Abstract
Fungal and actinobacterial communities were analyzed together with soil chemistry and enzyme activities in order to profile the microbial diversity associated with the economically important mushroom Tricholoma matsutake. Samples of mycelium-soil aggregation (shiro) were collected from three experimental sites where sporocarps naturally formed. PCR was used to confirm the presence and absence of matsutake in soil samples. PCR-denaturing gradient gel electrophoresis (DGGE) fingerprinting and direct sequencing were used to identify fungi and actinobacteria in the mineral and organic soil layers separately. Soil enzyme activities and hemicellulotic carbohydrates were analyzed in a productive experimental site. Soil chemistry was investigated in both organic and mineral soil layers at all three experimental sites. Matsutake dominated in the shiro but also coexisted with a high diversity of fungi and actinobacteria. Tomentollopsis sp. in the organic layer above the shiro and Piloderma sp. in the shiro correlated positively with the presence of T. matsutake in all experimental sites. A Thermomonosporaceae bacterium and Nocardia sp. correlated positively with the presence of T. matsutake, and Streptomyces sp. was a common cohabitant in the shiro, although these operational taxonomic units (OTUs) did not occur at all sites. Significantly higher enzyme activity levels were detected in shiro soil. These enzymes are involved in the mobilization of carbon from organic matter decomposition. Matsutake was not associated with a particular soil chemistry compared to that of nearby sites where the fungus does not occur. The presence of a significant hemicellulose pool and the enzymes to degrade it indicates the potential for obtaining carbon from the soil rather than tree roots.
INTRODUCTION
Tricholoma matsutake (S. Ito et Imai) is an ectomycorrhizal (ECM) fungus found in pine and spruce forests in the Northern Hemisphere (26, 46, 49). This fungus produces commercially valuable mushrooms that have been revered in Japan for their flavor, medicinal properties, and iconic significance for centuries (11). Nearly 3,000 tons of T. matsutake or closely related species is exported to Japan annually, with a retail value of approximately one billion U.S. dollars (39). This mushroom was known as T. nauseosum in Nordic countries until recently, when molecular techniques revealed its conspecificity with T. matsutake (1). Matsutake mushrooms are distributed patchily throughout Finland (20), where they became a commercially harvested mushroom in 2007. The new and growing value of this nonwoody forest product has received increasing attention in Nordic countries.
Matsutake persistently colonizes the rhizosphere of host trees and gradually grows as a concentric circle, forming a dense mat of fungal filaments adhered to host plant roots and soil particles. This unique and massive mycorrhizal mycelial aggregate is called shiro and is limited mostly to the B layer of the mineral soil prior to sporocarp production (11). In the shiro, T. matsutake dominates the ECM community (21).
Prior to fruiting, field observations suggest that many root tips that are mycorrhizal with T. matsutake become necrotic or die during the early summer (L.-M. Vaario et al., unpublished data). Gill et al. (8) reported a progressive darkening and subsequent necrosis from base to tip in all types of matsutake mycorrhizal roots. These natural phenomena imply that the fungus becomes somewhat disconnected from the host tree before sporocarps are produced. This observation has driven questions concerning the potential existence of an alternative carbon source during sporocarp formation. The decomposition of litter provides an important carbon source for soil microbes. Of the organic carbon found in litter, hemicelluloses are the most abundant of the relatively water-soluble carbohydrates. Hemicellulotic carbohydrates decrease rapidly during the early stages of decomposition (35). Moreover, matsutake mycelium produces β-glucosidase, an enzyme that hydrolyzes oligosaccharides having a β-1,4 linkage when incubated on pine bark or even in pure cultures (19, 44). These in vitro studies indicate that T. matsutake is able to directly degrade and mobilize certain sources of organic carbon. However, while its in vitro physiology has been characterized, soil enzyme activities in the shiro are unknown. In natural ecosystems, ECM fungal species usually coexist with many other fungi, and species interactions are manifold and poorly understood (41). Together with fungi, actinobacteria are the most important producers of exocellular enzymes (32) that catalyze the release of oligosaccharides in the environment. Such enzymes are vital for soil nutrient recycling. Some of the actinobacteria involved in the ectomycorrhizophere consistently promote mycorrhizal development and support the concept of commensal “mycorrhization” bacteria (5, 6). Thus, fungi and actinobacteria may play one or more roles in the shiro, but the microbial communities associated with T. matsutake have yet to be investigated from this perspective. Therefore, a better understanding of the microbial community in the shiro and how it relates to soil chemistry (particularly water-soluble organic carbon) could provide some insight into the ecology of matsutake fruitbody production.
In this study, we propose the following hypothesis: T. matsutake dominates but also coexists with a number of microbes in coniferous forest soil and can obtain carbon directly from the soil rather than from tree roots. We analyzed fungal and actinobacterial communities together with performing soil chemical and enzymatic analyses from the shiro (Shiro+) and nearby nonshiro (Shiro−) control spots. The organic layer immediately above these samples (Shiro+abv or Shiro−abv) was included in all analyses except the profiling of soil enzyme activity. The study was divided into three steps: (i) assessment of fungal and actinobacterial communities and their correlation with T. matsutake and other soil microbes, (ii) determination of exoenzyme activities and related organic carbon concentrations of shiro soil, and (iii) determination of soil chemical properties.
MATERIALS AND METHODS
Study sites.
From 2007 to 2010, three experimental sites were monitored for the presence of sporocarps. The first study site was a 100-m by 135-m plot in Nuuksio National Park in southern Finland (SF) (60°18′16″N, 24°31′10″E). This site contained a mixed stand of Pinus sylvestris, Picea abies, and Betula pendula. The other two sites were 10 m by 10 m in western Finland: Kourajärvi (WF-K) (62°10′13″N, 22°50′31″E) and Alkkia (WF-A) (62°9′14″N 22°50′26″E). Pinus sylvestris is the dominant tree species at both sites in WF. Sporocarps of T. matsutake were found during 2008 to 2009 (17 sporocarps in 2008 and 67 in 2009) in the SF site. However, they appeared only in 2007 and 2010 and no sporocaps were found during the sampling years (2008 and 2009) in the WF sites. Mean maximum and minimum temperatures in the SF and WF sites during 2008 to 2009 were 9.6 and 9.0°C and 3.8 and 2.3°C, respectively (annual means = 6.7°C and 5.7°C, respectively), and annual rainfall was about 800 mm and 700 mm, respectively (http://mesi.metla.fi/tuska/Venalainen_et_al._FMI_2005.pdf).
Soil sampling.
The sites were determined by the presence of T. matsutake sporocarps in 2007. In spring 2008, we sampled 2 ml of mineral soil from 10 spots in SF, nine spots in WF-A, and nine spots in WF-K where matsutake sporocarps were found in 2007. DNA was extracted from 0.25 g of each soil sample with the PowerSoil DNA kit according to the manufacturer's instructions, and the presence of live mycelia was confirmed by PCR amplification with specific primers (15) annealed at 52°C.
Soil sampling for microbial community analysis.
Soil sampling was performed during the 2008 harvest. Locations of the sampling spots were determined by the presence of sporocarps, but the presence of T. matsutake mycelium in the soil samples was also confirmed by PCR. Samples of mineral soil that yielded PCR targets were designated Shiro+ and organic soil samples taken directly above the Shiro+ samples as Shiro+abv. Correspondingly, matsutake-free controls (PCR target negative, designated Shiro− and Shiro−abv) were sampled 1 meter to the east from Shiro+ spots in all three study sites. In the SF site, 17 matsutake sporocarps were harvested. In the WF sites, because no matsutake sporocarps were found in autumn 2008, we sampled spots where matsutake DNA was confirmed by PCR amplification in spring 2008. If the sporocarp reappeared, we repeated soil sampling in the same SF spots as in autumn 2009. Soil samples (Table 1) were placed into 1.5-ml microcentrifuge tubes for further analysis.
Table 1.
Details of soil samples for microbial community analysis from three experimental sites
| Sample type | Yr | Sample no. used for fungal (actinobacterial) OTU analysis at: |
|||
|---|---|---|---|---|---|
| SF | WF-K | WF-A | Total | ||
| Shiro+a | 2008 | 17 (14) | 6 (4) | 7 (3) | 43 (34) |
| 2009 | 13 (13) | NAb | NA | ||
| Shiro− | 2008 | 9 (5) | 2 (2) | 2 (2) | 24 (16) |
| 2009 | 11 (7) | NA | NA | ||
| Shiro+abv | 2008 | 14 (11) | 7 (4) | 8 (3) | 41 (28) |
| 2009 | 12 (10) | NA | NA | ||
| Shiro−abv | 2008 | 2 (2) | 3 (3) | 2 (2) | 11 (15) |
| 2009 | 4 (8) | NA | NA | ||
Shiro+ samples in the SF site were confirmed by both sporocarps and PCR positivity; those in the WF sites were confirmed by PCR positivity only.
NA, not available.
In total, we screened 61 shiro soil samples with T. matsutake-specific internal transcribed spacer (ITS) primers and obtained 43 sets of Shiro+/Shiro+abv soil samples from the three study sites. Shiro+ samples were also confirmed by screening for T. matsutake hyphae with the aid of a light microscope. We screened 27 control samples with the same method from three sites and obtained 24 sets of Shiro−/Shiro−abv soil samples. After screening, a final set of 43 Shiro+, 41 Shiro+abv, 24 Shiro−, and 11 Shiro−abv samples were subjected to fungal community analysis, and a set of 34 Shiro+, 28 Shiro+abv, 16 Shiro−, and 15 Shiro−abv samples were subjected to actinobacterial community analysis (see Tables S1 and S2 in the supplemental material). There were two reasons for the unequal number of samples. First, Shiro−/Shiro−abv were sampled 1 meter from Shiro+/Shiro+abv spots, and several Shiro− samples later showed denaturing gradient gel electrophoresis (DGGE) bands that corresponded to T. matsutake and were discarded from subsequent analyses. Second, several samples only weakly amplified with PCR (and therefore DGGE), and computer scoring of the gels failed to detect any bands. In such cases, these soil samples were excluded from the microbial community analysis.
Soil sampling for enzyme activity assays.
We randomly sampled five Shiro+ spots and five Shiro− spots in the SF site in 2008. A 10-ml sample of the mineral layer soil from each spot was collected after sporocarps were harvested, and this was designated Shiro+ (PCR positive). Correspondingly, control (PCR negative, Shiro−) spots were sampled 1 meter to the east of Shiro+ spots.
Soil sampling for determination of total hemicellulotic carbohydrates.
In 2009, we found T. matsutake sporocarps in or within 10 cm of five sampling spots analyzed for enzymatic activity in the SF site. Immediately after harvesting, 15 to 20 ml of soil was taken from five sets of Shiro+/Shiro+abv samples and five sets of Shiro−/Shiro−abv samples for the quantification of total hemicellulotic carbohydrates in the adjacent soil.
Soil sampling for analysis of soil pH, moisture, nutrients, and trace elements.
Soil samples were taken for chemical analyses with a corer (diameter, 6 cm) from three study sites in late autumn 2008. Two plots known to contain T. matsutake (PCR positive) and two matsutake-free plots (PCR negative) were selected in the SF site. Both WF sites were sampled in the same manner except that only one PCR-positive and one PCR-negative plot were sampled. In total, four matsutake PCR-positive plots and four matsutake PCR-negative plots were selected. From each plot, we oriented four 20-cm transects along the main compass points. Transects intercepted at the center of each plot, and soil was sampled at 10- and 20-cm intervals from the center. Including the center, a total of nine spots were sampled, pooled, and subsequently divided into 5 subsamples for the final chemical analysis.
DNA extraction, PCR, and DGGE analysis.
Genomic DNA was extracted from 0.25 g of each soil sample with the PowerSoil DNA kit according to the manufacturer's instructions. For the fungal community analysis, the internal transcribed spacer (ITS) region of the ribosomal DNA (rDNA) was amplified with GC-clamped ITS1F (5′-CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG GCT TGG TCA TTT AGA GGA AGT AA-3′) and ITS2 primers (48). PCR amplification for DGGE analysis was performed with Biotools polymerase (B & M Laboratories, Madrid, Spain) as described by Korkama et al. (16).
For the actinobacterial community analysis, the 16S rRNA gene was amplified with GC-clamped S-C-Act-0235-a-S-20 and S-C-Act-0878-a-A-19 primers (12, 37) and the thermal cycling parameters described by Jaatinen et al. (12). A gradient of 48 to 62% of 6% (wt/vol) acrylamide-bisacrylamide (37:5:1) was prepared, and gels were run at 70 V and 60°C for 16 h (12).
The DCode denaturing gradient gel system (Bio-Rad, Hercules, CA) was used for both analyses. Control ladders with several known DGGE bands were included for comparison. DGGE gels were stained with SYBR gold, visualized under low-power UV light (Dark Reader transluminator; Clare Chemical Research, Dolores, CO), and photographed digitally. DGGE gel photographs were screened for the presence (1) or absence (0) of fungal and actinobacterial bands using Alphaimager 2.1 (Alpha Innotech Corp., CA). Based on their mobilities, two to four bands of similar mobility were selected for sequencing, excised, amplified, and run again in DGGE three to five times until a high-quality and single band was obtained. Finally, DGGE-PCR products were reamplified with 23 to 25 cycles and purified with the High-Pure PCR product purification kit (Roche, Germany).
Direct sequencing of the partial ITS (fungi) and partial 16S rRNA gene (actinobacteria) products was conducted by a commercial sequencing service (Macrogen Inc., South Korea) with the same primers (ITS1F [7] and ITS2) used in amplification of ITS and Act237F and Act876R for the 16S rRNA gene. Cycle sequencing of actinobacterial products consisted of, for the reverse primer, an initial denaturation at 96°C for 1 min and 40 cycles of 96°C for 20 s, 55°C for 20 s, and 60°C for 4 min; for the forward primer, an initial denaturation at 96°C for 1 min and 40 cycles of 96°C for 20 s, 59°C for 20 s, and 60°C for 4 min were used. Sequences were aligned using Geneious Pro (version 5.0.4) (Biomatters Ltd., New Zealand) and adjusted accordingly by eye.
Identification of fungal and actinobacterial OTUs.
Two to four replicates of each fungal or actinobacterial DGGE band were sequenced. All DGGE-derived sequences were aligned with those available in GenBank using the BLAST algorithm. At least 97% similarity was used as the limit for classifying an operational taxonomic unit (OTU). When the closest sequences were less than 97% similar, the highest BLAST score was chosen and noted accordingly.
Soil enzyme activity assays.
Eight enzyme assays were performed on 4-methylumbelliferone (MU) or 7-amino-4-methylcoumarin (AMC) substrates and one via photometric detection of 2,2′-azinobis-3-ethylbenzothiazoline-6-sulfonate (ABTS) reaction products. The substrates used were MU-phosphate (for the detection of the acid phosphatase, EC 3.1.3.2), MU-β-d-glucopyranoside (β-glucosidase, EC 3.2.1.3), MU-N-acetyl-β-d-glucosaminide (chitinase, EC 3.2.1.14), MU-β-d-glucuronide hydrate (glucuronidase, EC 3.2.1.31), MU-β-d-xylopyranoside (xylosidase, EC 3.2.1.37), MU-β-d-cellobioside (cellobiohydrolase, EC 3.2.1.91), l-leucine-AMC (leucine aminopeptidase, EC 3.4.11.1), and ABTS (laccase, EC 1.10.3.2). The assay was described in detail by Courty et al. (4), but we prepared soil samples from the field experiment with a centrifuge method (10a). Briefly, 500 μl of soil was placed into a microcentrifuge tube with 500 μl sterile distilled water, and the tube was centrifuged for 30 min at 16,000 × g. The supernatants from three tubes of each replicate were pooled and brought to a total volume of 5 ml with water. After mixing, the tube was stored on ice until analyzed. In contrast to the protocol described by Courty et al. (4), water in the reaction mix was replaced by soil solution.
Determination of hemicellulotic carbohydrates.
Total hemicellulotic carbohydrate was determined from 15 to 20 ml of dry soil sample. The concentration of neutral and acidic sugar units in noncellulosic polysaccharides was obtained by acid methanolysis followed by silylation and gas chromatography (GC) (38). The hemicellulose concentration was calculated as described by Merilä et al. (25).
Soil chemical analysis.
Soil pH was measured in a water suspension using fresh soil (1:1.7, vol/vol). Soil moisture was determined after drying at 105°C overnight, and organic matter (OM) content was determined as loss on ignition after combustion at 550°C for 4 h. Carbon and nitrogen were determined from air-dried soil with a LECO CHN-1000 analyzer according to ISO 10694 and ISO 13878, and concentrations of other elements (Al, Ca, Fe, K, Mg, Mn, Na, P, and S) were determined with an inductively coupled plasma atomic emission spectrometer (ICP-AES, ARL 3580) after dry ashing and dissolution with hydrochloric acid (HCl) according to SFS-EN ISO 11885. Arithmetic means for five soil samples were used in subsequent analyses.
Statistical analyses.
DGGE gel images were analyzed with the GelCompar II (ver. 5.1; Applied Maths BVBA, Belgium) and a binary matrix (presence/absence data) was produced with a band-matching optimization of 0% and band position tolerance of 1%. The relative frequency of each OTU was calculated as the proportion of all samples examined in which it was detected.
We used the Phi coefficient (9) as a measure of association between the presence of T. matsutake and other fungal and actinobacterial OTUs. The statistical significance of the association was tested using a two-sided Fisher exact test.
Variation in fungal and actinobacterial community compositions among Shiro+, Shiro−, Shiro+abv, and Shiro−abv samples was investigated by nonmetric multidimensional scaling (NMDS) using a Jaccard distance measure and two-dimensional solutions. Statistical analyses were carried out with vegan (29) in R (ver. 2.10.1) (34).
The respective impacts of shiro on soil enzyme activities and total hemicellulotic carbohydrates at the SF site were evaluated by analysis of variance (ANOVA). Soil layers were analyzed separately. The effect of shiro on soil chemical properties was evaluated using mixed-model analysis by pairwise comparisons; shiro, soil type and, shiro × soil type were treated as fixed effects and sample plots as a random effect (in total, there were 2 plots from the SF site and 2 plots from the WF sites). We performed variable transformation when necessary to obtain a normal distribution. Normality and homogeneity of the variance of the residuals were examined using scatter plots and Q-Q-plots using SPSS (version 15.0; SPSS Inc., Chicago, IL).
Nucleotide sequence accession numbers.
All sequence data generated in this study were deposited in GenBank (accession numbers HQ215859 to HQ215908 and HQ293015 for fungal OTUs and accession numbers HQ204333 to HQ204369 for actinobacterial OTUs).
RESULTS
Fungal and actinobacterial communities.
Since the sampling strategy was based on the presence of T. matsutake DNA in Shiro+ samples, the OTU identified as T. matsutake was found in all Shiro+ samples but also in 41.9% of the Shiro+abv samples. In total, 51 fungal OTUs were identified based on DGGE bands and represented three fungal phyla. The majority of sequences belonged to Basidiomycota (58.8%), 21.6% to Ascomycota, and 9.8% to Zygomycota, and 9.8% were unknown (see Table S1 in the supplemental material).
OTU identities were derived from closest matches in a BLAST search of GenBank sequences. The Phi coefficient was used to measure the association between the presence of T. matsutake and other fungal OTUs in Shiro+ or in Shiro+abv samples. Many fungal OTUs detected in Shiro+ samples, such as Basidiomycete sp. 2 (F56), Basidiomycete sp. 1 (F54), Tricholoma portentosum (F23), and Mortierella sp. 1 (F12), were negatively correlated with the presence of matsutake in both SF and WF sites (P = 0.000 to 0.088). Only Piloderma sp. (F37/F32) was positively correlated with the presence of T. matsutake in Shiro+ samples, although the relationship was quite weak (P = 0.081 to 0.092). In Shiro+abv samples, Tomentellopsis sp. (F38/F44) correlated positively with the presence of T. matsutake in the soil layer directly below (Shiro+) (P = 0.061 to 0.083) in both SF and WF sites. In WF sites, there were more fungal OTUs such as Cortinarius sp. (F13), Tylospora sp. (F19), and Trichoderma sp. (F46), positively correlated with the presence of T. matsutake (P = 0.017 to 0.085) (Table 2). Relative frequencies of all fungal OTUs (see Table S1 in the supplemental material) showed that about half of those cooccurring with T. matsutake belonged to genera known to form ectomycorrhiza.
Table 2.
Correlation between the presence of T. matsutake in Shiro+ soil and of other fungal OTUs in Shiro+ and Shiro+abv soilsa
| Site | Soil layer | OTU code | Fungal OTU | Phylum | Accession no. | Closest GenBank match | % Match | Significance by Fisher's exact test (2 sided) | Phi value |
|---|---|---|---|---|---|---|---|---|---|
| SF | Shiro+abv | F44 | Tomentellopsis sp. | Basidiomycota | HQ215891 | FJ554204 | 99 (326/330) | 0.083 | 0.306 |
| F13 | Cortinarius biformis | Basidiomycota | HQ215863 | AY669688 | 100 (261/261) | 0.073 | −0.413 | ||
| F41 | Hebeloma sp. | Ascomycota | HQ215888 | AF430291 | 99.6 (249/250) | 0.049 | −0.387 | ||
| Shiro+ | F37 | Piloderma sp. 4 | Basidiomycota | HQ215884 | FJ552742 | 99 (284/287) | 0.092 | 0.212 | |
| F43 | Clavulina cf. amethystina | Basidiomycota | HQ215890 | EU862204 | 99 (317/319) | 0.079 | 0.225 | ||
| F12 | Mortierella sp. 1 | Zygomycota | HQ215862 | GQ302682 | 99 (178/179) | 0.076 | −0.28 | ||
| F13 | Cortinarius biformis | Basidiomycota | HQ215863 | AY669688 | 100 (261/261) | 0.076 | −0.28 | ||
| F23 | Tricholoma portentosum | Basidiomycota | HQ215871 | EU186273 | 97 (341/352) | 0.041 | −0.303 | ||
| F30 | Piloderma sp. 2 | Basidiomycota | HQ215877 | FJ552742 | 99.7 (311/312) | 0.034 | −0.333 | ||
| F32 | Piloderma sp. 3 | Basidiomycota | HQ215879 | FJ552742 | 98 (307/312) | 0.001 | −0.453 | ||
| F36 | Cantharellales sp. 3 | Basidiomycota | HQ215883 | AF440673 | 90 (247/274)b | 0 | −0.693 | ||
| F47 | Atheliaceae | Basidiomycota | HQ215894 | FM992889 | 100 (256/256) | 0.015 | −0.375 | ||
| F48 | Uncultured Dermateaceae | Ascomycota | HQ215895 | FJ475808 | 98 (271/276) | 0.063 | −0.297 | ||
| F50 | Russula xerampelina | Basidiomycota | HQ215897 | FJ845433 | 99.5 (192/193) | 0.076 | −0.28 | ||
| F54 | Basidiomycete sp. 1 | Basidiomycota | HQ215900 | EF493313 | 100 (140/140) | 0.014 | −0.356 | ||
| F55 | Uncultured fungus 3 | HQ215901 | GQ160052 | 100 (189/189) | 0.004 | −0.408 | |||
| F56 | Basidiomycete sp. 2 | Basidiomycota | HQ215902 | EF493313 | 100 (160/160) | 0 | −0.584 | ||
| F60 | Polyporales | Basidiomycota | HQ215906 | FJ475705 | 100 (302/303) | 0.076 | −0.28 | ||
| WF | Shiro+abv | F13 | Cortinarius biformis | Basidiomycota | HQ215863 | AY669688 | 100 (261/261) | 0.017 | 0.601 |
| F19 | Tylospora sp. | Basidiomycota | HQ215868 | AB456677 | 98 (281/288) | 0.057 | 0.524 | ||
| F38 | Tomentellopsis submollis | Basidiomycota | HQ215885 | AJ438983 | 99 (291/293) | 0.061 | 0.419 | ||
| F46 | Trichoderma viride | Basidiomycota | HQ215893 | AF486010 | 99.6 (230/231) | 0.085 | 0.385 | ||
| F40 | Phialocephala fortinii | Ascomycota | HQ215887 | AY394915 | 99 (273/277) | 0.001 | −0.787 | ||
| Shiro+ | F32 | Piloderma sp. 3 | Basidiomycota | HQ215879 | FJ552742 | 98 (307/312) | 0.081 | 0.381 | |
| F12 | Mortierella sp. 1 | Zygomycota | HQ215862 | GQ302682 | 99 (178/179) | 0.058 | −0.414 | ||
| F15 | Cortinarius coleoptera | Basidiomycota | HQ215865 | EU266682 | 99.5 (211/212) | 0.058 | −0.414 | ||
| F18 | Pezizomycetes | Ascomycota | HQ293015 | GQ268570 | 100 (74/74) | 0.058 | −0.414 | ||
| F22 | Uncultured Herpotrichiellaceae | Ascomycota | HQ215870 | FJ475766 | 99.6 (278/279) | 0.058 | −0.414 | ||
| F23 | Tricholoma portentosum | Basidiomycota | HQ215871 | EU186273 | 97 (341/352) | 0.058 | −0.414 | ||
| F33 | Uncultured fungus 2 | HQ215880 | DQ309123 | 97 (204/211) | 0.047 | −0.482 | |||
| F40 | Phialocephala fortinii | Ascomycota | HQ215887 | AY394915 | 99 (273/277) | 0.012 | −0.618 | ||
| F54 | Basidiomycete sp. 1 | Basidiomycota | HQ215900 | EF493313 | 100 (140/140) | 0.088 | −0.372 | ||
| F56 | Basidiomycete sp. 2 | Basidiomycota | HQ215902 | EF493313 | 100 (160/160) | 0.003 | −0.713 |
Bold indicates a positive correlation.
Highest similarity from BLAST results but less than 97%.
In total, 37 actinobacterial OTUs were detected, including 29 OTUs belonging to the Actinomycetales order and eight sequences that matched A18 (uncultured actinobacterial clone) (see Table S2 in the supplemental material). The correlation analysis showed that there were clear differences between SF and WF sites. The Phi coefficient showed that 10 actinobacterial OTUs in Shiro+ samples of the SF site correlated negatively with the presence of T. matsutake, but only one was in a WF site. In contrast, two actinobacterial OTUs in Shiro+ samples of WF sites correlated positively with the presence of T. matsutake: Thermomonosporaceae bacterium 1 (A29) (P = 0.024) and Streptomyces sp. (A25) (P = 0.088) (Table 3). In Shiro+abv samples, Thermomonosporaceae bacterium 2 (A50) (P = 0.035), uncultured actinobacterium 4 (A31) (P = 0.06), and Nocardia sp. (A35) (P = 0.08) correlated positively with the presence of T. matsutake directly below the soil layer (Table 3). Four actinobacterial OTUs that were more common in Shiro+abv samples were absent from the Shiro−abv samples: uncultured actinobacterium 2 (A27), Nocardia sp. (A35), uncultured actinobacterium 4 (A31), and Actinospica sp. 2 (A39) (see Table S2 in the supplemental material).
Table 3.
Correlation between the presence of T. matsutake in Shiro+ soil and of actinobacterial OTUs in both Shiro+ and Shiro+abv soilsa
| Site | Soil layer | OTU code | Actinobacterial OTU | Family | Accession no. | Closest GenBank match | % Match | Significance by Fisher's exact test (2 sided) | Phi value |
|---|---|---|---|---|---|---|---|---|---|
| SF | Shiro+abv | A50 | Thermomonosporaceae bacterium 2 | Thermomonosporaceae | HQ204366 | FJ037190 | 98 (612/626) | 0.035 | 0.425 |
| Shiro+ | A9 | Leucobacter sp. | Microbacteriaceae | HQ204336 | EF138947 | 98 (635/645) | 0.024 | −0.433 | |
| A21 | Mycobacterium sp. 4 | Mycobacteriaceae | HQ204341 | GU358074 | 99 (649/653) | 0 | −0.626 | ||
| A24 | Uncultured actinobacterium 1 | HQ204343 | EF220614 | 98 (607/622) | 0.025 | −0.409 | |||
| A26 | Actinomycetales bacterium 1 | HQ204345 | X92703 | 100 (621/623) | 0.001 | −0.57 | |||
| A28 | Cellulomondaceae bacterium | Cellulomonadaceae | HQ204347 | FJ626810 | 97 (514/528) | 0.043 | −0.372 | ||
| A30 | Uncultured actinobacterium 3 | HQ204349 | FJ570524 | 99.5 (623/626) | 0.079 | −0.349 | |||
| A31 | Uncultured actinobacterium 4 | HQ204350 | FJ570359 | 99 (607/612) | 0.006 | −0.507 | |||
| A32 | Uncultured actinobacterium 5 | HQ204351 | EF220691 | 99.8 (614/615) | 0.004 | −0.497 | |||
| A37 | Goodfellowiella sp. | Pseudonocardiaceae | HQ204356 | DQ093349 | 97 (593/611) | 0.007 | −0.486 | ||
| A39 | Actinospica sp. 2 | Actinospicaceae | HQ204358 | AJ865862 | 99 (633/642) | 0.089 | −0.349 | ||
| WF | Shiro+abv | A31 | Uncultured actinobacterium 4 | HQ204350 | FJ570359 | 99 (607/612) | 0.061 | 0.69 | |
| A35 | Nocardia sp. | Nocardiaceae | HQ204354 | EF538741 | 97 (546/565) | 0.08 | 0.633 | ||
| Shiro+ | A25 | Streptomyces sp. | Streptomycetaceae | HQ204344 | EU080938 | 97 (546/565) | 0.088 | 0.607 | |
| A29 | Thermomonosporaceae bacterium 1 | Thermomonosporaceae | HQ204348 | EF663839 | 98 (619/632) | 0.024 | 0.81 | ||
| A30 | Uncultured actinobacterium 3 | HQ204349 | FJ570524 | 99.5 (623/626) | 0.039 | −0.624 |
Bold indicates a positive correlation.
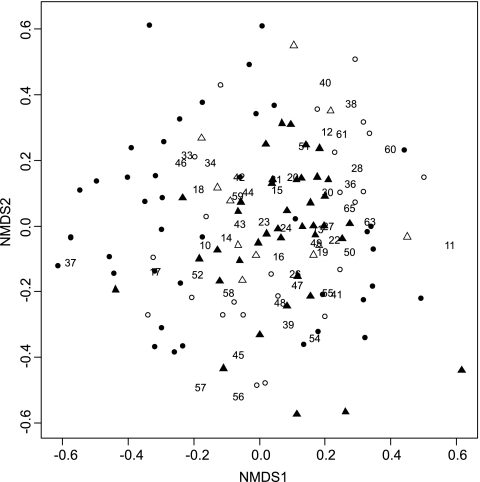
Most Shiro+ samples clustered at either end of axis 1 in the NMDS ordination of fungal community structure, but Shiro− samples did not separate as well. Moreover, Shiro+ samples from SF and WF were evenly scattered around the plots. Shiro−, Shiro+abv, and Shiro−abv samples clearly overlapped to some extent (Fig. 1).
Fig. 1.
Two-dimensional NMDS ordination of fungal communities in Shiro+ (solid circles), Shiro− (open circles), Shiro+abv (solid triangles), and Shiro−abv (open triangles) samples from the three experimental sites. T. matsutake OTU (F35) was excluded from the analysis. The stress value was 0.031. Numbers are fungal OTU codes.
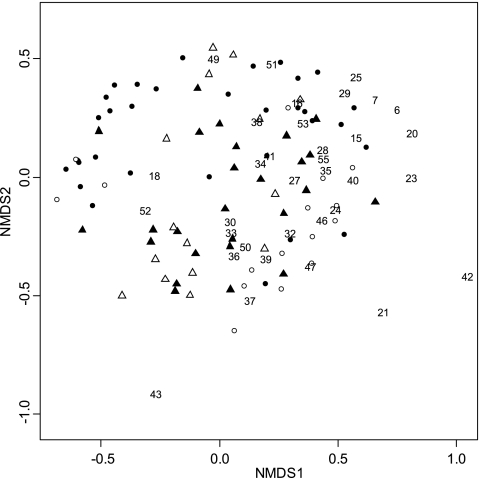
NMDS ordination of the actinobacterial community did not completely separate Shiro+ and Shiro− samples, but samples from SF and WF sites tended to separate in different clusters along axis 1. Similarly, Shiro− samples or their respective organic layers did not separate in NMDS ordination (Fig. 2).
Fig. 2.
Two-dimensional NMDS ordination of actinobacterial communities in Shiro+ (solid circles), Shiro− (open circles), Shiro+abv (solid triangles), and Shiro−abv (open triangles) samples from the three experimental sites. The stress value was 0.029. Numbers are actinobacterial OTU codes.
Enzymatic activity of soil dominated by T. matsutake.
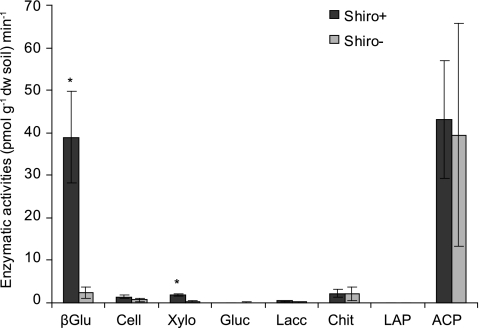
Significantly higher activities were detected for β-glucosidase in Shiro+ soil than in Shiro− soil (ANOVA, P = 0.010) (Fig. 3). In addition, higher activities of acid phosphatase were detected in both Shiro+ and Shiro− soil samples. However, differences in acid phosphatase between Shiro+ and Shiro− samples were not significant. Clear activities of cellobiohydrolase, xylosidase, and chitinase were detected, but only xylosidase (ANOVA, P = 0.002) was significantly higher in Shiro+ compared to Shiro− samples. No or negligible activity was recorded for glucuronidase, leucine aminopeptidase, or laccase (Fig. 3).
Fig. 3.
Enzyme activities in soil samples dominated by T. matsutake (Shiro+) and without T. matsutake (Shiro−). Mean values and standard errors (SEs) are shown; asterisks indicate significant differences between Shiro+ and Shiro− (ANOVA, P < 0.05; n = 5). βGlu, β-glucosidase; Cell, cellobiohydrolase; Xylo, xylosidase; Gluc, glucuronidase; Lacc, laccase; Chit, chitinase; LAP, leucine aminopeptidase; ACP, acid phosphatase.
Total hemicellulotic carbohydrates in soil dominated by T. matsutake.
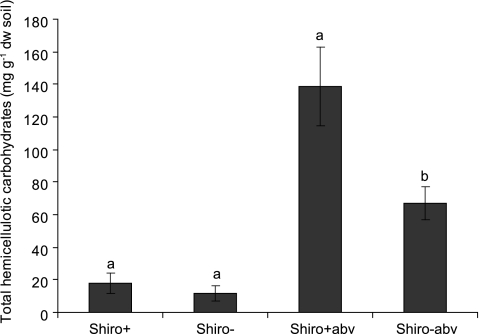
There was a significantly higher concentration of total hemicellulotic carbohydrates in Shiro+abv than in Shiro−abv samples (P = 0.026) (Fig. 4). In the mineral layer, the total hemicellulotic carbohydrates tended to be higher in Shiro+ samples, but the difference between Shiro+ and Shiro− samples was not significant (P = 0.460) (Fig. 4).
Fig. 4.
Total hemicellulotic carbohydrates in soil samples. Common letters indicate nonsignificant differences in each soil type (ANOVA, P < 0.05; n = 5). Bars are means and errors are SEs.
Soil chemical properties at the study sites.
No significant differences between Shiro+ and Shiro− samples or between Shiro+abv and Shiro−abv samples were detected among the measured soil properties or their interactions (Table 4). However, significantly different soil chemical properties were found between soil layers (data not shown). Soil moisture (P = 0.075), carbon content (P = 0.064), and nitrogen content (P = 0.064) tended to be higher in Shiro+abv than in Shiro−abv samples. Site effects were also detected for some chemical properties, e.g., soil organic matter, carbon, and nitrogen of Shiro+abv soil in SF were much higher than those in the WF sites (data not shown).
Table 4.
Mean values of soil chemical properties and significance of the shiro effect from three experimental sites
| Soil | Mean value (n = 20) |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pH | Moisture (%) | OM (%) | C (%) | N (%) | C/N | Al (mg g−1 organic matter) | Ca (mg g−1 organic matter) | Fe (mg g−1 organic matter) | K (mg g−1 organic matter) | Mg (mg g−1 organic matter) | Mn (mg kg−1 organic matter) | Na (mg kg−1 organic matter) | P (mg kg−1 organic matter) | S (mg g−1 organic matter) | NH4-N (mg kg−1 organic matter) | NO3-N (mg kg−1 organic matter) | |
| Shiro+ | 4.55 | 1.15 | 8.20 | 4.29 | 0.12 | 36.23 | 14.16 | 0.67 | 3.16 | 1.02 | 0.22 | 27.30 | 133.35 | 153.98 | 0.90 | 8.10 | 0.32 |
| Shiro− | 4.40 | 0.70 | 5.39 | 2.99 | 0.09 | 32.13 | 7.13 | 1.32 | 2.88 | 0.78 | 0.29 | 84.60 | 133.39 | 118.19 | 0.61 | 9.43 | 0.30 |
| P value | 0.177 | 0.169 | 0.553 | 0.527 | 0.628 | 0.108 | 0.484 | 0.145 | 0.798 | 0.160 | 0.372 | 0.451 | 0.998 | 0.374 | 0.132 | 0.675 | 0.581 |
| Shiro+abv | 4.20 | 4.06 | 47.51 | 28.91 | 0.92 | 32.32 | 0.70 | 1.55 | 0.11 | 0.95 | 0.31 | 53.06 | 76.18 | 127.67 | 0.19 | 9.62 | 0.08 |
| Shiro−abv | 4.11 | 2.82 | 33.16 | 18.71 | 0.57 | 34.02 | 0.77 | 2.03 | 0.22 | 0.84 | 0.39 | 162.05 | 91.29 | 156.91 | 0.18 | 8.35 | 0.07 |
| P value | 0.340 | 0.075 | 0.140 | 0.064 | 0.064 | 0.468 | 0.952 | 0.268 | 0.926 | 0.534 | 0.308 | 0.172 | 0.563 | 0.464 | 0.967 | 0.691 | 0.630 |
DISCUSSION
This study combined a molecular survey of microbial diversity together with soil chemical and enzyme analyses. We focused on shiro soil (mineral layer) as a unique and massive mycorrhiza-mycelium aggregate of host plant roots and soil particles where the T. matsutake sporocarp forms, as well as the organic layer immediately above. Our study yielded three key observations: (i) T. matsutake dominates in Shiro+ soil but also coexists with a high diversity of fungi and actinobacteria, (ii) higher activities of enzymes involved with the degradation of organic carbon were detected in shiro soil as well as a higher concentration of hemicellulotic carbohydrates in the soil immediately above, and (iii) soil chemical differences between shiro and nonshiro soil in both layers were not significant but there seems to be more organic matter and N in soil above the shiro.
Fungal and actinobacterial diversity in the shiro.
T. matsutake has been reported as a dominant species at root tips in the shiro (21). However, the mechanism by which T. matsutake and its extraradical mycelia achieve and maintain dominance in the shiro remains unclear. In line with previous studies (see, e.g., references 21 and 27), our study showed that T. matsutake inhabits mainly the mineral soil layer. Although matsutake is the dominant ECM fungus at root tips, 45 fungal OTUs were found in the shiro and 51 fungal OTUs in the organic soil immediately above it. Surprisingly, nearly half of the fungal OTUs were identified as ECM fungi. This DNA must originate from the extraradical mycelia of ECM fungi instead of colonized root tips, since most of the root tips in shiros were fully colonized by T. matsutake (21; Vaario et al., unpublished data). Many root tips showed signs of necrosis, which is a typical response of the host plant to colonization by T. matsutake (8, 26). This response may possibly result in a limited number of lateral roots becoming exposed to colonization by other fungi.
Taken together, our results suggest that T. matsutake dominates in the shiro and influences the surrounding microbial community. However, we must note that these OTUs were obtained using DGGE, which reveals only the dominant species (30) due to incomplete resolution of bands in the gel (33). Importantly, this study showed that certain fungal species in the shiro (e.g., Piloderma sp. and Clavulina cf. amethystine) and in the soil above (e.g., Tomentellopsis sp., Tylospora sp., and Trichoderma sp.) seemed to commonly coexist with T. matsutake, since a positive correlation among these species was found. In particular, positive correlations between T. matsutake and Piloderma sp. in Shiro+ samples and between T. matsutake and Tomentellopsis sp. in Shiro+abv samples were both found in two different geographical sites and may be general features of a matsutake-driven community. This result leads us to consider if there are functional connections between these microbes.
To our knowledge, this is the first study to document the actinobacterial community in the T. matsutake shiro. Actinomycetes constitute a significant component of the microbial population in many soil types. Our results showed that most of the actinobacterial OTUs that were present were negatively correlated with the presence of T. matsutake. Ohara and Hamada (28) reported that certain antibacterial compounds produced by T. matsutake ECM in the roots of Pinus densiflora led to the elimination of local bacteria and actinomycetes. This could explain the negative correlation between many actinobacterial OTUs and the presence of T. matsutake.
Several actinobacterial OTUs that were clearly more abundant in the Shiro+abv samples were missing in Shiro−abv samples, e.g., uncultured actinobacterium 2, Nocardia sp., uncultured actinobacterium 4, and Actinospica sp. 2. Notably, positive correlations between T. matsutake and Nocardia sp. and between T. matsutake and uncultured actinobacterium 4 were both significant. It has been reported that Nocardia species was able to produce antibiotics (36). At the moment, whether this actinobacterial OTU and other, more abundant actinobacterial OTUs in Shiro+abv soil have a functional connection with T. matsutake is unclear. The actinobacterial community structures at the SF and WF sites were clearly different and may have been influenced by a higher productivity of the SF site during the study period.
Enzyme activity, chemical properties, and microbial communities in shiro.
This study detected significantly higher activities of β-glucosidase and xylosidase in Shiro+ soil than in nearby Shiro− soil sampled immediately after sporocarps were harvested. Many root tips in Shiro+ soil showed signs of necrosis, which is a typical response of the host plant to colonization by T. matsutake (8, 26) and suggests degradation of root tissues. Vaario et al. (45) showed that T. matsutake mycelium invaded the xylem cell walls of pine sawdust and how this fungus was able to produce β-glucosidase when pine bark was used as a substrate in vitro (19, 44). Within Shiro+ soil, the products degraded from cellulose or hemicellulose by matsutake itself and other Shiro+ microbes would be important carbon sources for subsequent growth.
We observed a significantly higher concentration of hemicellulotic carbohydrates in Shiro+abv soil than in Shiro−abv soil. This invites questions concerning the importance of hemicelluloses to matsutake in situ. Dissolved organic matter derived from the litter layer has been suggested as an important energy and nutrient source for soil microbes (24), and it plays a key role in C and N dynamics of forest soils. Root and leaf litter (10, 31, 43) and root exudates (18) generate considerable quantities of dissolved organic matter in soil. Hemicellulose is a more soluble form of organic carbon than cellulose and has been found in root and leaf litter in relatively high concentrations (14). We found several fungal OTUs (e.g., Tomentellopsis sp., Tylospora sp., and Trichoderma sp.) that were positively correlated with the presence of T. matsutake in both the southern and western study sites. These fungi are believed to take part in litter or wood degradation. For example, mycorrhizal root tips of Tomentellopsis sp. expressed high levels of cellobiohydrolase and β-glucosidase (4), Tomentellopsis is suspected to be both a mycorrhizal and litter decay fungus (17), Tylospora sp. is believed to possess peroxidase-encoding genes (2), and species of Trichoderma are well known for their ability to produce enzymes that degrade cellulose, chitin (23), and hemicellulose (40). Piloderma sp. (F37/F32) in the sandy Shiro+ soil was weakly correlated with the presence of T. matsutake in both SF and WF sites. Piloderma sp. has been found to influence plant nutrient uptake and modify soil mineral weathering, especially in the ectomycorrhizosphere (13). Multiple laccase-like genes were also identified in Piloderma spp. (3). Whether there is any functional connection between Piloderma sp. and T. matsutake in sandy soil would be an interesting subject of future research.
There are a number of other possible explanations for the larger amount of hemicellulotic carbohydrates in Shiro+abv than in Shiro−abv soil. A higher input or different qualities of litter in specific Shiro+ spots could lead to localized concentrations of organic matter and available carbon. Decomposition of recalcitrant litter could be accelerated, and small molecules from incomplete decomposition of cellulose and/or lignin could leach down to the shiro. Matsutake can use cellobiose as its sole carbon source in vitro (22), so this is at least feasible. On the other hand, the large pool of hemicellulose in Shiro+abv soil suggests slower loss of hemicellulose, since many fungal OTUs correlate negatively with T. matsutake, especially in SF, where sporocarps were continuously produced. This observation implies that processes in the shiro inhibit the microbial breakdown of litter. However, we stress that functional connections between T. matsutake and its associated fungal community have yet to be demonstrated.
We did not find significant differences in soil chemical properties between Shiro+ and Shiro− soil in either soil layer. However, site effects seemed to exist. In agreement with earlier work, Tsutsuki et al. (42) compared soil chemistry between shiro and nonshiro soil in Gifu Prefecture, Japan, and reported that the pH and the concentrations of Ca2+ and K+ ions were slightly lower in shiro soil. However, no clear relationship between sporocarp formation and soil chemistry was found. More carbon and nitrogen in the soil immediately above the shiro might be related to the higher hemicellulose content and a common property of shiro soil, because P values were relatively low (= 0.064) compared to those for other soil constituents. Compared with Shiro+abv soil, differences between the soil chemistry of Shiro+ and Shiro− soils were much smaller.
Matsutake usually does not form rhizomorphs to transport nutrients over long distances to developing sporocarps, and extension through the substrate in vitro is rather limited compared to that for other ECM fungi (27, 47). The observation of apparently dead mycorrhizal root tips suggests that disconnection from the host plant may occur and calls into question the source of energy and nutrients used during sporocarp formation. Nutrient uptake from the shiro and superficial organic layer may be essential for sporocarp production, and our results support this hypothesis. Future research should test the saprobic potential of T. matsutake to utilize different forms of organic carbon (e.g., hemicelluloses and related compounds) and examine the effects of those positively correlating with the presence of T. matsutake in the shiro. Understanding the natural ecosystem of this fungus will help culture T. matsutake and determine the functional diversity of ECM fungi and their role in the environment.
In conclusion, T. matsutake appears to be a specialized member of the soil microbial community that, except for its carbon source, does not require specific soil conditions. We found that T. matsutake coexists with a diversity of fungal and actinobacterial species and observed a higher enzyme activity involved in the degradation of organic carbon in the shiro. The composition of soil organic matter and its dynamics above the shiro should be investigated to better understand the establishment and productivity of this fungus.
Supplementary Material
ACKNOWLEDGMENTS
We thank Eira-Maija Savonen, Tytti Sarjala, and Toyahiro Miyazawa for providing T. matsutake fruiting information in 2007 and for help with soil sampling and Tero Tuomivirta for his valuable comments concerning the molecular work. Minna Sinkkonen, Johanna Exell, Seija Vanhakoski, and Tuija Hytönen helped with PCR and sequencing, Satu Repo helped with the hemicellulose analysis, and Leena Hamberg, Mikko Peltoniemi, and Jaakko Heinonen kindly provided advice on statistical procedures. Michael Hardman checked the language, and Anne Siika helped with the illustrations. We also thank the reviewers for their help improving this study.
This research was fully supported by the Foundation for Research of Natural Resources in Finland.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 7 October 2011.
REFERENCES
- 1. Bergius N., Danell E. 2000. The Swedish matsutake (Tricholoma nauseosum syn. T. matsutake): distribution, abundance and ecology. Scand. J. Forest Res. 15:318–325 [Google Scholar]
- 2. Chambers S. M., Burke R. M., Brooks P. R., Cairney J. W. G. 1999. Molecular and biochemical evidence for managanese-dependent peroxidase activity in Tylospora fibrillosa. Mycol. Res. 103:1098–1102 [Google Scholar]
- 3. Chen D. M., Bastias B. A., Taylor A. F. S., Cairney J. W. G. 2003. Identification of laccase-like genes in ectomycorrhizal basidiomycetes and transcriptional requlation by nitrogen in Piloderma byssinum. New Phytol. 157:547–554 [DOI] [PubMed] [Google Scholar]
- 4. Courty P.-E., Pritsch K., Schloter M., Hartmann A., Garbaye J. 2005. Activity profiling of ectomycorrhiza communities in two forest soils using multiple enzymatic tests. New Phytol. 167:309–319 [DOI] [PubMed] [Google Scholar]
- 5. Frey-Klett P., Garbaye J., Tarkka M. 2007. The mycorrhiza helper bacteria revisited. New Phytol. 176:22–36 [DOI] [PubMed] [Google Scholar]
- 6. Garbaye J. 1994. Helper bacteria: a new dimension to the mycorrhizal symbiosis. New Phytol. 128:197–210 [DOI] [PubMed] [Google Scholar]
- 7. Gardes M., Bruns T. D. 1993. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol. Ecol. 2:113–118 [DOI] [PubMed] [Google Scholar]
- 8. Gill W. M., Guerin-Laguette A., Lapeyrie F., Suzuki K. 2000. Matsutake—morphological evidence of ectomycorrhiza formation between Tricholoma matsutake and host roots in a pure Pinus densiflora forest stand. New Phytol. 147:381–388 [Google Scholar]
- 9. Guilford J. P., Perry N. C. 1951. Estimation of other coefficients of correlation from the phi coefficient. Psychometrika 16:335–346 [Google Scholar]
- 10. Hasson K., Kleja D. B., Kalbitz K., Larsson H. 2010. Amounts of carbon mineralized and leached as DOC during decomposition of Norway spruce needles and fine roots. Soil Biol. Biochem. 42:178–185 [Google Scholar]
- 10a. Heinonsalo J., et al. Filter centrifugation as a sampling method for miniaturization of extracellular fungal enzyme activity measurements in solid media. Fungal Ecol., in press [Google Scholar]
- 11. Hosford D., Pilz D., Molina R., Amaranthus M. 1997. Ecology and management of the commercially harvested American matsutake. General technical report PNW-GTR-412. USDA Forest Service Pacific Northwest Research Station, Portland, OR [Google Scholar]
- 12. Jaatinen K., et al. 2008. Responses of aerobic microbial communities and soil respiration to water-level drawdown in a northern boreal fen. Environ. Microbiol. 10:339–353 [DOI] [PubMed] [Google Scholar]
- 13. Jongmans A. G., et al. 1997. Rock-eating fungi. Nature 389:682–683 [Google Scholar]
- 14. Kiikkilä O., Kitunen V., Smolander A. 2011. Properties of dissolved organic matter derived from silver birch and Norway spruce stands: degradability combined with chemical characteristics. Soil Biol. Biochem. 43:421–430 [Google Scholar]
- 15. Kikuchi K., Matsushita N., Gurein-Laguette A., Ohta A., Suzuki K. 2000. Detection of Tricholoma matsutake by specific ITS primers. Mycol. Res. 104:1427–1430 [Google Scholar]
- 16. Korkama T., Pakkanen A., Pennanen T. 2006. Ectomycorrhizal community structure varies among Norway spruce (Picea abies) clones. New Phytol. 171:815–824 [DOI] [PubMed] [Google Scholar]
- 17. Kotiranta H., Saarenoksa R., Kytövuori I. 2009. Aphyllophoroid fungi of Finland. A check-list with ecology, distribution, and threat categories. Norrlinia 19:1–223 [Google Scholar]
- 18. Kramer C., et al. 2010. Recent (<4 year old) leaf litter is not a major source of microbial carbon in a temperate forest mineral soil. Soil Biol. Biochem. 42:1028–1037 [Google Scholar]
- 19. Kusuda M., et al. 2006. Detection of β-glucosidase as a saprotrophic ability from an ectomycorrhizal mushroom, Tricholoma matsutake. Mycoscience 47:184–189 [Google Scholar]
- 20. Kytövuori I. 1988. The Tricholoma caligatum group in Europe and North Africa. Karstenia 28:65–77 [Google Scholar]
- 21. Lian C., Narimatsu M., Nara K., Hogetsu T. 2006. Tricholoma matsutake in a natural Pinus densiflora forest: correspondence between above- and below-ground genets, association with multiple host trees and alteration of existing ectomycorrhizal communities. New Phytol. 171:825–836 [DOI] [PubMed] [Google Scholar]
- 22. Lun Z.-M., Li Y.-H., Vaario L.-M. 2004. Ability of ectomycorrhizal fungus Tricholoma matsutake to utilize cellobiose. Mycosystema 23:563–567 [Google Scholar]
- 23. Mandels M., Reese E. T. 1957. Induction of cellulase in Trichoderma viride as influenced by carbon sources and metals. J. Bacteriol. 73:269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marchner B., Kalbitz K. 2003. Controls of bioavailability and biodegradability of dissolved organic matter in soils. Geoderma 113:211–235 [Google Scholar]
- 25. Merilä P., et al. 2010. Soil organic matter quality as a link between microbial community structure and vegetation composition along a successional gradient in a boreal forest. Appl. Soil Ecol. 46:259–267 [Google Scholar]
- 26. Ogawa M. 1975. Microbial ecology of mycorrhizal fungus—Tricholoma matsutake (Ito et Imai) Sing in pine forest. II. Mycorrhiza formed by T. matsutake. Bull. Gov. For. Exp. Stat. 278:21–80 [Google Scholar]
- 27. Ogawa M. 1978. The biology of matsutake mushroom. Tsukiji Shokan, Tokyo, Japan: (In Japanese.) [Google Scholar]
- 28. Ohara H., Hamada M. 1967. Disappearance of bacteria from the zone of active mycorrhizas in Tricholoma matsutake (S. Ito et Imai) Singer. Nature 213:528–529 [Google Scholar]
- 29. Oksanen J., et al. 2009. Vegan: community ecology package. R package version 1.15-2. [Google Scholar]
- 30. Ovaskainen O., et al. 2010. Identifying wood-inhabiting fungi with 454 sequencing—what is the probability that BLAST gives the correct species? Fungal Ecol. 3:274–283 [Google Scholar]
- 31. Park J. H., Kalbitz K., Matzner E. 2002. Resource control on the production of dissolved organic carbon and nitrogen in a deciduous forest floor. Soil Biol. Biochem. 34:813–822 [Google Scholar]
- 32. Paul E. A., Clark F. E. 1996. Soil microbiology and biochemistry. Academic Press, San Diego, CA [Google Scholar]
- 33. Pennanen T., Paavolainen L., Hantula J. 2001. Rapid PCR-based method for the direct analysis of fungal communities in complex environmental samples. Soil Biol. Biochem. 33:697–699 [Google Scholar]
- 34. R Development Core Team 2009. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 35. Šnajdr J., et al. 2011. Transformation of Quercus petraea litter: successive changes in litter chemistry are reflected in differential enzyme activity and changes in the microbial community composition. FEMS Microbiol. Ecol. 75:291–303 [DOI] [PubMed] [Google Scholar]
- 36. Southern P. M., Jr., Kutscher A. E., Ragsdale R., Luttrell B. 1987. Susceptibility in vitro of Nocardia species to antimicrobial agents. Diagn. Microbiol. Infect. Dis. 8:119–122 [DOI] [PubMed] [Google Scholar]
- 37. Stach J. E. M., Maldonado L. A., Ward A. C., Goodfellow M., Bull A. T. 2003. New primers for the class Actinobacteria: application to marine and terrestrial environments. Environ. Microbiol. 5:828–841 [DOI] [PubMed] [Google Scholar]
- 38. Sundberg A., Sundberg K., Lillandt C., Holmbom B. 1996. Determination of hemicelluloses and pectins in wood and pulp fibres by acid methanolysis and gas chromatography. Nord. Pulp Pap. Res. J. 11:216–219 [Google Scholar]
- 39. Suzuki K. 2005. Ectomycorrhizal ecophysiology and the puzzle of Tricholoma matsutake. J. Jpn. For. Soc. 87:90–102 (In Japanese with English summary.) [Google Scholar]
- 40. Tenkanen M., Puls J., Poutanen K. 1992. Two major xylanases of Tricholoma reesei. Enzyme Microb. Technol. 14:566–574 [Google Scholar]
- 41. Timonen S., Marschner P. 2006. Mycorrhizosphere concept, p. 155–172 In Mukerji K. G., Manoharachary C., Singh J. (ed.) Microbial activity in the rhizosphere. Springer-Verlag, Berlin, Germany [Google Scholar]
- 42. Tsutsuki K., Kawakata T., Tani M., Kondo R. 2005. Characteristics of Tricholoma matsutake growing soil, p. 26 In Proceedings of the annual meeting of the Japanese Society of Soil Science and Plant Nutrition, vol. 51 (In Japanese.) [Google Scholar]
- 43. Uselman S. M., Qualls R. G., Lilienfein J. 2009. Production of total potentially solube organic C, N, and P across an ecosystem chronosequence: root versus leaf litter. Ecosystems 12:240–260 [Google Scholar]
- 44. Vaario L.-M., Guerin-Laguette A., Matsushita N., Suzuki K., Lapeyrie F. 2002. Saprobic potential of Tricholoma matsutake: growth over pine bark treated with surfactants. Mycorrhiza 12:1–5 [DOI] [PubMed] [Google Scholar]
- 45. Vaario L.-M., Gill W. M., Samejima M., Suzuki K. 2003. Detection of the ability of Tricholoma matsutake to utilize sawdust in aspetic culture. Symbiosis 34:43–52 [Google Scholar]
- 46. Vaario L.-M., Pennanen T., Sarjala T., Savonen E., Heinonsalo J. 2010. Ectomycorrhization of Tricholoma matsutake and two main forest tree species in Finland—an assessment of in vitro mycorrhiza formation. Mycorrhiza 20:511–518 [DOI] [PubMed] [Google Scholar]
- 47. Wang Y. 1995. Tricholoma matsutake. Ph.D. thesis University of Otago, Dunedin, New Zealand [Google Scholar]
- 48. White T. J., Bruns T., Lee S., Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315–322 In Innis M. A., Gelfaud D. H., Sninsky J. J., White T. J. (ed.). PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA [Google Scholar]
- 49. Yamada A., Maeda K., Ohmasa M. 1999. Ectomycorrhiza formation of Tricholoma matsutake isolates on seedlings of Pinus densiflora in vitro. Mycoscience 40:455–463 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.