Abstract
Soil nitrification is an important process for agricultural productivity and environmental pollution. Though one cultivated representative of ammonia-oxidizing Archaea from soil has been described, additional representatives warrant characterization. We describe an ammonia-oxidizing archaeon (strain MY1) in a highly enriched culture derived from agricultural soil. Fluorescence in situ hybridization microscopy showed that, after 2 years of enrichment, the culture was composed of >90% archaeal cells. Clone libraries of both 16S rRNA and archaeal amoA genes featured a single sequence each. No bacterial amoA genes could be detected by PCR. A [13C]bicarbonate assimilation assay showed stoichiometric incorporation of 13C into Archaea-specific glycerol dialkyl glycerol tetraethers. Strain MY1 falls phylogenetically within crenarchaeal group I.1a; sequence comparisons to “Candidatus Nitrosopumilus maritimus” revealed 96.9% 16S rRNA and 89.2% amoA gene similarities. Completed growth assays showed strain MY1 to be chemoautotrophic, mesophilic (optimum at 25°C), neutrophilic (optimum at pH 6.5 to 7.0), and nonhalophilic (optimum at 0.2 to 0.4% salinity). Kinetic respirometry assays showed that strain MY1's affinities for ammonia and oxygen were much higher than those of ammonia-oxidizing bacteria (AOB). The yield of the greenhouse gas N2O in the strain MY1 culture was lower but comparable to that of soil AOB. We propose that this new soil ammonia-oxidizing archaeon be designated “Candidatus Nitrosoarchaeum koreensis.”
INTRODUCTION
Eutrophication of terrestrial and aquatic systems, caused by industrial production and use of artificial nitrogen fertilizers worldwide, has led to a host of environmental problems (13). Autotrophic nitrification is a microbially mediated process that converts ammonia to nitrate and thus plays an essential role in soil nitrogen cycles. In agricultural systems, nitrification results in substantial loss of soil fertilizer nitrogen (50 to 70%) (75) due to metabolic coupling with denitrification (13) and anaerobic ammonia oxidation (33) that discharges nitrogen as dinitrogen gas. Soil nitrification from increased agricultural activities contributes significantly to global warming, since nitrification is a major source of the strong greenhouse gas nitrous oxide (N2O), which has a ca. 300-times-higher warming impact than CO2 (66). Nitrous oxide emissions are also responsible for ozone depletion in the stratosphere (19).
The first step of nitrification, oxidation of ammonia, long thought to be exclusive to the domain Bacteria (10), was recently tied to the archaeal domain: metagenomic analysis showed that a soil fosmid clone harbored both archaeal 16S rRNA and amoA-like genes (86). In an independent study, archaeal amoA-like genes were also recovered from surface waters of the Sargasso Sea (90). There is growing evidence that these archaeal amoA genes occur widely in a variety of environments, including soils and marine habitats (49, 97). Further, critical evidence for autotrophic ammonia-oxidizing archaea (AOA) was obtained by the characterization of the cultivated mesophilic Crenarchaea (recently proposed as Thaumarchaea [12]) from marine (group I.1a) (40, 59, 97) and hot spring (group I.1b and thermophilic AOA lineage) environments (21, 28).
The contribution of AOA to the nitrification of soil is still controversial. Quantitative analysis of amoA gene copies has indicated that AOA can predominate over ammonia-oxidizing bacteria (AOB) in various soils (16, 49). Also, copy numbers of archaeal amoA genes were found to increase and 13CO2 was incorporated into genomic DNA of the AOA during ammonia oxidation (99). Additionally, expression of archaeal amoA was elevated in ammonia-amended soils (49, 86). In contrast, there have been several recent reports demonstrating that growth of AOB (not AOA) can be coupled with ammonia oxidation in soils. For example, 13CO2 was incorporated mainly into the DNA of AOB, and AOB abundance was correlated with ammonia oxidation activity (35). In other reports (23, 24), inorganic-fertilizer amendment increased the copy numbers of amoA of AOB, and this increase was eliminated by added dicyanodiamide (DCD) (an ammonia oxidation inhibitor), while AOA abundance did not respond to the same fertilizer amendment. The study by Schauss et al. (72) showed that both AOA and AOB were active ammonia oxidizers in soil amended with organic fertilizer. Clearly, both domains have the potential to carry out ammonia oxidation.
To date, only a limited number of AOA have been isolated and/or enriched in laboratory culture. They have predominantly been from nonterrestrial (nonsoil) habitats: seawater aquarium filters, hot springs, and marine sediment (21, 28, 40, 59, 97). Despite the common retrieval of archaeal 16S rRNA gene and amoA gene sequences from various terrestrial environments, we are aware of only two soil-derived AOA, both members of the crenarchaeal group I.1b lineage: “Candidatus Nitrososphaera viennensis” strain EN76 was isolated from garden soil (85). This narrow field of characterized AOA has impaired our ability to advance our understanding of AOA in soil environments. In the present study, we were able to obtain a highly enriched culture of an ammonia-oxidizing archaeon from an agricultural soil and to compare the ammonia oxidation properties of this AOA to those of AOB and, hence, to obtain clues about the role of AOA in the nitrogen cycle for soil environments.
MATERIALS AND METHODS
Soil sampling site.
We collected soil samples from plots planted with Caragana sinica at the experimental agricultural station of Chungbuk National University of South Korea (36°37′N, 127°27′E). The soils were loamy sand (sand, 82.4%; silt, 6.1%; clay, 11.5%) and did not receive nitrogen fertilizers due to C. sinica's ability to carry out nitrogen fixation. Bulk and rhizosphere soils collected at depths of 0 to 30 cm, respectively, were combined into a single 100-g sample and transported to the laboratory to be stored at 4°C before being used for inoculation.
Cultivation of AOA.
One gram of soil was inoculated into an artificial freshwater medium (AFM), and the cultures were grown aerobically in the dark without shaking at 25°C. The AFM contained the following components per liter of culture medium: 0.4 g MgCl2 · 6H2O, 0.5 g KCl, 0.2 g KH2PO4, 1.0 g NaCl, and 0.1 g CaCl2 · 2H2O. After autoclaving 1 ml nonchelated trace element solution (95), 1 ml NaFeEDTA solution (7.5 mM) and 3 ml NaHCO3 (1 M) were added aseptically per liter of media. Unless stated otherwise, each starting batch culture was supplemented with 1 mM ammonium chloride as a sole energy source. After the oxidation of ammonia (typically after 3 weeks), 1% of the culture volume was transferred to fresh AFM. The pH of the medium remained almost constant (6.8 to 7.0) during the culture cycle. The concentrations of nitrite and nitrate were determined with an ion chromatograph (ICS-1500; Dionex, Sunnyvale, CA) with an OnGuard II Ag cartridge (Dionex). The ammonia concentration was determined colorimetrically (77). After 2 years of triweekly transfers, the enrichment culture was 10-fold serially diluted in Hungate tubes and incubated under the same conditions used for the enrichment culture. The Hungate tube showing nitrification activity in most diluted series was checked for culture purity using microscopy and molecular techniques. The nitrification activity was assessed spectrophotometrically by determining the decrease in the ammonia concentration in the Hungate tube.
The activity of ammonia oxidation by Archaea was analyzed in the presence of various antibiotics: streptomycin (100 μg/ml), kanamycin (50 μg/ml), ampicillin (50 μg/ml), penicillin-G (50 μg/ml), gentamicin (20 μg/ml), mitomycin C (20 μg/ml), and lincomycin (50 μg/ml). The cultures were incubated for 3 weeks, as described for enrichment cultures. Ammonia oxidation activity and growth of Archaea were determined by spectrophotometry and by PCR amplification of archaeal 16S rRNA genes, respectively.
To compare the ammonia oxidation traits of strain MY1 to those of Nitrosomonas europaea ATCC 19718, the AFM medium and cultivation conditions described above were used. Inocula (mid-log-phase cultures) were at initial cell densities of 1.2 × 106 ± 0.1 × 106 and 3.3 × 106 ± 0.3 × 106 cells/ml for strain MY1 and N. europaea, respectively. No nitrite was formed in the control that was not inoculated.
Quantification of gene copy numbers using real-time PCR.
In order to determine AOA and AOB in the cultures, the copy numbers of the amoA and 16S rRNA genes were determined using primers described in Table 1. Cells were harvested from a 50-ml culture by centrifugation and immediately frozen and stored at −70°C until further analysis. DNA was isolated from frozen cells using the Genomic DNA Prep Kit (Solgent, South Korea) according to the manufacturer's instructions. The concentration of DNA was determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE).
Table 1.
Primers used for PCR amplification for library construction and real-time quantification
| Target gene and primer | Application | Sequence (5′ to 3′) | Position | Note | Reference |
|---|---|---|---|---|---|
| 16S rRNA | |||||
| 519F | Archaeal quantification | CAGCMGCCGCGGTAA | 519–533a | 59, 94 | |
| 727R | GCTTTCRTCCCTCACCGT | 712–727a | |||
| Bac518F | Bacterial quantification | CCAGCAGCCGCGGTAAT | 518–534a | 6, 55 | |
| Bac786R | CTACCAGGGTATCTAATC | 786–803a | |||
| 20F | Archaeal clone library | TTCCGGTTGATCCYGCCRG | 2–20a | 22 | |
| 1492R | TACGGYTACCTTGTTACGACTT | 1510–1492a | 94 | ||
| 27F | Bacterial clone library | AGAGTTTGATCMTGGCTCAG | 8–27a | With 1492R | 94 |
| NIT2R | Nitrobacter spp. | CGGGTTAGCGCACCGCCT | 1433–1450a | With 27F | 91 |
| NIT3R | CCTGTGCTCCATGCTCCG | 1035–1048a | |||
| Ntspa 662R | Nitrospira spp. | GGAATTCCGCGCTCCTCT | 662–679a | With 27F | 20 |
| Ntspa 712R | CGCCTTCGCCACCGGCCTTCC | 712–732a | |||
| βAMOF | PCR (β-AOB) | TGGGGRATAACGCAYCGAAAG | 143–163a | 53 | |
| βAMOR | AGACTCCGATCCGGACTACG | 1296–1315a | |||
| amoA | |||||
| AamoAF | Archaeal clone library | STAATGGTCTGGCTTAGACG | 19–38b | 25 | |
| AamoAR | ACATACAGATGGATGGCCGC | 582–601b | |||
| I.1a-amoAF | Archaeal quantification | TGTACWCACTACTTRTTCATA | 239–270c | This study | |
| I.1a-amoAR | GARTGYTTRTTYTTCTTTGT | 590–610c | |||
| amoA1F | PCR (β-AOB) | GGGGTTTCTACTGGTGGT | 332–349d | 69 | |
| amoA2R | CCCCTCKGSAAAGCCTTCTTC | 820–822d | |||
| A189F | PCR (γ-AOB) | GGNGACTGGGACTTCTGG | 172–189e | 57 | |
| A682R | GAASGCNGAGAAGAASGC | 665–682e | |||
| nirK | |||||
| March_nirKF | PCR (archaeal nirK) | TCTGGTGTTAAACTAATTGGT | 46–66f | This study | |
| March_nirKR | GTTSCTGCGGATTGTACT | 540–557f | |||
| nirK1F | PCR (bacterial nirK) | GGMATGGTKCCSTGGCA | 526–542g | 11 | |
| nirK5R | GCCTCGATCAGRTTRTGG | 1021–1040g | |||
| norB | |||||
| norB1F | PCR (bacterial norB) | CGNGARTTYCTSGARCARCC | 400–419h | 15 | |
| norB8R | CRTADGCVCCRWAGAAVGC | 1051–1069h |
Numbering is based on the 16S rRNA gene of E. coli.
The numbering of the gene is based on the gene of a metagenomic clone from the Sargasso Sea (AACY01435967).
Numbering of each gene is based on the gene of “Ca. Nitrosopumilus maritimus” (NC 010085).
Numbering is based on the bacterial amoA of N. europaea (L08050).
Numbering is based on the bacterial amoA of Nitrosococcus oceani (AF047705).
Numbering is based on the archaeal nirK-like gene of “Ca. Nitrosopumilus maritimus” (NC 010085).
Numbering is based on the bacterial nirK gene of Alcaligenes faecalis (D13155).
Numbering is based on the bacterial norB gene of Pseudomonas stutzeri (Z28384).
All quantitative real-time PCR experiments were carried out using a MiniOpticon real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA) and Opticon Monitor software version 3.1 (Bio-Rad Laboratories, Hercules, CA). For the amplification of all genes, the following thermal-cycling parameters were used: 15 min at 95°C and 40 cycles of 20 s at 95°C, 20 s at 55°C, and 20 s at 72°C. Readings were taken between cycles. Standard curves were generated to obtain the relationship between the known copy numbers of reference genes and cycle threshold (CT) values as previously described (61). Standard curves were prepared in each run using standards of reference genes (archaeal 16S rRNA gene, DQ831586; bacterial 16S rRNA gene, FJ656473; archaeal amoA, EF534487; archaeal nirK, HQ631404; bacterial nirK and norB, JF737970 and JF737971, respectively) at abundances ranging from 0 to 109 gene copies per reaction. These curves were used to estimate gene abundance in the enrichment samples. The specificity of real-time PCRs was tested by analyzing melting curves, checking the sizes of reaction products using gel electrophoresis, and sequencing of the reaction products. Information about the PCR amplification primers for each target gene is provided in Table 1.
Phylogenetic analysis of 16S rRNA, amoA, and nirK genes.
Archaeal 16S rRNA and archaeal amoA and nirK genes were amplified by PCR using the primers listed in Table 1. The PCR conditions for both the 16S rRNA and amoA genes were as follows: 5 min at 94°C; 30 cycles of 30 s at 94°C, 30 s at 55°C, and 45 s at 72°C; and 7 min at 72°C. The PCR conditions for the nirK gene amplification were 5 min 94°C; 35 cycles of 30 s at 94°C, 30 s at 50°C, and 45 s at 72°C; and 7 min at 72°C. Clone libraries were constructed using the PCR products, which were purified using the PCR Purification System Kit (Solgent, South Korea), ligated using the T&A Cloning Vector Kit (Real Biotech Corporation, Taiwan), and then transformed into Escherichia coli DH5α cells according to the manufacturer's instructions. Both strands of the gene from the positive clone were sequenced using corresponding PCR primers (Table 1). For phylogenetic analysis, the gene sequences of related taxa were obtained from the GenBank database. Multiple alignments of 16S rRNA gene sequences were performed using SILVA (http://www.arb-silva.de/aligner) (64) and considering the secondary structure of the rRNA gene. Shared regions of the amoA (560 bp) and nirK (460 bp) gene sequences were aligned using Clustal X (83). Phylogenetic analyses were conducted using MEGA version 4.0 (43), and the neighbor-joining tree was constructed using Kimura 2-parameter correction (39) with 1,000 replicates to produce bootstrap values.
Bicarbonate incorporation analysis.
In order to determine bicarbonate incorporation by AOA during ammonia oxidation, a known amount of [13C]bicarbonate (99% 13C; Cambridge Isotope Laboratories, Andover, MA) was applied, resulting in a 6.0% labeling of total bicarbonate. A control culture was incubated under the same conditions without the addition of [13C]bicarbonate.
Total cells were harvested by centrifugation and freeze-dried for archaeal glycerol dialkyl glycerol tetraether (GDGT) membrane lipid analysis using the following procedures described by Pitcher et al. (62). Briefly, the freeze-dried cells were extracted by a modified Bligh-Dyer extraction, and the intact polar lipids were acid hydrolyzed to release the core GDGTs. To measure the degree of 13C incorporation in archaeal GDGTs, they were analyzed on an Agilent (Palo-Alto, CA) 1100 series LC/MSD SL using selective ion monitoring as described in detail elsewhere (73). m/z 1302 to 1312 and m/z 1292 to 1302 were measured in selected time intervals.
Determination of the kinetics of ammonia oxidation and oxygen uptake.
Kinetic studies of oxygen uptake and ammonia oxidation were performed with the strain MY1 culture as described elsewhere (52) with appropriate modifications. Oxygen uptake was measured in an Oxygraph system (Hansatech Instruments Ltd., England) equipped with an S1 Clark-type polarographic oxygen electrode disc, a 2-ml DW1 borosilicate glass reaction/sample vessel, magnetic stir bars, and the supplied Oxygraph Plus software. Oxygen microsensors were polarized continuously for >3 h before use. All measurements were done in a recirculated water bath at 25°C. Activity measurements were carried out with late-exponential or early-stationary-phase cells by monitoring ammonia oxidation and nitrite production by strain MY1 cells. Aliquots (10 ml) were removed from cultures and immediately transferred to prewarmed 25-ml glass tubes in a 25°C water bath. Subsamples were then used to fill the 2-ml DW1 electrode glass vessel assembled with an oxygen electrode disc and plunger assembly before being carefully sealed. Oxygen uptake was monitored continuously after an initial equilibration of at least 40 min. Ammonia was added as necessary from a stock solution using a Hamilton syringe. The apparent half-saturation constant (Km) for oxygen and ammonia and the maximum velocity (Vmax) were determined from plots of oxygen uptake rates.
Determination of N2O yield.
Production of N2O was measured in serum bottles (120 ml with a 70-ml headspace; sealed with a butyl rubber stopper) that contained 50 ml of AFM inoculated with strain MY1 or N. europaea. We used two different oxygen tensions for cultivation; one was fully aerated, and the other was oxygen limited. For the oxygen-limited condition, we removed oxygen from the medium by purging with N2 gas and then added only 2 ml of air to the headspaces of bottles. The cultures were incubated at 25°C for 2 weeks under dark conditions. Triplicate samples were prepared and incubated for determination of the N2O yield. The concentration of N2O gas in the headspace was measured with a gas chromatograph with an electron capture detector (GC/ECD) (Agilent 6890A). Sample gas was taken from the headspaces of bottles using a 5-ml gas-tight syringe. The GC was fitted with a 1-ml sample loop and a 4-m stainless steel column packed with Porapak Q (Restek). The oven was isothermal (50°C), and N2O concentrations were determined three times for each sample. We used six certified reference gas mixtures of N2O in nitrogen—0.198, 0.331, 0.550, 0.798, 12.0, and 94.9 μmol N2O/mol—to calibrate the GC/ECD. These reference gas mixtures were gravimetrically prepared by the Korea Research Institute for Standards and Science and verified by international comparisons, CCQM-K68, in 2010 (46). The uncertainty in the N2O concentration was ca. 7%, caused by the nonlinearity of the ECD and the sampling procedure. Oxygen in the headspace was determined by using GC/thermal conductivity detector (TCD) as described previously (47).
FISH and electron microscopy analysis.
Fluorescence in situ hybridization (FISH) was carried out on paraformaldehyde-fixed samples as described by Amann et al. (4). Cells from a 10-ml culture volume were harvested and resuspended in phosphate-buffered saline (PBS) (130 mM NaCl, 10 mM sodium phosphate, pH 7.5), fixed by the addition of 3 volumes of cold paraformaldehyde solution (4% in PBS), and stored at 4°C for 16 h. Cells were harvested and washed 3 times with PBS solution to remove residual fixation solution and concentrated 10-fold by centrifugation in PBS solution. The fixed cell suspension was mixed with an equal volume of cold absolute ethanol, and the mixture was stored at −20°C. For the FISH analysis of Bacteria and Archaea, paraformaldehyde-fixed samples were hybridized with Cy3-labeled Archaea-specific probe (Arc915) (2) and 6-carboxyfluorescein (FAM)-labeled Bacteria-specific probe (EUB338) (3). DAPI (4′,6-diamidino-2-phenylindole) was used to visualize total cells. Samples were observed with a Nikon 80i fluorescence microscope (Nikon, Tokyo) with an oil immersion objective.
For scanning electron microscopy (SEM), cells were harvested and immersed in 4% (vol/vol) glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.2) for 24 h at 4°C and dehydrated through a graded ethanol series (70 to 100%). The specimens were examined with a Zeiss DSM 940 electron microscope (Carl Zeiss). To enable transmission electron microscopy (TEM) analyses of AOA from the enrichment cultures, the cells from a 10-ml culture were fixed in a 2.5% paraformaldehyde-1.5% glutaraldehyde mixture buffered with 0.1 M phosphate (pH 7.2) for 2 h at 4°C, postfixed in 1% osmium tetroxide in the same buffer for 1 h, dehydrated in a graded ethanol series (70% to 100%), transferred to propylene oxide, and embedded in Epon-812 (TAAB, England) (50). Ultrathin sections generated with an Ultracute (Leica, Austria) ultramicrotome were stained with uranyl acetate and lead citrate and examined with a CM 20 electron microscope (Philips, Netherlands) (68).
Nucleotide sequence accession numbers.
The sequences obtained in this study have been deposited in GenBank (NCBI) under accession numbers HQ331116 to HQ331117, HQ631403 to HQ631405, and JF737930 to JF37971.
RESULTS
Establishment of an ammonia-oxidizing enrichment culture.
Soil samples were gathered from field plots where the leguminous plant C. sinica was growing and were used for enrichment cultivation of ammonia-oxidizing microorganisms. The properties of the soil were as follows: sandy loam texture; pH, 5.6; organic C, 16 g/kg; total N, 0.5 g/kg; ammonia N, 3 mg/kg; nitrate N, 6 mg/kg; and total P, 150 mg/kg. After the initial soil inoculum (1 g in 100 ml AFM) catalyzed conversion of ammonia to nitrate, the nitrifying culture was repeatedly transferred (1% [vol/vol]) to fresh AFM triweekly. Early signs that the culture carried active Archaea were derived from sequences of PCR-amplified archaeal 16S rRNA (primer set 20F-1492R) and amoA (primer set AamoAF-AamoAR) genes (data not shown). The series of 1% enrichment transfers continued for 2 years. Finally, the culture was serially diluted to extinction in 10-fold steps, and the highest dilution showing nitrifying activity was selected for further characterization.
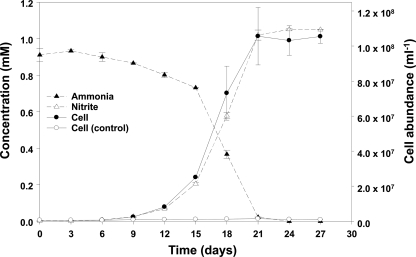
Characterization of the microbial community showed that the initially complex archaeal community became refined and eventually seemed uniarchaeal, as revealed by a single band in denaturing gradient gel electrophoresis (DGGE) analysis (data not shown). Ammonia oxidation by the final (serially diluted) culture is shown in Fig. 1. After inoculation (initial cell concentration, ca. 106 cells/ml), ammonia was stoichiometrically converted to nitrite over a period of about 3 weeks. Concomitantly, growth of archaeal cells (determined by Archaea-specific FISH of filtered culture samples) occurred only in the inoculated treatment (Fig. 1). In medium without ammonia as an electron donor, no archaeal growth was observed.
Fig. 1.
Ammonia oxidation by the enrichment culture. The concentrations of ammonia and nitrite and the cell abundance of Archaea are indicated. FISH of filtered culture samples was used for determination of archaeal cell numbers. The control culture contained nitrate instead of ammonia. The error bars represent the standard deviations from triplicate experiments.
Bacterial 16S rRNA gene sequence analysis revealed that all sequences of Bacteria cooccurring in the Archaea-dominated enrichment culture belonged to the Proteobacteria and Bacteroidetes/Chlorobi groups (see Fig. S1 in the supplemental material). Three clone sequences of Proteobacteria were dominant (82.5%). The closest relatives to the cloned sequences were Acinetobacter calcoaceticus, Pseudomonas knackmussii, and Variovorax boronicumulans, which are catabolically diverse heterotrophs generally viewed as contributing to mineralization of organic compounds in typical soils (see Fig. S1 in the supplemental material).
Quantitative purity of the ammonia-oxidizing archaeal enrichment and partial characterization of its key autotrophic member, “strain MY1,” using a 13CO2 incorporation assay.
The degree to which the ammonia-oxidizing culture was enriched in Archaea was determined by microscopy of cell preparations, using domain-specific FISH probes. Ninety-one percent of the cells were archaeal, as determined by the ratio of cell counts/ml (1.4 × 108 ± 1.1 × 107Archaea versus 1.3 × 107 ± 2.1 × 106 Bacteria). This was confirmed with real-time PCR of domain-specific 16S rRNA genes: Archaea represented 90% of the rRNA genes (archaeal [1.1 × 108 ± 0.4 × 108 copies/ml] versus bacterial [1.0 × 107 ± 0.2 × 107 copies/ml]). The yield of Archaea from ammonia oxidation was estimated at 1.1 × 105 cells/ml/μM ammonia consumed; this is lower than the yield reported for an acidophilic AOA, “Candidatus Nitrosotalea devanaterra” (4.5 × 105 cells/ml/μM ammonia consumed) (48) but higher than that of the marine AOA “Candidatus Nitrosopumilus maritimus” (5.0 × 104 cells/ml/μM ammonia consumed) (40).
The bacterial cells present in the ammonia-oxidizing enrichment could not be eliminated by treatment with various antibiotics (streptomycin, kanamycin, ampicillin, penicillin G, gentamicin, mitomycin C, or lincomycin); this was because the antibiotics inhibited both the Bacteria and the Archaea (see Fig. S2 in the supplemental material).
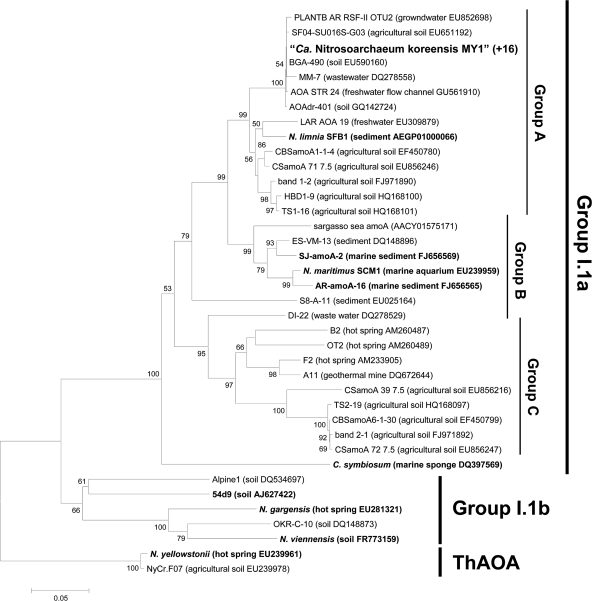
After DNA extraction, attempts to amplify bacterial amoA genes and 16S rRNA genes of gamma- and betaproteobacterial AOB using specific primers (bacterial amoA, amoA1F-amoA2R and A189F-A682R; bacterial 16S rRNA gene, βAMOF-βAMOR) failed. However, nitrite-oxidizing bacteria (NOB) related to Nitrobacter vulgaris DSM 10236T (97.9% similarity [89]) and Nitrospira sp. (95.7% similarity [32]) were initially detected by PCR assays in the culture. NOB were eventually eliminated from the culture by using 10 μM chlorate, a NOB inhibitor. In contrast to the bacterial amoA genes, archaeal amoA genes could be amplified using specific primers (Table 1). Archaeal amoA gene amplicons, all sharing identical sequences, were obtained from 16 randomly selected library clones from the enrichment culture; they fell within “group A” of known archaeal amoA genes (Fig. 2). The uniformity of the archaeal amoA gene library indicated a high-purity enrichment.
Fig. 2.
Phylogenetic analysis of the archaeal amoA gene sequence (ca. 560 bp) obtained from strain MY1. Archaeal amoA genes were amplified using primers AamoAF and AamoAR. The amoA gene sequences from 16 clones randomly selected from the library were identical. Branching patterns supported by more than 50% bootstrap values (1,000 iterations) by means of neighbor joining are denoted by their respective bootstrap values. Cluster groups are denoted at the right based on the origins of reference sequences. “ThAOA” indicates thermophilic AOA lineage. The scale bar represents 5% estimated sequence divergence. Enriched or isolated AOA among the reference sequences are indicated in boldface.
PCR amplification and cloning of the archaeal 16S rRNA genes from the highly purified enrichment culture yielded a library of 20 identical sequences. Analysis of the 16S rRNA gene showed a sequence, here referred to as “strain MY1,” that is phylogenetically affiliated with group I.1a within Crenarchaea, composed mostly of sequences from soil (Fig. 3). The 16S rRNA gene tree (Fig. 3) shows two soil clusters in the I.1a group that are distinct from the marine cluster; the latter contains mostly marine sequences, including the first cultivated mesophilic ammonia-oxidizing archaeon, “Ca. Nitrosopumilus maritimus” (40). “Soil cluster 1” and “soil cluster 2” contain mostly sequences from terrestrial environments. The similarities between the 16S rRNA gene sequence of strain MY1 and those of “Ca. Nitrosopumilus maritimus,” “Ca. Nitrososphaera viennensis,” and “Ca. Nitrosoarchaeum limnia” (9) were 96.9%, 84.8%, and 99.1%, respectively, while the 16S rRNA gene sequence similarity between strains MY1 and 54d9, a fosmid clone of crenarchaeal group I.1b previously found in soil (86), was only 83.1% (Fig. 3).
Fig. 3.
Phylogenetic analysis of the archaeal 16S rRNA gene sequence (ca. 1.3 kbp) obtained from strain MY1. Archaeal 16S rRNA genes were amplified using primers 20F and 1492R. The 16S rRNA gene sequences from 20 randomly selected clones from the library were identical. Branching patterns supported by more than 50% bootstrap values (1,000 iterations) by means of neighbor joining are denoted by their respective bootstrap values. Cluster groups are denoted at the right based on the origins of reference sequences. “ThAOA” indicates thermophilic AOA lineage. The scale bar represents 2% estimated sequence divergence. Enriched or isolated AOA among the reference sequences are indicated in boldface.
The phylogeny of strain MY1's amoA sequence was congruent with that of strain MY1's16S rRNA gene sequence (compare Fig. 2 and 3); this is consistent with MY1's affiliation with the soil cluster of crenarchaeal group I.1a. The similarities of the amoA (AmoA) sequences of strain MY1 to those of “Ca. Nitrosopumilus maritimus,” “Ca. Nitrososphaera viennensis,” and “Ca. Nitrosoarchaeum limnia” were 89.2% (96.7%), 70.0% (82.0%), and 94.5% (99%), respectively. The similarity of the amoA (AmoA) sequence of strain MY1 with that of 54d9 was 74.8% (83.7%) (Fig. 2).
To further confirm that the active agent (strain MY1) in our highly purified enrichment culture was, in fact, an autotrophic archaeon, we investigated its carbon fixation capability with a labeling experiment. We completed ammonia oxidation assays in [13C]bicarbonate-amended treatments (6.0% 13C enriched) and subsequently extracted glycerol dialkyl glycerol tetraethers (GDGTs), known to be characteristic of Archaea (62, 76). As determined by liquid chromatography/mass spectrometry (LC/MS), the major core GDGTs of strain MY1 contained no rings (GDGT-0) or crenarchaeol; this is the characteristic lipid distribution of AOA (59, 62, 76). The shift in molecular mass of the GDGTs by ∼6 Da for GDGT-0 (see Fig. S3A and B in the supplemental material) and crenarchaeol (see Fig. S3C and D in the supplemental material) indicated complete incorporation of the 13C label into these AOA membrane lipids, providing evidence for autotrophic growth of strain MY1.
The influence of environmental variables on ammonia oxidation by strain MY1.
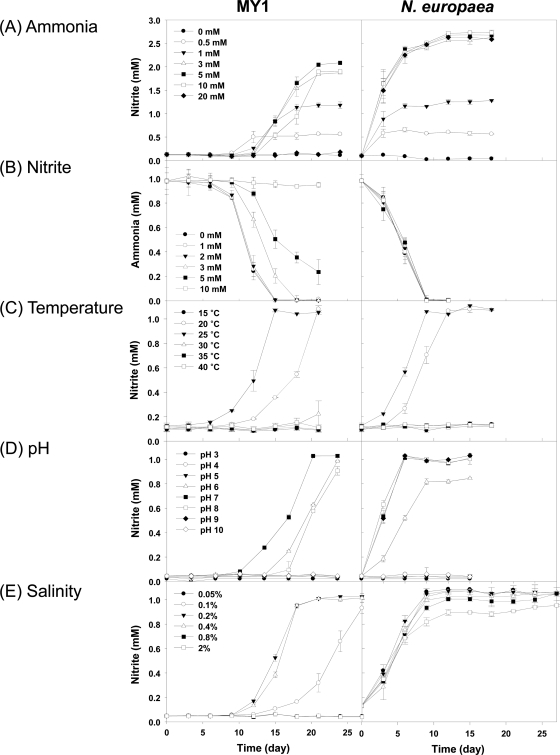
In an attempt to understand the physiological capabilities that may have allowed strain MY1 to function in terrestrial environments, the traits of strain MY1 were compared to those of an ammonia-oxidizing bacterium in soil, N. europaea ATCC 19718. Treatment with known ammonia oxidation inhibitors (10 μM nitrapyrin, 50 μM chlorite, or 500 μM DCD) strongly inhibited ammonia oxidation by both strain MY1 and N. europaea. However, allylthiourea (up to 500 μM) did not inhibit archaeal ammonia oxidation by strain MY1, while N. europaea was very sensitive to allylthiourea (<10 μM) (see Fig. S4 in the supplemental material). The typical MIC of allylthiourea for ammonia oxidation by AOB is ca. 1 to 10 μM (8).
The abilities of strain MY1 and N. europaea ATCC 19718 to convert ammonia to nitrite were also compared. Under our culture conditions, oxidation of ammonia by strain MY1 only started after 9 to 12 days of inoculation, while no lag time was detected in N. europaea. Ammonia oxidation by strain MY1 was not significantly inhibited in the presence of up to 5 mM ammonia and was still observed at 10 mM (Fig. 4A). In the presence of 20 mM ammonia, ammonia oxidation by strain MY1 was completely inhibited, while no inhibition of ammonia oxidation was observed at this concentration for N. europaea. This result indicates that, like other AOA (51, 59), strain MY1 has a lower tolerance for ammonia than AOB.
Fig. 4.
Effects of ammonia (A), nitrite (B), temperature (C), pH (D), and salinity (E) on the ammonia oxidation activities of strain MY1 and N. europaea. The same AFM and cultivation conditions were used for strain MY1 and N. europaea. Ammonia oxidation activity was indicated by the amount of nitrite accumulated. In the study on nitrite inhibition, ammonia consumption was determined. The error bars represent standard deviations from triplicate experiments.
Nitrite produced by ammonia oxidation is toxic and may suppress the growth of ammonia oxidizers (79, 81). Strain MY1 was more sensitive to nitrite than N. europaea (Fig. 4B). Above 2 mM nitrite, ammonia oxidation by strain MY1 was inhibited. Although ammonia oxidation by strain MY1 was still observed with up to 5 mM nitrite, it was completely inhibited with 10 mM nitrite. However, ammonia oxidation by N. europaea was not affected by 10 mM nitrite and possibly even higher nitrite concentrations (Fig. 4B).
The responses to temperature were similar for strain MY1 and N. europaea (Fig. 4C). Strain MY1 showed maximum ammonia oxidation at 25°C, with a slightly decreased rate at 20°C. However, the ammonia oxidation rate for strain MY1 was significantly reduced at 15°C; it took 6 months for complete oxidation of 1 mM ammonia (data not shown).
Strain MY1 showed optimal ammonia oxidation at pH 7.0. At pH 6 and 8, the lag time was slightly increased. N. europaea showed an optimum pH range of 7 to 9 (Fig. 4D). At pH 6, the growth of N. europaea was depressed, and oxidation of 1 mM ammonia was not complete after prolonged incubation. This result indicates that the optimal pHs for ammonia oxidation were slightly basic and neutral for N. europaea and strain MY1, respectively.
Growth of strain MY1 was in the range of 0.1 to 0.4% salinity and was severely inhibited at <0.1 and >0.4% salinity (Fig. 4E). N. europaea was not sensitive to salinity up to 2%, although the strain was isolated from a soil environment. The properties demonstrated here indicate that strain MY1 is mesophilic, neutrophilic, and nonhalophilic.
Kinetics of ammonia oxidation and oxygen uptake.
The ammonia oxidation rate was determined during batch growth experiments with strain MY1 (Fig. 1). The specific oxidation rate of ammonia was 2.5 fmol cell−1 day−1, which is much lower than that of AOB (36). Strain MY1's rate was only slightly lower than that reported for “Ca. Nitrosopumilus maritimus” (4.0 fmol cell−1 day−1 [40]) and than for AOA enrichment cultures from marine sediments originating in the East Sea, South Korea (2.8 fmol cell−1 day−1 [59]), the Arctic Sea (3.0 fmol cell−1 day−1 [59]), and the North Sea (4.3 fmol cell−1 day−1 [97]). Ammonia oxidation by strain MY1 was significantly inhibited by shaking, which was also observed in “Ca. Nitrosopumilus maritimus” (51) and in AOA enriched from marine sediments (59).
Oxygen uptake coupled with ammonia oxidation was respirometrically measured in order to determine the affinities (Km) for oxygen and ammonia of strain MY1. The kinetics of ammonia oxidation and oxygen consumption by strain MY1 followed Michaelis-Menten-type kinetics (Table 2). The affinities for oxygen and ammonia (and maximum rates of oxygen uptake and ammonia oxidation) for strain MY1 were slightly lower than those of “Ca. Nitrosopumilus maritimus” (51) and AOA enriched from marine sediments (59). However, the affinities for oxygen and ammonia of strain MY1 were much higher than those of AOB, and the maximum rates of oxygen uptake and ammonia oxidation in strain MY1 were much lower than those of AOB (Table 2). The Km range of AOB is broad, from ca. 30 to 2,000 μM (42, 45, 63, 78). These results indicate that, as reported for marine AOA (51, 59), strain MY1 is physiologically adapted to low levels of ammonia and oxygen.
Table 2.
Comparison of kinetics of ammonia oxidation and oxygen uptake for strain MY1 and previously characterized AOA or cultures
| Strain | Ammoniaa,b |
Oxygenb |
Reference | ||
|---|---|---|---|---|---|
| Km (μM) | Vmax (μmol N mg protein−1 h−1) | Km (μM) | Vmax (μmol O2 mg protein−1 h−1) | ||
| AOA | |||||
| MY1 | 0.69 (0.04) | 11c | 10.38 (1.08) | 20c | This study |
| AR enrichment culture | 0.61 (0.02) | 14c | 2.01 (0.45) | 11c | 59 |
| “Ca. Nitrosopumilus maritimus” | 0.13 (0.04) | 24 | 3.90 (0.60) | 36 | 51 |
| AOB | |||||
| N. europaea C-31 (ATCC 25978) | 1,300 | 36 | 186 | 129 | 59 |
| N. europaea (ATCC 19718) | 553 | 122 | NDd | ND | 51 |
| Activated sludge | 59 | ND | 15.6 | ND | 29 |
The values are given for NH4+ and NH3.
The numbers in parentheses indicate the standard deviation from triplicate experiments.
We assumed that the protein content per cell of AOA (∼10 fg/cell) equals that of “Ca. Nitrosopumilus maritimus.”
ND, not determined.
N2O production.
Microbial N2O production arises during nitrification (via chemical decomposition of hydroxylamine and nitrite) and during denitrification via the direct action of nitric oxide reductase (NorB) on NO (18). We monitored N2O production by strain MY1 and N. europaea in batch cultures with various ammonia and oxygen concentrations (Table 3). The total amount of N2O accumulated by strain MY1 was substantial but far lower than that produced by N. europaea: at 2 mM ammonia, strain MY1's N2O production was 18% of that of N. europaea (Table 3, column 2); when normalized to the nitrite yield (Table 3, column 4), the yield of N2O by strain MY1 was ca. 15% of that of N. europaea. In tests at 4 mM ammonia, the trend toward relatively low (∼14 to 16%) N2O production by strain MY1 recurred (Table 3). As expected for an aerobic process, reduction of the oxygen tension drastically reduced the proportion of ammonia oxidized (0.3 to 0.4 mM in the 2 mM ammonia treatments) for both strain MY1 and N. europaea; corresponding yields of N2O when normalized to nitrite increased about 2-fold for both cultures (Table 3).
Table 3.
Nitrous oxide production from strain MY1 and N. europaeaa
| Strain and conditionsc | Production |
||
|---|---|---|---|
| N2O (N2O-N) (μmol) | NO2− (NO2−-N) (μmol) | N2O-N/NO2−-N (%) | |
| MY1 | |||
| 2 mM NH4+ | 0.135 (0.065) | 100.6 (10.2) | 0.13 |
| 4 mM NH4+ | 0.231 (0.022) | 105.5 (21.1) | 0.22 |
| 2 mM NH4+ (low oxygen)b | 0.039 (0.003) | 17.0 (5.3) | 0.23 |
| N. europaea | |||
| 2 mM NH4+ | 0.750 (0.241) | 81.3 (5.2) | 0.92 |
| 4 mM NH4+ | 1.534 (0.717) | 116.4 (11.4) | 1.32 |
| 2 mM NH4+ (low oxygen) | 0.493 (0.012) | 21.2 (10.6) | 2.07 |
The data are mean values of triplicate experiments, and the numbers in parentheses indicate the standard deviations. Both cultures were incubated in AFM in sealed serum bottles for 2 weeks prior to N2O determination in headspace gases.
After the headspace was replaced by N2 gas, 2 ml air was injected. Headspace oxygen was 0.34% (0.03%) (vol/vol) under low-oxygen conditions, which is equivalent to 0.4 mM dissolved oxygen. The control (without inoculation) showed about 372 (23.5) ppb N2O, which corresponds to the atmospheric N2O concentration in the laboratory.
The initial archaeal and bacterial cell concentrations in the culture were 1.7 × 106 and 1.5 × 106 cells/ml, respectively.
The nirK gene (encoding nitrite reductase) confers nitrite tolerance in nitrifiers and is involved in nitrifier denitrification, reducing nitrite to NO, which can then be further reduced to N2O (96). nirK was successfully PCR amplified from strain MY1; the gene showed 85.8% and 40.4% nucleotide sequence similarities with those of “Ca. Nitrosopumilus maritimus” and the group I.1b soil fosmid clone 54d9, respectively (see Fig. S5 in the supplemental material). We found expression of the archaeal nirK and archaeal amoA genes during oxidation of ammonia and simultaneous N2O production (see Fig. S6 and Table S1 in the supplemental material); as expected, in the presence of the ammonia oxidation inhibitor (50 μM chlorite) or nitrite (without ammonia), neither N2O production nor archaeal nirK gene expression was observed (data not shown). To address the possibility that denitrifying Bacteria in the coculture might contribute to N2O production, we simultaneously monitored bacterial nirK and norB gene expression: neither was expressed (see Fig. S6 and Table S1 in the supplemental material). We acknowledge that the high specificity of reverse transcription (RT)-PCR assays may fail to detect related mRNA targets (in this case, expressed bacterial nirK and norB); nonetheless, the assay is broadly accepted as state of the art for assessing expression of genes involved in N2O production by denitrification pathways (11, 15). Thus, we have assembled four independent lines of evidence that argue against the possibility that minor denitrifying bacterial populations in the enrichment are responsible for N2O production in the strain MY1 culture: (i) assays were clearly aerobic (ammonia oxidation and N2O production were impaired at reduced O2 [see above]), (ii) the AFM had only NH3 as an electron donor, (iii) an ammonia oxidation inhibitor (which does not inhibit denitrification) completely inhibited the production N2O, and (iv) neither of the two bacterial genes that might potentially participate in N2O production by Bacteria present in the coculture was expressed. Thus, by weight of evidence, we conclude that the archaeal strain MY1 is the agent in N2O production.
Ultrastructural analysis.
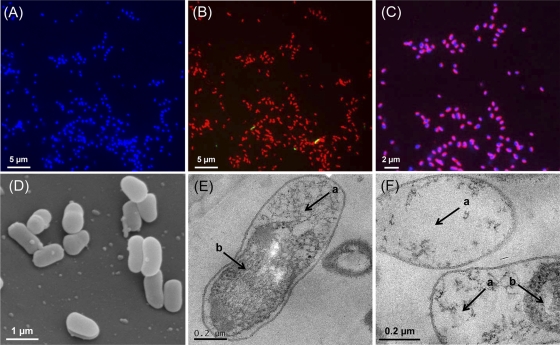
FISH analysis showed archaeal dominance over bacteria in the strain MY1 culture (Fig. 5A and B). Unlike other rod-shaped mesophilic archaeal cells (40, 48), ribosomes of strain MY1 were not largely concentrated at the poles. As observed in Fig. 5C, the chromosomes of strain MY1 were mostly localized at the sides of the cells. The cells were Gram negative. SEM analysis revealed short-rod morphology (diameter, 0.3 to 0.5 μm, and length, 0.6 to 1.0 μm) with a slightly curved peanut shape (Fig. 5D). Thin-section TEM analysis showed that strain MY1 appeared to have a significant intracellular volume with low electron density (Fig. 5E and F). Such a structure, consisting of inner membrane and periplasmic space, is unusual in the archaeal domain and has previously been described only in the thermophilic crenarchaeal genus Ignicoccus (44, 65). It is not clear if the electron-dense space is enveloped by a membrane.
Fig. 5.
Morphology of strain MY1. Shown is FISH analysis of the strain MY1 culture. (A) Epifluorescence micrographs of images from cells stained by DAPI (blue). (B) Merged image of Cy3-labeled Archaea-specific probe (Arc915; red) and FAM-labeled Bacteria-specific probe (EUB338; green). (C) Merged image of Cy3-labeled Archaea-specific probe (Arc915) and DAPI. Magenta indicates the archaeal cells. (D) Scanning electron micrograph of MY1 cells. (E and F) Transmission electron micrographs of ultrathin sections of MY1 cells. The arrows indicate electron-light areas (a) and electron-dense areas (b).
DISCUSSION
This study revealed that an autotrophic ammonia-oxidizing archaeon affiliated with the soil cluster of crenarchaeal group I.1a was enriched from an agricultural soil. The medium composition and cultivation conditions for the enrichment were not drastically different from those for typical AOB isolated from soil. We speculate that Archaea, not Bacteria, were enriched because of a combination of the native archaeal populations in the soil inoculum and the biochemical traits (especially ammonia and oxygen affinities [Table 2]) that were fortuitously suited to our incubation conditions. We were unable to eliminate cocultured bacteria, despite efforts utilizing serial dilution cultures and/or antibiotic treatments. The vast majority of prior reports of Archaea carrying out ammonia oxidation have been derived from enriched cocultures (21, 28, 59, 97); the notable exceptions are the pure culture, “Ca. Nitrosopumilus maritimus” (40), and “Ca. Nitrososphaera viennensis” (85). Oxidation of ammonia by strain MY1 began after a substantial lag period, while N. europaea showed no observable lag period under our culture conditions (see Fig. S3 in the supplemental material). Such lag periods have been frequently observed (28, 59) before initiation of ammonia oxidation by AOA. Also, Park et al. (59) demonstrated that growth of sulfur-oxidizing bacteria was a prerequisite for the enrichment culture of AOA from marine sediment. Despite significant effort in the present study to obtain a pure archaeal culture (e.g., see Fig. S1 and S2 in the supplemental material), we found that three typical terrestrial heterotrophic bacteria among the Proteobacteria (Acinetobacter, Pseudomonas, and Variovorax) persisted in the nitrifying culture. This observation leads us to raise the possibility that interdomain population interactions may contribute to the successful cultivation of AOA; further investigation is warranted.
Ammonia concentration is known to select for distinct groups of AOB in soils (93). Tolerance of ammonia by AOB has been documented up to 600 mM (41, 80). Tolerance for ammonia toxicity by strain MY1 was much lower than that of N. europaea (this study) and slightly lower than that of “Ca. Nitrososphaera viennensis” (15 mM [85]). In contrast, strain MY1's tolerance for ammonia was slightly higher than that of other AOA, including the marine “Ca. Nitrosopumilus maritimus” (51), AOA enrichment culture AR (4.0 mM [59]), and “Candidatus Nitrososphaera gargensis” (3.1 mM [28]). This tolerance by strain MY1 is consistent with generally higher ammonia concentrations that occur in soil pore waters than in marine environments (<0.03 to 1 μM in the open ocean and <0.03 to 100 μM in coastal waters [14]).
Strain MY1 was also more sensitive to nitrite than N. europaea (Fig. 4B). Thus, tight metabolic coupling of AOA and NOB might be essential for the activity of AOA in soil niches. Strain MY1 showed active nitrification in a typical pH range of neutral soil (pH 6 to 8). The activity of AOB has been found to be severely repressed below pH 7 and thus is considered more favored in neutral and basic environments (1, 36). Although ammonia oxidation by AOA was optimal at 20 to 25°C, it was still observed at 15°C with severe repression. In contrast, although “Ca. Nitrososphaera viennensis” was isolated from temperate soil, the optimal growth temperature was 37°C (85). We also documented that strain MY1 has a relatively high affinity for ammonia and oxygen (Table 2). These traits have been shown in other AOA to provide important competitive advantages (51, 59) and likely provide similar advantages to strain MY1-like Archaea in soil. AOA's preference for oligotrophic environments may correspond to the general capacity of archaeal groups to dominate and outcompete Bacteria under environmental conditions of chronic energy stress (87). Taken together, our results imply that strain MY1 and its relatives are adapted to carry out autotrophic ammonia oxidation in temperate soils.
Inhibitors of ammonia oxidation significantly repressed ammonia oxidation by both strain MY1 and N. europaea. However, tolerance for inhibition of allylthiourea of strain MY1 was much higher than that of N. europaea. It was reported that “Ca. Nitrososphaera gargensis,” enriched at 46°C from a hot spring, showed some degree of tolerance for 100 μM allylthiourea (28). Differential tolerance for ammonia oxidation inhibitors could be employed to assess the relative contributions of AOA versus AOB to nitrification in soils. For example, Taylor et al. (82) recently used the absence of inhibition of ammonia oxidation by allylthiourea to argue that AOA may exhibit nitrification activity. In addition, Santoro and Casciotti (71) observed partial inhibition of marine archaeal ammonia-oxidizing activity by allylthiourea at 86 μM, a concentration known to completely inhibit cultivated AOB (31) and environmental AOB (26).
Autotrophic nitrification is known to be a significant source of N2O in soil environments (17, 36, 37, 74). Recently, Santoro et al. (70) reported that marine AOA (crenarchaeal group I.1a) can produce N2O. Since the predominance of archaeal over bacterial amoA genes in various soils was demonstrated (49, 56, 72, 99), N2O produced by AOA has been suspected to be an overlooked but significant source of greenhouse gas emissions from soils. In this study, we observed that strain MY1 clearly produced N2O during ammonia oxidation (Table 3); the N2O yield was 5- to 6-fold lower than that from N. europaea under the same conditions. However, the N2O production rates are expected to be variable, depending on the ammonia concentration and dissolved oxygen concentration, as reported elsewhere (27, 34). It is accepted that the values for molar yield of N2O on the basis of nitrite produced are more robust for relative comparisons (74). The yield of N2O from strain MY1 on the basis of nitrite produced (0.13 to 0.23%) falls in the same order of magnitude as AOB: Nitrosospira spp. and Nitrosomonas spp. (0.03 to 0.7% [74 ]), N. europaea strain 28 (0.05 to 1.95% [67]), Nitrosomonas sp. strain ATCC 25978 (0.2%), Nitrosospira sp. strains (0.05 to 1% [36]), and other AOB (0.05 to 3.3% [17]). Oxygen-limited conditions increased the yield of N2O at 2 mM ammonia, and this type of response has typically been observed in AOB (17). Overall, these results indicate that N2O production from soil AOA should be considered a significant source of greenhouse gases emitted from soils.
Two different physiological pathways of N2O production by nitrifiers have been proposed: (i) during ammonia oxidation, chemical decomposition of the metabolites hydroxylamine and nitrite, and (ii) during respiratory reduction of nitrite to dinitrogen (involving nirK and norB), N2O is a key metabolite (5, 96). The presence of the nirK gene was documented in strain MY1 (see Fig. S5 in the supplemental material); in “Ca. Nitrosopumilus maritimus” (92), and, likewise, in the AOA enrichment cultures SJ and AR; and in several environmental metagenomic studies (7, 86, 90). Here, we showed in strain MY1 that the archaeal amoA and nirK genes were highly expressed during ammonia oxidation (see Fig. S6 and Table S1 in the supplemental material). In the genome of strain MY1 (38), we have not been able to recognize the canonical hydroxylamine oxidoreductase or nitric oxide reductase (nor) gene. Thus, the mechanism by which strain MY1 produces N2O is unclear. The biochemical and genetic mechanisms involved in the production of N2O by AOA should be explored in the future.
The sequences of the 16S rRNA and amoA genes related to those of strain MY1 have been widely detected in previous reports examining various types of terrestrial environments, including agricultural soils (56, 58, 84, 99), lake littoral rhizosphere (30), contaminated ground water (98), and water treatment plants (60, 88) (see also the reference sequences in Fig. 2 and 3). These phylogenetic analyses imply that MY1-like AOA may be cosmopolitan ammonia-oxidizing archaea in terrestrial environments. Particularly interesting is the match between strain MY1's 16S rRNA and amoA gene sequences and several previously reported clones from soil microcosm studies: 16S rRNA gene clone CS16S_52_7.5 (97.6% similarity [56]) and amoA gene clones CSamoA_71_7.5 (95.4% [56]), CBSamoA1-1-4 (95.1% [84]), band 11-2 (94.5% [58]), and HBD1-9 and TS1-16 (94.1% and 94.3%, respectively [99]). Signals from all of these sequences showed enhanced intensity during ammonia oxidation in agricultural plots and thus suggest the likely involvement of MY1-like organisms in soil nitrification.
Strain MY1 is a new mesophilic ammonia-oxidizing archaeon cultivated from soil. Based on the results of this study, we propose, according to Murray and Stackebrandt (54), provisional classification of the novel strain as “Candidatus Nitrosoarchaeum koreensis.”
Taxonomy.
The short description of “Canditatus Nitrosoarchaeum koreensis” sp. nov is as follows: Ko.re.en′sis. L. masc. adj. koreensis, of or belonging to Korea. Locality. Temperate soils. AOA with nearly identical 16S rRNA and amoA gene sequences have been detected in various terrestrial environments, including soils, freshwater and groundwater. Diagnostics. Short rod-shaped cells with a size of 0.3 to 0.5 by 0.6 to 1.0 μm. Periplasmic space is extraordinarily wide with inner membrane-like structure. Gram negative. Aerobic chemolithotrophic metabolism with oxidation of ammonia to nitrite. Carbon dioxide is used as the sole carbon source. Nonmarine. Growth range between 15°C and 30°C with an optimum at 25°C. Growth pH range between 6 and 8 with an optimum at 7.0. Nitrite was not toxic up to 2 mM with a tolerance limit of 5 mM. Half-saturation constants (affinity) for oxygen and ammonia are 10.38 and 0.69 μM, respectively. Adapted to ammonia concentrations up to 5 mM with a tolerance limit of 10 mM. N2O was produced. Produces characteristic GDGT membrane lipids, including crenarchaeol. Phylogeny. Phylogenetically assigned to the genus Nitrosoarchaeum of the phylum Crenarchaea based on 16S rRNA gene (HQ331116), amoA gene (HQ221117), and genomic sequence (AFPU00000000). Not isolated.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Basic Science Research Program (2009-0087901) and the Mid-Career Researcher Program (2010-0014384) through the National Research Foundation of MEST (Ministry of Education, Science and Technology), South Korea. E.L.M. was supported by NSF grant no. DEB-0841999.
We thank E. C. Hopmans for technical support.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 14 October 2011.
REFERENCES
- 1. Allison S., Prosser J. I. 1991. Survival of ammonia oxidising bacteria in air-dried soil. FEMS Microbiol. Lett. 79:65–68 [Google Scholar]
- 2. Alm E. W., Oerther D. B., Larsen N., Stahl D. A., Raskin L. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557–3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amann R. I., et al. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Amann R. I., Ludwig W., Schleifer K. H. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arp D. J., Stein L. Y. 2003. Metabolism of inorganic N compounds by ammonia-oxidizing bacteria. Crit. Rev. Biochem. Mol. Biol. 38:471–495 [DOI] [PubMed] [Google Scholar]
- 6. Baker G. C., Smith J. J., Cowan D. A. 2003. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55:541–555 [DOI] [PubMed] [Google Scholar]
- 7. Bartossek R., Nicol G. W., Lanzen A., Klenk H. P., Schleper C. 2010. Homologues of nitrite reductases in ammonia-oxidizing archaea: diversity and genomic context. Environ. Microbiol. 12:1075–1088 [DOI] [PubMed] [Google Scholar]
- 8. Bédard C., Knowles R. 1989. Physiology, biochemistry, and specific inhibitors of CH4, NH4+, and CO oxidation by methanotrophs and nitrifiers. Microbiol. Rev. 53:68–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blainey P. C., Mosier A. C., Potanina A., Francis C. A., Quake S. R. 2011. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS One 6:e16626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bock E., Wagner M. 2006. Oxidation of inorganic nitrogen compounds as an energy source, p. 457–495 In Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E. (ed.), The Prokaryotes. Springer, New York, NY [Google Scholar]
- 11. Braker G., Fesefeldt A., Witzel K. P. 1998. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 64:3769–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brochier-Armanet C., Boussau B., Gribaldo S., Forterre P. 2008. Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microbiol. 6:245–252 [DOI] [PubMed] [Google Scholar]
- 13. Canfield D. E., Glazer A. N., Falkowski P. G. 2010. The evolution and future of Earth's nitrogen cycle. Science 330:192–196 [DOI] [PubMed] [Google Scholar]
- 14. Capone D. G. 2000. The marine microbial nitrogen cycle, p. 455–494 In Kirchman D. L. (ed.), Microbial ecology of the oceans. Wiley-Liss, New York, NY [Google Scholar]
- 15. Casciotti K. L., Ward B. B. 2005. Phylogenetic analysis of nitric oxide reductase gene homologues from aerobic ammonia-oxidizing bacteria. FEMS Microbiol. Ecol. 52:197–205 [DOI] [PubMed] [Google Scholar]
- 16. Chen X. P., Zhu Y. G., Xia Y., Shen J. P., He J. Z. 2008. Ammonia-oxidizing archaea: important players in paddy rhizosphere soil? Environ. Microbiol. 10:1978–1987 [DOI] [PubMed] [Google Scholar]
- 17. Colliver B. B., Stephenson T. 2000. Production of nitrogen oxide and dinitrogen oxide by autotrophic nitrifiers. Biotechnol. Adv. 18:219–232 [DOI] [PubMed] [Google Scholar]
- 18. Conrad R. 1996. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol. Rev. 60:609–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Crutzen P. J. 1981. Atmospheric chemical processes of the oxides of nitrogen, including N2O, p. 17–44 In Delwiche C. C. (ed.), Denitrification, nitrification, and atmospheric nitrous oxide. Wiley, New York, NY [Google Scholar]
- 20. Daims H., Nielsen J. L., Nielsen P. H., Schleifer K. H., Wagner M. 2001. In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl. Environ. Microbiol. 67:5273–5284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. de la Torre J. R., Walker C. B., Ingalls A. E., Konneke M., Stahl D. A. 2008. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ. Microbiol. 10:810–818 [DOI] [PubMed] [Google Scholar]
- 22. DeLong E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. U. S. A. 89:5685–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Di H. J., et al. 2010. Ammonia-oxidizing bacteria and archaea grow under contrasting soil nitrogen conditions. FEMS Microbiol. Ecol. 72:386–394 [DOI] [PubMed] [Google Scholar]
- 24. Di H. J., et al. 2009. Nitrification driven by Bacteria and not Archaea in nitrogen-rich grassland soils. Nat. Geosci. 2:621–624 [Google Scholar]
- 25. Francis C. A., Roberts K. J., Beman J. M., Santoro A. E., Oakley B. B. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U. S. A. 102:14683–14688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ginestet P., Audic J. M., Urbain V. V., Block J. C. 1998. Estimation of nitrifying bacterial activities by measuring oxygen uptake in the presence of the metabolic inhibitors allylthiourea and azide. Appl. Environ. Microbiol. 64:2266–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goreau T. J., et al. 1980. Production of NO2− and N2O by nitrifying bacteria at reduced concentrations of oxygen. Appl. Environ. Microbiol. 40:526–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hatzenpichler R., et al. 2008. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc. Natl. Acad. Sci. U. S. A. 105:2134–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Henze M., Gujer W., Mino T., van Loosdrecht M. 2000. Activated sludge models ASM1, ASM2, ASM2d and ASM3. IWA Publishing, London, United Kingdom [Google Scholar]
- 30. Herrmann M., Saunders A. M., Schramm A. 2008. Archaea dominate the ammonia-oxidizing community in the rhizosphere of the freshwater macrophyte Littorella uniflora. Appl. Environ. Microbiol. 74:3279–3283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hooper A. B., Terry K. R. 1973. Specific inhibitors of ammonia oxidation in Nitrosomonas. J. Bacteriol. 115:480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hovanec T. A., Taylor L. T., Blakis A., Delong E. F. 1998. Nitrospira-like bacteria associated with nitrite oxidation in freshwater aquaria. Appl. Environ. Microbiol. 64:258–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Humbert S., et al. 2010. Molecular detection of anammox bacteria in terrestrial ecosystems: distribution and diversity. ISME J. 4:450–454 [DOI] [PubMed] [Google Scholar]
- 34. Hynes R. K., Knowles R. 1984. Production of nitrous oxide by Nitrosomonas europaea: effects of acetylene, pH, and oxygen. Can. J. Microbiol. 30:1397–1404 [Google Scholar]
- 35. Jia Z., Conrad R. 2009. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ. Microbiol. 11:1658–1671 [DOI] [PubMed] [Google Scholar]
- 36. Jiang Q. Q., Bakken L. R. 1999. Comparison of Nitrosospira strains isolated from terrestrial environments. FEMS Microbiol. Ecol. 30:171–186 [DOI] [PubMed] [Google Scholar]
- 37. Kester R. A., De Boer W., Laanbroek H. J. 1997. Production of NO and N2O by pure cultures of nitrifying and denitrifying bacteria during changes in aeration. Appl. Environ. Microbiol. 63:3872–3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim B. K., et al. 2011. Genome sequence of an ammonia-oxidizing soil archaeon, “Candidatus Nitrosoarchaeum koreensis” MY1. J. Bacteriol. 193:5539–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111–120 [DOI] [PubMed] [Google Scholar]
- 40. Könneke M., et al. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546 [DOI] [PubMed] [Google Scholar]
- 41. Koops H. P., Bottcher B., Moller U. C., Pommerening-Roser A., Stehr G. 1991. Classification of eight new species of ammonia-oxidizing bacteria: Nitrosomonas communis sp. nov., Nitrosomonas ureae sp. nov., Nitrosomonas aestuarii sp. nov., Nitrosomonas marina sp. nov., Nitrosomonas nitrosa sp. nov., Nitrosomonas eutropha sp. nov., Nitrosomonas oligotropha sp. nov. and Nitrosomonas halophila sp. nov. J. Gen. Microbiol. 137:1689–1699 [Google Scholar]
- 42. Koops H. P., Pommerening Röser A. 2001. Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol. Ecol. 37:1–9 [Google Scholar]
- 43. Kumar S., Tamura K., Nei M. 2004. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5:150–163 [DOI] [PubMed] [Google Scholar]
- 44. Küper U., Meyer C., Muller V., Rachel R., Huber H. 2010. Energized outer membrane and spatial separation of metabolic processes in the hyperthermophilic archaeon Ignicoccus hospitalis. Proc. Natl. Acad. Sci. U. S. A. 107:3152–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Laanbroek H. J., Bodelier P. L. E., Gerards S. 1994. Oxygen consumption kinetics of Nitrosomonas europaea and Nitrobacter hamburgensis grown in mixed continuous cultures at different oxygen concentrations. Arch. Microbiol. 161:156–162 [Google Scholar]
- 46. Lee J. B., et al. 2011. Final report on international comparison CCQM-K68: nitrous oxide in synthetic air. Metrologia 48:08004 [Google Scholar]
- 47. Lee J. S., et al. 2010. Final report on international key comparison CCQM-K53: oxygen in nitrogen. Metrologia 47:08005 [Google Scholar]
- 48. Lehtovirta-Morley L. E., Stoecker K., Vilcinskas A., Prosser J. I., Nicol G. W. 2011. Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc. Natl. Acad. Sci. U. S. A. 108:15892–15897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Leininger S., et al. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809 [DOI] [PubMed] [Google Scholar]
- 50. Luft J. H. 1961. Improvements in epoxy resin embedding methods. J. Biophys. Biochem. Cytol. 9:409–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Martens-Habbena W., Berube P. M., Urakawa H., de la Torre J. R., Stahl D. A. 2009. Ammonia oxidation kinetics determine niche separation of nitrifying archaea and bacteria. Nature 461:976–979 [DOI] [PubMed] [Google Scholar]
- 52. Martens-Habbena W., Stahl D. A. 2011. Nitrogen metabolism and kinetics of ammonia-oxidizing archaea. Methods Enzymol. 496:465–487 [DOI] [PubMed] [Google Scholar]
- 53. McCaig A. E., Embley T. M., Prosser J. I. 1994. Molecular analysis of enrichment cultures of marine ammonia oxidisers. FEMS Microbiol. Lett. 120:363–367 [DOI] [PubMed] [Google Scholar]
- 54. Murray R. G., Stackebrandt E. 1995. Taxonomic note: implementation of the provisional status Candidatus for incompletely described procaryotes. Int. J. Syst. Bacteriol. 45:186–187 [DOI] [PubMed] [Google Scholar]
- 55. Muyzer G., de Waal E. C., Uitterlinden A. G. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nicol G. W., Leininger S., Schleper C., Prosser J. I. 2008. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ. Microbiol. 10:2966–2978 [DOI] [PubMed] [Google Scholar]
- 57. Nold S. C., Zhou J., Devol A. H., Tiedje J. M. 2000. Pacific Northwest marine sediments contain ammonia-oxidizing bacteria in the beta subdivision of the Proteobacteria. Appl. Environ. Microbiol. 66:4532–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Offre P., Prosser J. I., Nicol G. W. 2009. Growth of ammonia-oxidizing archaea in soil microcosms is inhibited by acetylene. FEMS Microbiol. Ecol. 70:99–108 [DOI] [PubMed] [Google Scholar]
- 59. Park B. J., et al. 2010. Cultivation of autotrophic ammonia-oxidizing archaea from marine sediments in co-culture with sulfur-oxidizing bacteria. Appl. Environ. Microbiol. 76:7575–7587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Park H. D., Wells G. F., Bae H., Criddle C. S., Francis C. A. 2006. Occurrence of ammonia-oxidizing archaea in wastewater treatment plant bioreactors. Appl. Environ. Microbiol. 72:5643–5647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Park S. J., Park B. J., Rhee S. K. 2008. Comparative analysis of archaeal 16S rRNA and amoA genes to estimate the abundance and diversity of ammonia-oxidizing archaea in marine sediments. Extremophiles 12:605–615 [DOI] [PubMed] [Google Scholar]
- 62. Pitcher A., et al. 2010. Crenarchaeol dominates the membrane lipids of Candidatus Nitrososphaera gargensis, a thermophilic group I.1b archaeon. ISME J. 4:542–552 [DOI] [PubMed] [Google Scholar]
- 63. Prosser J. I. 1989. Autotrophic nitrification in Bacteria. Adv. Microb. Physiol. 30:125–181 [DOI] [PubMed] [Google Scholar]
- 64. Pruesse E., et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rachel R., Wyschkony I., Riehl S., Huber H. 2002. The ultrastructure of Ignicoccus: evidence for a novel outer membrane and for intracellular vesicle budding in an archaeon. Archaea 1:9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rahn T., Wahlen M. 1997. Stable isotope enrichment in stratospheric nitrous oxide. Science 278:1776–1778 [DOI] [PubMed] [Google Scholar]
- 67. Remde A., Conrad R. 1990. Production of nitric oxide in Nitrosomonas europaea by reduction of nitrite. Arch. Microbiol. 154:187–191 [Google Scholar]
- 68. Reynolds E. S. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17:208–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rotthauwe J. H., Witzel K. P., Liesack W. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Santoro A. E., Buchwald C., McIlvin M. R., Casciotti K. L. 2011. Isotopic signature of N2O produced by marine ammonia-oxidizing archaea. Science 333:1282–1285 [DOI] [PubMed] [Google Scholar]
- 71. Santoro A. E., Casciotti K. L. 12 May 2011. Enrichment and characterization of ammonia-oxidizing archaea from the open ocean: phylogeny, physiology and stable isotope fractionation. ISME J. doi:10.1038/ismej.2011.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Schauss K., et al. 2009. Dynamics and functional relevance of ammonia-oxidizing archaea in two agricultural soils. Environ. Microbiol. 11:446–456 [DOI] [PubMed] [Google Scholar]
- 73. Schouten S., Huguet C., Hopmans E. C., Kienhuis M. V., Sinninghe Damsté J. S. 2007. Analytical methodology for TEX86 paleothermometry by high-performance liquid chromatography/atmospheric pressure chemical ionization-mass spectrometry. Anal. Chem. 79:2940–2944 [DOI] [PubMed] [Google Scholar]
- 74. Shaw L. J., et al. 2006. Nitrosospira spp. can produce nitrous oxide via a nitrifier denitrification pathway. Environ. Microbiol. 8:214–222 [DOI] [PubMed] [Google Scholar]
- 75. Singh S. N., Verma A. 2007The potential of nitrification inhibitors to manage the pollution effect of nitrogen fertilizers in agricultural and other soils: a review. Environ. Practice 9:266–279 [Google Scholar]
- 76. Sinninghe Damsté J. S., et al. 2002. Linearly concatenated cyclobutane lipids form a dense bacterial membrane. Nature 419:708–712 [DOI] [PubMed] [Google Scholar]
- 77. Solorzano L. 1969. Determination of ammonia in natural waters by the phenolhypochlorite method. Limnol. Oceanogr. 14:799–801 [Google Scholar]
- 78. Stehr G., Böttcher B., Dittberner P., Rath G., Koops H. P. 1995. The ammonia-oxidizing nitrifying population of the River Elbe estuary. FEMS Microbiol. Ecol. 17:177–186 [Google Scholar]
- 79. Stein L. Y., Arp D. J. 1998. Loss of ammonia monooxygenase activity in Nitrosomonas europaea upon exposure to nitrite. Appl. Environ. Microbiol. 64:4098–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Suwa Y., Imamura Y., Suzuki T., Tashiro T., Urushigawa Y. 1994. Ammonia-oxidizing bacteria with different sensitivities to (NH4)2SO4 in activated sludges. Water Res. 28:1523–1532 [Google Scholar]
- 81. Tan N. C., et al. 2008. Physiological and phylogenetic study of an ammonium-oxidizing culture at high nitrite concentrations. Syst. Appl. Microbiol. 31:114–125 [DOI] [PubMed] [Google Scholar]
- 82. Taylor A. E., Zeglin L. H., Dooley S., Myrold D. D., Bottomley P. J. 2010. Evidence for different contributions of Archaea and Bacteria to the ammonia-oxidizing potential of diverse Oregon soils. Appl. Environ. Microbiol. 76:7691–7698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tourna M., Freitag T. E., Nicol G. W., Prosser J. I. 2008. Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ. Microbiol. 10:1357–1364 [DOI] [PubMed] [Google Scholar]
- 85. Tourna M., et al. 2011. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc. Natl. Acad. Sci. U. S. A. 108:8420–8425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Treusch A. H., et al. 2005. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ. Microbiol. 7:1985–1995 [DOI] [PubMed] [Google Scholar]
- 87. Valentine D. L. 2007. Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nat. Rev. Microbiol. 5:316–323 [DOI] [PubMed] [Google Scholar]
- 88. van der Wielen P. W., Voost S., van der Kooij D. 2009. Ammonia-oxidizing bacteria and archaea in groundwater treatment and drinking water distribution systems. Appl. Environ. Microbiol. 75:4687–4695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Vanparys B., et al. 2007. The phylogeny of the genus Nitrobacter based on comparative rep-PCR, 16S rRNA and nitrite oxidoreductase gene sequence analysis. Syst. Appl. Microbiol. 30:297–308 [DOI] [PubMed] [Google Scholar]
- 90. Venter J. C., et al. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74 [DOI] [PubMed] [Google Scholar]
- 91. Wagner M., Rath G., Koops H. P., Flood J., Amann R. 1996. In situ analysis of nitrifying bacteria in sewage treatment plants. Water Sci. Technol. 34:237–244 [Google Scholar]
- 92. Walker C. B., et al. 2010. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc. Natl. Acad. Sci. U. S. A. 107:8818–8823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Webster G., Embley T. M., Freitag T. E., Smith Z., Prosser J. I. 2005. Links between ammonia oxidizer species composition, functional diversity and nitrification kinetics in grassland soils. Environ. Microbiol. 7:676–684 [DOI] [PubMed] [Google Scholar]
- 94. Weisburg W. G., Barns S. M., Pelletier D. A., Lane D. J. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Widdel F., Bak F. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p. 3352–3378 In Balows A., Trüper M., Dworkin M., Harder W., Schleifer K.-H. (ed.). The prokaryotes, 2nd ed., vol. IV Springer-Verlag, New York, NY [Google Scholar]
- 96. Wrage N., Velthof G. L., van Beusichem M. L., Oenema O. 2001. Role of nitrifier denitrification in the production of nitrous oxide. Soil Biol. Biochem. 33:1723–1732 [Google Scholar]
- 97. Wuchter C., et al. 2006. Archaeal nitrification in the ocean. Proc. Natl. Acad. Sci. U. S. A. 103:12317–12322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yagi J. M., Neuhauser E. F., Ripp J. A., Mauro D. M., Madsen E. L. 2010. Subsurface ecosystem resilience: long-term attenuation of subsurface contaminants supports a dynamic microbial community. ISME J. 4:131–143 [DOI] [PubMed] [Google Scholar]
- 99. Zhang L. M., et al. 2010. Autotrophic ammonia oxidation by soil Thaumarchaea. Proc. Natl. Acad. Sci. U. S. A. 107:17240–17245 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.