was checked by morphology and sequencing of the internal transcribed spacer (ITS) regions according to the method of de la Peña et al. (11). The uninfected roots were also checked for the absence of nematodes by the same method.
Root and gall DNA extraction and PCR.
All PCR experiments were run using DNA preparations consisting of 100 mg of pooled root and gall extracts for each plant and replication block separately. Thus, 18 DNA extractions from and PCRs of 12 root samples (6 infected plants and 6 uninfected plants) and 6 gall samples were carried out. For each sample, total DNA was extracted from the frozen material (the average root length was 18 cm, and an average of 13 nematode galls were used in each case) using a DNeasy plant minikit by following the manufacturer's recommendations (Qiagen). The root or gall samples were placed into 2-ml screw-cap propylene tubes, and the DNA extracts were obtained by disrupting roots or galls with a sterile, disposable micropestle in liquid nitrogen. The DNA was resuspended in 20 μl of water.
Several dilutions of extracted DNA (1/10, 1/50, and 1/100) were prepared, and 2 μl was used as the template. Partial small-subunit (SSU) rRNA gene fragments were amplified using nested PCR with the universal eukaryotic primers NS1 and NS4 (40). PCR was carried out in a final volume of 25 μl using Ready-To-Go PCR beads (Amersham Pharmacia Biotech), a 0.2 μM concentration of the deoxynucleoside triphosphates (dNTPs), and a 0.5 μM concentration of each primer with the following PCR conditions: 94°C for 3 min and then 30 cycles at 94°C for 30 s, 40°C for 1 min, and 72°C for 1 min, followed by a final extension period at 72°C for 10 min.
Two-microliter samples of several dilutions (1/10, 1/20, 1/50, and 1/100) from the first PCR were used as template DNAs in a second PCR performed using the specific primers AML1 and AML2 (20). PCRs were carried out in a final volume of 25 μl using Ready-To-Go PCR beads (Amersham Pharmacia Biotech), a 0.2 μM concentration of the dNTPs, and a 0.5 μM concentration of each primer with the following PCR conditions: 94°C for 3 min and then 30 cycles of 1 min of denaturation at 94°C, 1 min of primer annealing at 50°C, and 1 min of extension at 72°C, followed by a final extension period of 10 min at 72°C. Positive and negative controls using PCR-positive products and sterile water, respectively, were also included in all amplifications. All of the PCRs were run on a Perkin Elmer Cetus DNA thermal cycler. Reaction yields were estimated by using a 1.2% agarose gel containing ethidium bromide.
Molecular analysis.
The PCR products were purified using a gel extraction kit (Qiagen), cloned into pGEM-T Easy (Promega), and transformed into Escherichia coli (XL1-Blue). Forty putatively positive transformants were screened in each resulting SSU rRNA gene library by using 0.7 units of REDTaq DNA polymerase (Sigma) and a reamplification with AML1 and AML2 primers under the same conditions as those described above. Product quality and size were checked in agarose gels as described above. All clones having inserts of the correct size in each library were sequenced.
Clones were grown in liquid culture, and the plasmid was extracted using the QIAprep Spin miniprep kit (Qiagen). The sequencing was done by the Sistemas Genómicos laboratory (Valencia, Spain) using the universal primers SP6 and T7. Sequence editing was done using the program Sequencher, version 4.1.4 (Gene Codes Corporation).
Sequence similarities were determined using the BLASTn sequence similarity search tool (2) provided by GenBank. Phylogenetic analysis was carried out on the sequences obtained in this study and on those that were the closest matches in GenBank. Sequences were aligned with other published glomalean sequences using the program ClustalX (38), and the alignment was adjusted manually in GeneDoc (22). Neighbor-joining (NJ) (28) and maximum likelihood (ML) phylogenetic analyses were performed with the programs PAUP, version 4.08b (36), and RAxML, v.7.0.4 (34), respectively. Distances for the NJ tree were computed using the default parameters. For the ML analysis, a generalized time-reversible γ (GTR-γ) model of evolution was used. A total of 200 independent bootstrap analyses were performed to provide nodal support. The ML bootstrap values were calculated for 1,000 replicates with the same substitution model. Endogone pisiformis Link and Mortierella polycephala Coem were used as the outgroups.
AMF sequence types, or phylotypes, were defined as groups of closely related sequences, usually with a high level of bootstrap support in the phylogenetic analyses (higher than 80%) and sequence similarity of ≥97%. The pairwise analysis within clusters was carried out using MEGA software, version 4 (37).
The presence or absence of AMF phylotypes in each root and gall sample was used to construct the sampling effort curves (with 95% confidence intervals) using the software EstimateS 8.0 (8). The sample order was randomized by 100 replications.
Statistical analysis.
The Shannon (H′) index was calculated as an additional measure of diversity, as it combines two components of diversity, i.e., species richness and evenness. It is calculated from the equation H′ = −[summ]pi(ln pi), where pi is the proportion of individuals found in the ith species (in a sample, the true value of pi is unknown but is estimated as ni/n [here and throughout, ni is the number of individuals in the ith species]).
We applied a general log-linear analysis (SPSS, version 19.0) to test whether the compositions of AMF communities differed between the three experimental cases. A correspondence analysis (CA) with presence and/or absence data for all AMF sequence types from three experimental cases was performed, and the results were summarized in an ordination diagram. CA is a multivariate statistical method that allows comparisons of AM fungal community compositions between all experimental cases.
Nucleotide sequence accession numbers.
Of the 263 clones generated in this study, 102 representative sequences have been deposited in the National Center for Biotechnology Information (NCBI) GenBank database (http://www.ncbi.nlm.nih.gov) under the accession numbers FR847980 to FR848081.
RESULTS
Phylogeny.
From the 18 clone libraries, a total of 720 clones were screened by PCR; out of these, 263 contained the small-subunit rRNA gene fragment and were subsequently sequenced. All 263 clones corresponded to AMF sequences. The phylogenetic tree constructed using homologous sequences of AMF species from GenBank and our sequences made possible the recognition of 22 different AMF sequence types or phylotypes (Fig. 1), 17 of which belonged to the genus Glomus, 3 to the genus Paraglomus, 1 to the genus Scutellospora, and 1 to the genus Acaulospora. Since identical sequences were detected, the clones producing the same sequences for each experimental case were represented once in the alignment for clarity (unpublished data).
Fig. 1.
Neighbor-joining (NJ) phylogenetic tree showing AM fungal sequences isolated from the roots and galls of M. incognita-infected plants and from the roots of uninfected plants as well as reference sequences from GenBank. All bootstrap values of >80% are shown (1,000 replicates). Numbers above branches indicate the bootstrap values of the NJ analysis; numbers below branches indicate the bootstrap values of the maximum likelihood analysis. Sequences obtained in the present study are shown in bold type. They are identified by the different tissues from which they were obtained (galls [G], roots of infected plants [IP], and roots of noninfected plants [NIP]) and by the clone identity number. Group identifiers (for example, Glo G1) are AM fungal sequence types found in our study. Endogone pisiformis and Mortierella polycephala were used as outgroups.
Eight sequence types corresponded to morphologically defined species (Pa1 corresponded to Paraglomus laccatum, Pa2 to Paraglomus brasilianum, Glo G1 to the Glomus intraradices/Glomus irregulare group, Glo G4 to Glomus sinuosum, Glo G13 to Glomus mosseae, Glo G10 to Glomus indicum, Scu1 to Scutellospora cerradensis, and Aca1 to the Acaulospora scrobiculata/Acaulospora laevis group), and one sequence type (Glo G15) was not related to any sequences found in the database. The remaining sequence types were related to uncultured glomalean species sequences in GenBank (Fig. 1).
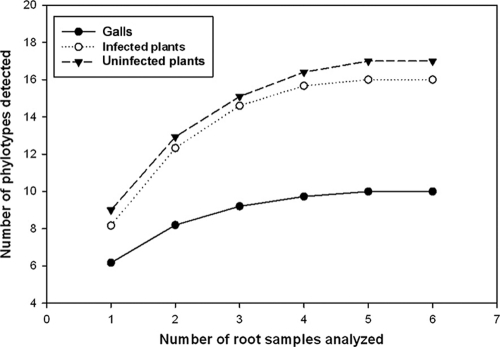
The sampling effort curves (Fig. 2) showed stabilization if the AMF sequence types found and the number of samples analyzed are considered.
Fig. 2.
Sampling effort curves for galls, M. incognita-infected roots, and uninfected roots. The sample order was randomized by 100 replications in EstimateS, version 8.0 (8).
AMF community composition.
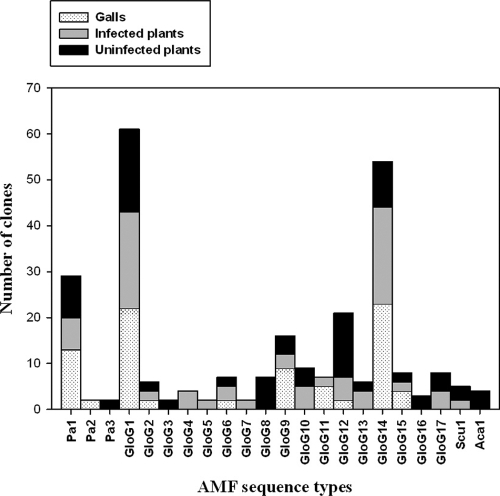
Uninfected plant roots harbored a mean number of AMF sequence types per root sample that was similar to that of infected roots (5.8 ± 0.4 and 5.5 ± 0.3, respectively). The number of AMF sequence types per gall sample showed the lowest mean value (3.7 ± 0.3). The compositions of the AMF communities from roots and galls were determined based on the numbers of clones of the 22 AMF sequence types detected (Fig. 3). A comparative analysis of AMF diversity did not show significant differences between blocks (F = 0.643, P = 0.540).
Fig. 3.
Proportional distribution of the total number of clones detected for each AMF sequence type in galls, the roots of M. incognita-infected plants, and the roots of uninfected plants.
The Shannon diversity index and the total number of AMF sequence types were slightly higher for uninfected than for infected plant roots (the H′ was 2.22 and there were 17 AMF sequence types for uninfected plants, compared with an H′ of 2.05 and 16 AMF sequence types for infected plants). However, their AMF communities were clearly different (P < 0.05) (Fig. 3). Also, the composition of the AMF communities colonizing the galls differed significantly with respect to both infected and uninfected roots (P < 0.001). The galls had both the lowest number of AMF sequence types (10) and the lowest diversity index (H′ = 1.55).
Eight AMF sequence types (Pa1, Glo G1, Glo G2, Glo G6, Glo G9, Glo G12, Glo G14, and Glo G15) occurred in all three experimental cases (Fig. 3). Of these, five (Pa1, Glo G1, Glo G9, Glo G12, and Glo G14) represented the highest numbers of clones, accounting for 68.30% of the AMF clones analyzed. Also, it is noteworthy that the largest number of clones of Pa1, Glo G1, Glo G9, Glo G14, and Glo G15 occurred in the galls. Some AMF sequence types were found exclusively in particular experimental cases analyzed: thus, Pa2 appeared exclusively in galls, while Pa3, Glo G3, Glo G8, Glo G16, and Aca1 were found only in uninfected roots, Glo G4, Glo G5, and Glo G7 were specific to infected plant roots, and Glo G11 appeared only in the galls and roots of infected plants (Fig. 4).
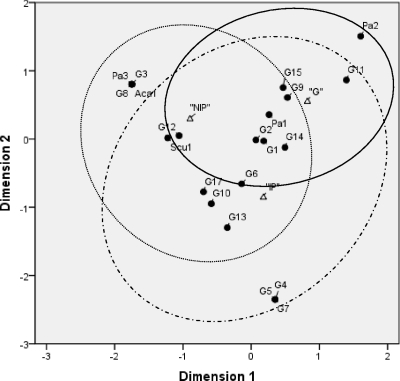
Fig. 4.
Correspondence analysis of the AM fungal community composition found in galls, the roots of M. incognita-infected plants, and the roots of uninfected plants. The eigenvalues of the x and y axes in the two-dimensional ordination diagram are as follows: 0.51 (first dimension) and 0.35 (second dimension). Small black circles represent the respective AMF sequence types, and triangles represent the experimental cases. Ovals drawn with solid, dotted, and dashed lines represent the distributions of AMF diversity in galls (G), the roots of noninfected plants (NIP), and the roots of M. incognita-infected plants (IP), respectively.
DISCUSSION
This study shows, for the first time with a molecular approach, that the galls produced in P. persica roots by M. incognita infection are colonized by a characteristic AMF community that differs clearly from both the root AMF community from which the galls were collected and the AMF community of uninfected roots.
The AMF communities associated with the roots of infected and uninfected plants were also different from each other (Fig. 3). Rodríguez-Echevarría et al. (27) also found that nematodes altered the compositions of the AMF communities inside Ammophila arenaria roots, although they did not identify the AMF species. We found similar numbers of AMF sequence types in infected and uninfected roots (16 and 17, respectively); however, the compositions of the two AMF communities clearly differed, with only 12 fungal sequence types being shared. Although the reason for this association is not known, it could be due to the fact that some AMF species are more sensitive to nematodes than others (19). Changes in root physiology after the nematode infection might have altered root exudation or chemistry (5, 6). In fact, it has been reported that root exudates are fundamental in stimulating the growth of microorganisms due to the release of organic compounds, such as carboxylic acids, and enzymes, such as acid phosphatases (15, 26). Thus, these processes could also affect, in some way, the ability of certain AMF to colonize infected roots.
The galls produced on the infected roots were colonized by two families of glomalean fungi (Paraglomeraceae and Glomeraceae). Between them, 10 AMF sequence types were found; the two most abundant types were Glo G1 and Glo G14. They were also the most abundant sequence types in the infected plant roots from which the galls were collected. Glo G1 is related to the G. intraradices/G. irregulare group, which includes the most-generalistic AMF found in the molecular diversity studies conducted so far (16, 23). Glo G14 matched database sequences belonging to uncultured Glomus, which had been reported previously from Phytolacca americana and Perilla frutescens roots (21). The third-most-abundant sequence type in galls, appearing also in infected and uninfected roots, was Pa1, which corresponds to Paraglomus laccatum. This fungal type was not related to any sequence type in the database obtained from environmental samples. Only the fungal type Pa2, related to Paraglomus brasilianum and reported previously from Panax japonicus roots (20), was found exclusively in galls, and it was represented by two clones, each clone belonging to a different plant.
The galls showed a lower Shannon diversity index (H′ = 1.55) than both infected and uninfected roots (H′ = 2.05 and H′ = 2.22, respectively). Although the results are not directly comparable, Sheublin et al. (29) also found that legume root nodules had less AMF diversity than the roots from which they were collected, corroborating the idea that AMF communities may vary among the different parts of a root system (29). The AMF diversity found in the galls is surprisingly high if we suppose that a greater surface area favors greater colonization by different AMF sequence types, since the gall samples analyzed had a surface area that was around 12 times lower than that of roots with the same weight (71 mm2 per 100 mg for galls and 850 mm2 per 100 mg for roots).
AMF colonization of the galls of nematode-infected roots has not been studied previously. The ecological and physiological roles of these symbionts in root knots are not clear. Root-feeding nematodes stimulate the production of galls (root knots) on the roots of their host plants. The galls disturb the roots' ability to absorb water and nutrients and also can serve as entry points for pathogens, such as fungi and bacteria, which cause plant diseases (4, 5, 26). This could have been the mechanism which enabled the AMF to colonize the galls to a high degree. The highly diverse AMF can compete successfully for space and nutrient sources with other endophytes, which can also use the openings in galls produced by a nematode infection; thus, AMF might protect the galls from opportunistic attack by pathogens.
In conclusion, the galls produced in P. persica roots by M. incognita infection were colonized by an AMF community, belonging to the families Paraglomeraceae and Glomeraceae, that was different from the AMF community detected in the roots. Although the function of the AMF in the galls is still unknown, we hypothesize that the AMF may act as protective agents against opportunistic pathogens. This study was carried out only with P. persica roots; therefore, more research has to be done to test whether the galls produced in other plant species are colonized by AMF, since the outcomes of AMF-nematode interactions are influenced by many factors, including physical, physiological, and temporal factors (17), and functional differences between different AMF taxa (18).
ACKNOWLEDGMENT
M.D.M.A. was supported by the Ramón and Cajal program (Ministerio de Educación y Ciencia, Spain).
Footnotes
Published ahead of print on 7 October 2011.
REFERENCES
- 1. Reference deleted.
- 2. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderson I. C., Cairney J. W. G. 2004. Diversity and ecology of soil fungal communities: increased understanding through the application of molecular techniques. Environ. Microbiol. 6:769–779 [DOI] [PubMed] [Google Scholar]
- 4. Back M. A., Haydock P. P. J., Jenkinson P. 2002. Disease complexes involving plant parasitic nematodes and soilborne pathogens. Plant Pathol. 51:683–697 [Google Scholar]
- 5. Back M., Jenkinson P., Deliopoulos T., Haydock P. 2010. Modifications in the potato rhizosphere during infestations of Globodera rostochiensis and subsequent effects on the growth of Rhizoctonia solani. Eur. J. Plant Pathol. 128:459–471 [Google Scholar]
- 6. Bardgett R. D., Cook R., Yeates G. W., Denton C. S. 1999. The influence of nematodes on below-ground processes in grassland ecosystems. Plant Soil 212:23–33 [Google Scholar]
- 7. Borowicz V. A. 2001. Do arbuscular mycorrhizal fungi alter plant-pathogen relations? Ecology 82:3057–3068 [Google Scholar]
- 8. Colwell R. 2005. EstimateS: statistical estimation of species richness and shared species from samples, version 8.0. User's guide. University of Connecticut, Storrs, CT: http://viceroy.eeb.uconn.edu/estimates [Google Scholar]
- 9. Crozzoli R., Vargas G. 1989. Reacción de trece patrones de durazno a infestaciones de Meloidogyne javanica. Fitopatol. Venez. 2:16–18 [Google Scholar]
- 10. Crozzoli R. 2002. Especies de nematodos fitoparasíticos en Venezuela. Interciencia 27:354–364 [Google Scholar]
- 11. de la Peña E., Karssen G., Moens M. 2007. Diversity and distribution of root-lesion nematodes (Pratytlenchus spp.) associated with Ammophila arenaria L. (Link) in coastal dunes of Western Europe. Nematology 9:881–901 [Google Scholar]
- 12. de la Peña E., Rodriguez-Echeverria S., van der Putten W. H., Freitas H., Moens M. 2006. Mechanism of control of root-feeding nematodes by mycorrhizal fungi in the dune grass Ammophila arenaria. New Phytol. 169:829–840 [DOI] [PubMed] [Google Scholar]
- 13. Duhoux E., et al. 2001. Angiosperm Gymnostoma trees produce root nodules colonized by arbuscular mycorrhizal fungi related to Glomus. New Phytol. 149:115–125 [DOI] [PubMed] [Google Scholar]
- 14. Elsen A., Gervacio D., Swennen R., De Waele D. 2008. AMF-induced biocontrol against plant parasitic nematodes in Musa sp.: a systemic effect. Mycorrhiza 18:251–256 [DOI] [PubMed] [Google Scholar]
- 15. Grayston S. J., Vaughan D., Jones D. 1997. Rhizosphere carbon flow in trees, in comparison with annual plants: the importance of root exudation and its impact on microbial activity and nutrient availability. Appl. Soil Ecol. 5:29–56 [Google Scholar]
- 16. Helgason T., Merryweather J. W., Young J. P. W., Fitter A. H. 2007. Specificity and resilience in the arbuscular mycorrhizal fungi of a natural woodland community. J. Ecol. 95:623–630 [Google Scholar]
- 17. Hol W. H. G., Cook R. 2005. An overview of arbuscular mycorrhizal fungi-nematode interactions. Basic Appl. Ecol. 6:489–503 [Google Scholar]
- 18. Klironomos J. N. 2003. Variation in plant response to native and exotic arbuscular mycorrhizal fungi. Ecology 84:2292–2301 [Google Scholar]
- 19. Klironomos J. N., McCune J., Moutoglis P. 2004. Species of arbuscular mycorrhizal fungi affect mycorrhizal responses to simulated herbivory. Appl. Soil Ecol. 26:133–141 [Google Scholar]
- 20. Lee J., Lee S., Young J. P. W. 2008. Improved PCR primers for the detection and identification of arbuscular mycorrhizal fungi. FEMS Microbiol. Ecol. 65:339–349 [DOI] [PubMed] [Google Scholar]
- 21. Long L. K., et al. 2010. Molecular community analysis of arbuscular mycorrhizal fungi associated with five selected plant species from heavy metal polluted soils. Eur. J. Soil Biol. 46:288–294 [Google Scholar]
- 22. Nicholas K. B., Nicholas H. B. J. 1997. GeneDoc: a tool for editing and annotating multiple sequence alignments. Distributed by the authors. http://www.citeulike.org/user/gwallau/article/6113940
- 23. Öpik M., Moora M., Liira J., Zobel M. 2006. Composition of root-colonizing arbuscular mycorrhizal fungal communities in different ecosystems around the globe. J. Ecol. 94:778–790 [Google Scholar]
- 24. Pinochet J., Calvet C., Camprubi A., Fernández C. 1996. Interactions between migratory endoparasitic nematodes and arbuscular mycorrhizal fungi in perennial crops: a review. Plant Soil 185:183–190 [Google Scholar]
- 25. Pozo M. J., Azcon-Aguilar C. 2007. Unraveling mycorrhiza-induced resistance. Curr. Opin. Plant Biol. 10:393–398 [DOI] [PubMed] [Google Scholar]
- 26. Raaijmakers J. M., Paulitz T. C., Steinberg C., Alabouvette C., Moënne-Loccoz Y. 2009. The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321:341–361 [Google Scholar]
- 27. Rodríguez-Echevarría S., de la Peña E., Monees M., Freitas H., van der Putten W. H. 2009. Can root-feeders alter the composition of AMF communities? Experimental evidence from the dune grass Ammophila arenaria. Basic Appl. Ecol. 10:131–140 [Google Scholar]
- 28. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 29. Scheublin T. R., Ridgway K. P., Young J. P. W., van der Heijden M. G. A. 2004. Nonlegumes, legumes, and root nodules harbor different arbuscular mycorrhizal fungal communities. Appl. Environ. Microbiol. 70:6240–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scheublin T. R., van der Heijden M. G. A. 2006. Arbuscular mycorrhizal fungi colonize nonfixing root nodules of several legume species. New Phytol. 172:732–738 [DOI] [PubMed] [Google Scholar]
- 31. Singh R., Adholeya A., Mukerij K. G. 2000. Mycorrhiza in control of soil-borne pathogens, p. 173–196 In Mukerij K. G., Chamalo B. P., Singh J. (ed.), Mycorrhizal biology. Academic-Plenum Publishers, New York, NY [Google Scholar]
- 32. Smith G. S., Kaplan D. T. 1988. Influence of mycorrhizal fungus, phosphorus, and burrowing nematode interactions on growth of rough lemon citrus seedlings. J. Nematol. 20:539–544 [PMC free article] [PubMed] [Google Scholar]
- 33. Soil Survey Staff 2006. Soil taxonomy. A basic system of soil classification for making and interpreting soil surveys. U.S. Department of Agriculture agricultural handbook no. 436. U.S. Department of Agriculture, Washington, DC [Google Scholar]
- 34. Stamatakis A. 2006. RaxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 35. St. Arnaud M., Elsen A. 2005. Interaction or arbuscular-mycorrhizal fungi with soil-borne pathogens and non-pathogenic rhizosphere micro-organisms, p. 217–231 In Declerck S., Strullu D.-G., Fortin J. A. (ed.), In vitro culture of mycorrhizas. Springer-Verlag, Berlin, Germany [Google Scholar]
- 36. Swofford D. L. 2002. PAUP: phylogenetic analysis using parsimony (and other methods), version 4.08b10. Sinauer Associates Inc., Sunderland, MA [Google Scholar]
- 37. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 38. Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vaast P., Cashwell-Chen E. P., Zasoski R. J. 1998. Influences of a root-lesion nematode, Pratylenchus coffeae, and two arbuscular mycorrhizal fungi, Acaulospora mellea and Glomus clarum on coffee (Coffea arabica L.). Biol. Fertil. Soils 26:130–135 [Google Scholar]
- 40. White T. J., Bruns T., Lee S., Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315–322 In Innis M. A., Gelfand D. H., Sninsky J. J., White T. J. (ed.), PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA [Google Scholar]
- 41. Zarina B., Shahina F. 2010. Research work carried out on the management of root-knot nematode diseases in Pakistan. Pak. J. Nematol. 28:153–239 [Google Scholar]