Abstract
In this study, we report a whole-genome single nucleotide polymorphism (SNP)-based evolutionary approach to study the epidemiology of a multistate outbreak of Salmonella enterica subsp. enterica serovar Montevideo. This outbreak included 272 cases that occurred in 44 states between July 2009 and April 2010. A case-control study linked the consumption of salami made with contaminated black and red pepper to the outbreak. We sequenced, on the SOLiD System, 47 isolates with XbaI PFGE pattern JIXX01.0011, a common pulsed-field gel electrophoresis (PFGE) pattern associated with isolates from the outbreak. These isolates represented 20 isolates collected from human sources during the period of the outbreak and 27 control isolates collected from human, food, animal, and environmental sources before the outbreak. Based on 253 high-confidence SNPs, we were able to reconstruct a tip-dated molecular clock phylogeny of the isolates and to assign four human isolates to the actual outbreak. We developed an SNP typing assay to rapidly discriminate between outbreak-related cases and non-outbreak-related cases and tested this assay on an extended panel of 112 isolates. These results suggest that only a very small percentage of the human isolates with the outbreak PFGE pattern and obtained during the outbreak period could be attributed to the actual pepper-related outbreak (20%), while the majority (80%) of the putative cases represented background cases. This study demonstrates that next-generation-based SNP typing provides the resolution and accuracy needed for outbreak investigations of food-borne pathogens that cannot be distinguished by currently used subtyping methods.
INTRODUCTION
Whole-genome sequencing has been demonstrated to provide a high-resolution view of the epidemiology and microevolution of pathogenic bacteria, such as the transmission (13) and phylogeography (12) of methicillin-resistant Staphylococcus aureus and the role of recombination in the evolution of Streptococcus pneumoniae lineages (6). The use of genome sequencing methods to investigate outbreaks of food-borne bacterial diseases is relatively new and holds great promise, as it can help to identify the temporal, geographical, and evolutionary origin of an outbreak. At the same time, whole-genome data can be used to rapidly develop single nucleotide polymorphism (SNP) assays that can be used to clarify the epidemiology of an outbreak and discriminate between outbreak-related and sporadic clinical cases during and after the period of the outbreak. A prerequisite to this approach is a thorough understanding of the population structure of the bacterial subtype involved in the outbreak, which can only be accomplished by an educated sampling effort of the background population.
In this study, we report a whole-genome SNP-based evolutionary approach to study the epidemiology of a multistate outbreak of Salmonella enterica subsp. enterica serovar Montevideo (S. Montevideo). This outbreak included 272 reported human cases from 44 states and lasted from July 2009 to April 2010 (5). A case-control study linked the outbreak to the consumption of salami made with contaminated black and red pepper (5). The XbaI pulsed-field gel electrophoresis (PFGE) pattern of the outbreak strain (JIXX01.0011) is the most common among 1,225 S. Montevideo isolates in the PulseNet database (5). Thus, PFGE typing was not useful in discriminating outbreak-associated isolates from the background population. The origin of the S. Montevideo strain that caused the outbreak is ambiguous. Initial reports suggested that the outbreak strain was imported with the pepper from Asia (5), but a later study that used whole-genome phylogenetic methods to link human isolates to the contaminated food concluded that the strain was of domestic origin (16). Our study focused on the temporal population structure of human clinical isolates isolated during the period of the outbreak and a period of 5 years before the start of the outbreak period. We demonstrate that this approach allows improved discrimination and phylogenetic characterization of this highly clonal pathogen.
MATERIALS AND METHODS
Isolates.
Forty-seven S. Montevideo isolates with XbaI PFGE pattern JIXX01.0011 were obtained from the Food Safety Laboratory at Cornell University, the New York State Department of Health, the New York City Department of Health and Mental Health, and Washington State University (Table 1). These isolates were selected to represent 20 putative case isolates collected from human sources during the period of the outbreak and 27 control isolates collected from human, food, animal, and environmental sources before the outbreak. A putative case isolate was defined as an isolate of S. Montevideo with XbaI PFGE pattern JIXX01.0011 that was collected from human sources in the United States between June 2009 and January 2010. Epidemiological information and food consumption history were not available a priori for the patients from whom these isolates were obtained. Given the high prevalence of background cases with identical XbaI PFGE patterns, a considerable proportion of these putative case isolates were expected to not be associated with this outbreak. Limited food exposure data (Table 1) were obtained from state health departments after completion of full genome sequencing. Control isolates were defined as isolates of S. Montevideo with XbaI PFGE pattern JIXX01.0011, collected from human, food, animal, or environmental sources before 1 June 2009. The control isolates were stratified by year to obtain a distribution of isolates between isolation years 2004 and 2009 that was as even as possible given the available isolates.
Table 1.
Strains and genome sequences used in this study
| Strain | Source | Yr | Moa | Originb | Pre-genome sequencing putative case/control assignment (temporal match with salami outbreak)c | Genome sequence-based classification | Reported consumption of salamid |
|---|---|---|---|---|---|---|---|
| Strains newly sequenced in this study | |||||||
| FSL S5-457 | Human | 2004 | 4 | NYSDOH | Control | Part of putative outbreak cluster 1 | – |
| FSL R8-4924 | Human | 2004 | 5 | NYSDOH | Control | Part of putative outbreak cluster 1 | – |
| FSL R8-4925 | Human | 2004 | 5 | NYSDOH | Control | Part of putative outbreak cluster 1 | – |
| FSL S5-470 | Human | 2004 | 5 | NYSDOH | Control | – | |
| FSL R8-4926 | Human | 2004 | 6 | NYSDOH | Control | Part of putative outbreak cluster 1 | – |
| FSL R8-4927 | Human | 2004 | 8 | NYSDOH | Control | Part of putative outbreak cluster 1 | – |
| FSL R8-4928 | Human | 2005 | 5 | NYSDOH | Control | – | |
| FSL R8-5050 | Human | 2005 | 7 | NYSDOH | Control | Part of putative outbreak cluster 3 | – |
| FSL R8-4929 | Human | 2005 | 8 | NYSDOH | Control | – | |
| FSL R8-5054 | Human | 2005 | 10 | NYCDOHMH | Control | – | |
| FSL R8-4930 | Human | 2005 | 12 | NYSDOH | Control | Part of putative outbreak cluster 3 | – |
| FSL R8-5057 | Human | 2006 | 2 | NYCDOHMH | Control | – | |
| FSL R8-4931 | Human | 2006 | 5 | NYSDOH | Control | – | |
| FSL R8-4916 | Farm environment | 2006 | N/A | WAU | Control | – | |
| FSL R8-4932 | Human | 2007 | 6 | NYSDOH | Control | Part of pistachio clade | – |
| FSL R8-4918 | Human | 2007 | N/A | WAU | Control | – | |
| FSL R8-4933 | Human | 2008 | 4 | NYSDOH | Control | – | |
| FSL R8-2533 | Human | 2008 | 8 | NYSDOH | Control | – | |
| FSL R8-2868 | Bovine | 2008 | 10 | NY_Warnick | Control | – | |
| FSL R8-4934 | Human | 2008 | 10 | NYSDOH | Control | Part of putative outbreak cluster 2 | – |
| FSL R8-4935 | Human | 2008 | 11 | NYSDOH | Control | Part of putative outbreak cluster 2 | – |
| FSL R8-4936 | Human | 2008 | 11 | NYSDOH | Control | Part of putative outbreak cluster 2 | – |
| FSL R8-4919 | Human | 2008 | N/A | WAU | Control | Part of pistachio clade | – |
| FSL R8-4920 | Human | 2008 | N/A | WAU | Control | Part of pistachio clade | – |
| FSL R8-4921 | Human | 2009 | 2 | WAU | Control | Part of pistachio clade | – |
| FSL R8-3515 | Pistachio | 2009 | 3 | N/A | Control | Part of pistachio clade | – |
| FSL R8-4922 | Human | 2009 | 5 | WAU | Control | Part of pistachio clade | – |
| FSL R8-4838 | Human | 2009 | 6 | NYSDOH | Putative case | Unlikely outbreak case | – |
| FSL R8-4464 | Human | 2009 | 7 | NYSDOH | Putative case | Unlikely outbreak case | – |
| FSL R8-4839 | Human | 2009 | 8 | NYSDOH | Putative case | Unlikely outbreak case | – |
| FSL R8-4887 | Human | 2009 | 8 | NYCDOHMH | Putative case | Unlikely outbreak case | – |
| FSL R8-4888 | Human | 2009 | 8 | NYCDOHMH | Putative case | Unlikely outbreak case | – |
| FSL R8-4889 | Human | 2009 | 8 | NYCDOHMH | Putative case | Unlikely outbreak case | – |
| FSL R8-4840 | Human | 2009 | 9 | NYSDOH | Putative case | Unlikely outbreak case | – |
| FSL R8-4673 | Human | 2009 | 11 | NYSDOH | Putative case | Unlikely outbreak case | – |
| FSL R8-4841 | Human | 2009 | 11 | NYSDOH | Putative case | Definitive outbreak case | Yes |
| FSL R8-4842 | Human | 2009 | 11 | NYSDOH | Putative case | Unlikely outbreak case | No |
| FSL R8-4890 | Human | 2009 | 11 | NYCDOHMH | Putative case | Unlikely outbreak case | Yes |
| FSL R8-4923 | Human | 2009 | 11 | WAU | Putative case | Definitive outbreak case | Yes |
| FSL R8-4679 | Human | 2009 | 12 | NYSDOH | Putative case | Unlikely outbreak case | – |
| FSL R8-4843 | Human | 2009 | 12 | NYSDOH | Putative case | Definitive oubreak case | No |
| FSL R8-4844 | Human | 2009 | 12 | NYSDOH | Putative case | Unlikely outbreak case | Yes |
| FSL R8-4891 | Human | 2009 | 12 | NYCDOHMH | Putative case | Definitive outbreak case | Yes |
| FSL R8-4893 | Human | 2009 | 12 | NYCDOHMH | Putative case | Unlikely outbreak case | – |
| FSL R8-4845 | Human | 2010 | 1 | NYSDOH | Putative case | Unlikely outbreak case | – |
| FSL R8-4846 | Human | 2010 | 1 | NYSDOH | Putative case | Unlikely outbreak case | – |
| FSL R8-4892 | Human | 2010 | 1 | NYCDOHMH | Putative case | Unlikely outbreak case | Yes |
| Strains used in Lienau et al. (16) | |||||||
| FDA_2010_142_Pistachio-3 | Pistachio | 2009 | N/A | FDA | |||
| FDA_2010_144_Black Pepper-6 | Black pepper | N/A | N/A | FDA | |||
| FDA_2010_145_Black Pepper-5 | Black pepper | N/A | N/A | FDA | |||
| FDA_2010_146_Black Pepper-7 | Black pepper | N/A | N/A | FDA | |||
| FDA_2010_147_Black Pepper-3 | Black pepper | 2010 | N/A | FDA | |||
| FDA_2010_148_Black Pepper-4 | Black pepper | 2010 | N/A | FDA | |||
| FDA_2010_149_Pistachio-2 | Pistachio | N/A | N/A | FDA | |||
| FDA_2010_222_Clinical-NC-4 | Human | 2009 | N/A | FDA | |||
| FDA_2010_155_Clinical-NC-5 | Human | 2009 | N/A | FDA | |||
| FDA_2010_156_Clinical-OH-3 | Human | 2009 | N/A | FDA | |||
| FDA_2010_157_Clinical-CA | Human | 2009 | N/A | FDA | |||
| FDA_2010_158_Clinical-MD | Human | 2009 | N/A | FDA | |||
| FDA_2010_204_Chicken | Chicken | N/A | N/A | FDA | |||
| FDA_2010_209_Romaine | Romaine lettuce | 2010 | N/A | FDA | |||
| FDA_2010_210_Mozzarella | Mozzarella | 2007 | N/A | FDA | |||
| FDA_2010_211_Perch | Fish | 2006 | N/A | FDA | |||
| FDA_2010_212_Sea Trout | Fish | 2007 | N/A | FDA | |||
| FDA_2010_213_King Fish | Fish | 2007 | N/A | FDA | |||
| FDA_2010_214_Black Pepper-1 | Black pepper | 2009 | N/A | FDA | |||
| FDA_2010_215_Red Pepper-2 | Red pepper | 2009 | N/A | FDA | |||
| FDA_2010_216_Black Pepper-2 | Black pepper | 2009 | N/A | FDA | |||
| FDA_2010_217_Drain_swab | Drain swab | 2009 | N/A | FDA | |||
| FDA_2010_219_Red Pepper-1 | Red pepper | 2010 | N/A | FDA | |||
| FDA_2010_220_Clinical-NC-3 | Human | 2009 | N/A | FDA | |||
| FDA_2010_221_Clinical-NC-2 | Human | 2009 | N/A | FDA | |||
| FDA_2010_155_Clinical-NC-5 | Human | 2009 | N/A | FDA | |||
| FDA_2010_223_Clinical-NC-1 | Human | 2009 | N/A | FDA | |||
| FDA_2010_224_Clinical-OH-2 | Human | 2009 | N/A | FDA | |||
| FDA_2010_225_Clinical-OH-1 | Human | 2009 | N/A | FDA | |||
| FDA_2010_227_Pistachio-1 | Pistachio | 2009 | N/A | FDA | |||
| FDA_2010_236_Clinical-IA-1 | Human | 2009 | N/A | FDA | |||
| FDA_2010_237_Lunch Meat-IA-1 | Lunch meat | 2010 | N/A | FDA | |||
| FDA_2010_238_Lunch Meat-IA-3 | Lunch meat | 2010 | N/A | FDA | |||
| FDA_2010_239_Lunch Meat-IA-2 | Lunch meat | 2010 | N/A | FDA | |||
| FDA_2010_240_Lunch Meat-IA-4 | Lunch meat | 2010 | N/A | FDA | |||
| FDA_2010_242_Lunch Meat-IA-5 | Lunch meat | 2010 | N/A | FDA |
N/A, not available. In the case the exact month of isolation was not available, June (6) of that year was used for the calibration of the tip-dated tree search.
NYSDOH, New York State Department of Health; NYCDOHMH, New York City Department of Health and Mental Hygiene; WAU, Washington State University; NY_Warnick, Warnick lab, Cornell University, Ithaca, NY; FDA, Food and Drug Administration, College Park, MD.
A putative case isolate was defined as an isolate of S. Montevideo with XbaI PFGE pattern JIXX01.0011 that was collected from human sources in the United States between June 2009 and January 2010.
This information was obtained by interviewing the patients. –, no interview data were available.
Whole-genome sequencing and SNP detection.
Genomic DNA was isolated from S. Montevideo strains using the Qiagen DNeasy blood and tissue kit (Qiagen Inc., Valencia, CA). Barcoded SOLiD system fragment libraries were prepared from 3 μg of each of these genomic DNA samples using the SOLiD fragment library construction kit (Life Technologies, Foster City, CA) with the SOLiD fragment library barcoding kit module 1-16 (Life Technologies) by following the manufacturer's instructions. Ten libraries were loaded onto each quad of a SOLiD System flowcell and sequenced according to the manufacturer's instructions.
SOLiD System data were analyzed using BioScope version 1.2. Specifically, reads were mapped to the draft assembly (34 scaffolds) of S. Montevideo strain 515920-2, isolated from a black pepper sample associated with the 2009–2010 outbreak (GenBank accession no. AESM01000000) (16). Coverage depth ranged from 20× to 300× (median, 56.2×). SNPs were called using the diBayes module and were used to calculate a consensus genome for each strain. All positions with coverage depths less than 8× were masked with N characters; all but two strains had at least 97% of bases with coverage depths of 8 or greater. SNPs called by diBayes were included in the consensus genome only when homozygous with a P value of less than 0.1 and a novel allele quality value (QV) of at least 12. Otherwise, the positions were masked with N to indicate uncertainty about the consensus call. The SOLiD System is for research use only and is not intended for any animal or human therapeutic or diagnostic uses.
The consensus genomes for each of the 47 strains and the 35 S. Montevideo draft assemblies available in GenBank were aligned to the strain 515920-2 reference genome (GenBank accession number AESM01000000) (16). Contigs were aligned to the reference using nucmer, and SNPs were identified using show-snps, both of which are software components of the MUMmer package (15).
Phylogenetic analyses.
To infer the relationship between the 47 isolates sequenced in this study and previously sequenced outbreak isolates (16), we performed a neighbor-joining phylogenetic analysis in PAUP* version 1.010b (22) with 1,000 bootstrap replicates (10) (see Fig. S1 in the supplemental material and Table 1). Ambiguously called and singleton SNPs were excluded from this analysis, because they are not phylogenetically informative (i.e., they cannot be used to infer putative phylogenetic relationships between isolates).
To infer the population structure of the isolates of S. Montevideo with XbaI PFGE pattern JIXX01.0011 in a temporal context, Bayesian phylogenetic analyses were performed in BEAST version 1.6.1 (8). An ascertainment bias correction model was used to account for the use of only variable sites instead of complete sequences (see Gray et al. [12] for more information). In this analysis, only isolates sequenced in this study were used, as these analyses require quality values and sequence reads for the identification of high-confidence singleton SNPs. Sequence quality information was not available for the publicly available draft assemblies (as of September 2011), so confidence levels could not be assigned to the SNPs in these genomes. High-confidence singletons should be included in this kind of molecular-clock-based analysis, because they are informative for the mutation rate estimations. We used the HKY model of nucleotide evolution for all analyses, and the date of isolation of each isolate was used to calibrate the tree. To select the most appropriate molecular clock model and model of population size through time for this data set, we performed four different analyses. Analyses were run with a strict molecular clock (assuming a constant substitution rate for the entire tree) or a log-normal relaxed molecular clock, which allows for different mutation rates on different branches. We further tested both the constant population coalescent model and the extended Bayesian skyline (EBSP) model. Each run (four in total) was run for 100 million generations, and model parameter values and trees were sampled every 10,000th generation. The analysis with the EBSP and log-normal relaxed molecular clock was run nine times in addition to the initial run to obtain a proper sampling of the parameter values of the model. Results were visualized in Tracer version 1.5 to assess convergence and proper sampling and to identify the burn-in period. In the case of the EBSP and log-normal relaxed molecular clock analysis, the results of eight convergent runs were combined and used for further analysis. Tracer version 1.5 was also used to calculate Bayes factors (BF) for model comparison. Recombination in the SNP data sets was evaluated using the pairwise homoplasy index (PHI) test (2) as implemented in Splitstree version 4.8 (14). In the absence of genome sequence data for food isolates from a possible outbreak source, a putative outbreak cluster was defined as a clade with posterior probability greater than 0.95 and with no more than 2 years between the estimated median age of the most recent common ancestor (MRCA) and the isolation date for the most recent isolate of the clade.
PFGE.
PFGE subtyping was performed with XbaI, SpeI (Roche Molecular Diagnostics, Pleasanton, CA), and NotI (New England BioLabs, Ipswich, MA). PFGE for XbaI and SpeI was performed according to the CDC PulseNet protocol (19). For NotI, PFGE was performed with a modified protocol using 4 units of enzyme per plug, a run time of 20.5 h, with an initial switch time of 2 s and a final switch time of 20 s. BioNumerics version 5.1 (Applied Maths, Austin, TX) was used to analyze the PFGE patterns.
Epidemiological data.
Interview data for eight patients representing putative outbreak cases were retrospectively available from case-control studies performed as part of the outbreak investigation by the New York State Department of Health and the New York City Department of Health and Mental Health. We focused specifically on the question of whether or not the patients had consumed Italian-style meats, such as salami, as this was the presumed vehicle of transmission for this outbreak (5) (Table 1).
SNP-based detection assay for discrimination of the outbreak strain.
Three biallelic SNPs were each found to be completely discriminatory for the outbreak strains versus all other isolates in this study. TaqMan SNP genotyping assays, comprising two allele-specific probes, were designed against these three SNPs using an in-house version of the Primer Express software (Life Technologies). All assays were run on a ViiA 7 real-time PCR instrument (Life Technologies). The reaction mixtures contained 12.5 μl of TaqMan universal PCR master mix (Life Technologies), 1.25 μl 20× working stock of SNP genotyping assay mix, and 11.25 μl of DNA template. Data were analyzed using TaqMan Genotyper software (Life Technologies). Two assays were run against all 47 isolates used for whole-genome sequencing and a panel of 65 Salmonella isolates representing 53 additional S. Montevideo isolates (40 isolates from human cases and 13 isolates from animal clinical cases), 10 Salmonella serovars commonly involved in human clinical cases, and two Salmonella isolates of which the serovar was unknown (see Table S1 in the supplemental material), while one assay was used only for preliminary evaluation of 20 isolates. Nine of the additional 53 S. Montevideo isolates in this panel were isolated during the outbreak period. Five of these nine isolates represented human clinical isolates with XbaI PFGE pattern JIXX01.0011 and were therefore putatively linked to the outbreak isolates.
Nucleotide sequence accession number.
Sequence reads have been deposited in the NCBI sequence read archive under accession number SRA046270.
RESULTS AND DISCUSSION
Short-term evolution of S. Montevideo follows a model that assumes a relaxed log-normal molecular clock.
Analysis of newly generated genome sequences for 47 S. Montevideo isolates along with previously reported genome sequences for 35 S. Montevideo isolates (16) allowed for a comprehensive characterization of the epidemiology and phylogeny of a highly clonal set of Salmonella isolates. Overall, 969 SNP positions were found among the 82 isolates analyzed. A 6-kb aberrant region containing 293 SNPs present only in three outlier genomes was removed, leaving a total of 676 SNPs (see File S1 in the supplemental material). Further removal of SNP positions that contained ambiguous SNPs resulted in 410 SNPs. A total of 253 of these SNPs were found among the 47 isolates sequenced in this study. Compared to PFGE, which grouped all 47 isolates into the same XbaI and SpeI PFGE patterns (see Fig. S2 in the supplemental material), genome sequencing was considerably more discriminative. Interestingly, PFGE analyses of these isolates with a third enzyme (NotI) also showed further discrimination (into 25 types) compared to that with XbaI and SpeI PFGE, although these data did not correlate well with the SNP-based phylogeny (see below) and did not appear to correlate with isolation date or other epidemiological data.
Comparison of four population genetic models by means of the Bayes factor (20) showed strong evidence against a strict clock rate (BF > 30), indicating significant differences in SNP mutation rates between branches (see Table S2 in the supplemental material). Evidence against a model of constant effective population size over time was moderate (BF > 3). Hence, the results of the model that assumes a population size that varies through time and a relaxed log-normal molecular clock will be presented in the remainder of this paper. The inferred SNP mutation rate has a median of 7.25 × 10−5 nucleotide substitutions per SNP per year, which is about 25% slower than the mutation rate inferred over a longer time period (ca. 40 years) by Gray et al. (12) for S. aureus. The PHI test found no evidence for recombination for the SNP data set after exclusion of the 6-kb aberrant region (see above).
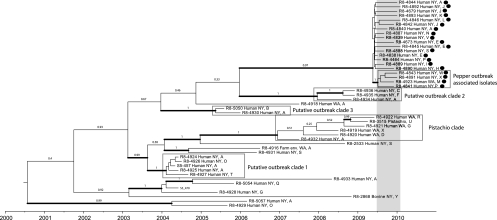
Only 4 out of 20 putative case isolates clustered with the pepper isolates.
All 20 putative case isolates obtained between June 2009 and January 2010 form a clade in the tip-dated phylogenetic analysis (Fig. 1). The posterior probability for this clade is high (>0.95), indicating that these isolates share a most recent common ancestor. While the 20 isolates in this clade all represent the same XbaI and SpeI PFGE patterns, they represent 10 different NotI patterns, including some NotI patterns that were also found in isolates obtained before June 2009 (Fig. 1). Phylogenetic comparison of the genome sequences reported here with genome sequences of additional isolates linked to the 2009-2010 S. Montevideo outbreak (16) identified four human isolates in our data set that very closely cluster with outbreak-associated isolates (bootstrap support of 95%; see Fig. S1 in the supplemental material), indicating that they are part of the pepper-associated outbreak. The tip-dated phylogeny reported for the genomes sequenced here also grouped these four outbreak-specific human isolates into a well-supported clade, further supporting the linkage of these isolates to this outbreak. This clade will be referred to as the “pepper-associated outbreak clade.” This clade comprises 3 isolates from New York State (FSL R8-4841, FSL R8-4843, and FSL R8-4891) and one isolate from the state of Washington (FSL R8-4923). XbaI and SpeI PFGE patterns were identical for these four isolates, while NotI PFGE patterns were different for each of the isolates, making it impossible to use PFGE subtyping to discriminate this clade from the background population. The most recent common ancestor of the four isolates was inferred to have originated in May 2009 (95% highest posterior probability density [HPD], March 2009 to October 2009), just 2 months before the first outbreak case was reported in July. While this cluster is distinct from the background population sampled during the period of the outbreak, we cannot draw any conclusions about the origin of the outbreak strain, whether domestic as suggested in reference 16 or imported with spices as suggested in reference 5. The current study and previous study (16) contain only isolates from the United States, so a global sampling of S. Montevideo isolates with XbaI PFGE pattern JIXX01.0011 is necessary to understand the population structure and thus origin of different lineages within this PFGE type.
Fig. 1.
Tip-dated maximum clade credibility tree based on SNP data for 47 S. Montevideo strains inferred under a coalescent model of a population size that varies over time and a relaxed molecular clock. Isolates that were obtained during the time period of the S. Montevideo outbreak are marked with a black dot, and the period of the outbreak has been marked with a gray bar. The alphabetical NotI pattern designation (see Fig. S2 in the supplemental material) is indicated after the geographical origin of each isolate. The gray boxes indicate putative outbreak clades, as referred to in the text. Thickened branches received a posterior probability of 95% or more.
After completion of genome sequencing, epidemiological data were obtained for eight patients: three that clustered into the pepper-associated outbreak clade and five that did not. Two of the three patients linked to the pepper outbreak-associated clade and four of the five patients with no link to the outbreak reported consumption of salami. In the previously published case-control study for this outbreak (5), 55% of the case patients reported having consumed salami, while 15.4% of the controls reported having eaten salami. As the epidemiological data for the isolates characterized here are limited and not comprehensive, one has to be careful in the interpretation of these data; it is tempting, however, to speculate that some patients in this outbreak may have been infected through other vehicles, e.g., secondary transmission, which is well documented in some salmonellosis outbreaks (9, 11). In addition, it is not surprising that at least some patients will report consumption of salami even though they were infected by S. Montevideo strains that do not seem to represent the outbreak subtype. While our data clearly indicate that whole-genome sequencing provides discriminatory power that can be critical for investigations of outbreaks caused by highly clonal bacteria, detailed epidemiological data will continue to be critical for the detection of outbreaks and identification of outbreak sources.
SNP data identify human isolates that are closely related to S. Montevideo linked to contaminated pistachios.
Four isolates from human clinical cases (FSL R8-4919, FSL R8-4920, FSL R8-4921, FSL R8-4922) from Washington State and one from New York State (FSL R8-4932) fall into a distinct clade that also includes isolates from contaminated pistachio nuts (isolate FSL R8-3515 in Fig. 1 and Fig S1 in the supplemental material; isolates FDA_2010_142_Pistachio-3 and FDA_2010_149_Pistachio-2 in Fig. S1 in the supplemental material). This group is referred to as the “pistachio clade.” These isolates were associated with a recall, in the United States, of pistachio nuts that were contaminated with Salmonella Montevideo (as well as several other Salmonella serovars) during the months before March 2009 (4). Only one patient, who was infected with a Salmonella strain with a PFGE pattern matching a strain of the contaminated products, was reported to have consumed a pistachio-containing product (4); identification of human cases in this instance was also complicated by the fact that the PFGE type of the outbreak strain was the same common PFGE type that was subsequently involved in the pepper outbreak discussed above. However, full genome sequencing data reported here discriminated the pistachio isolates from most other S. Montevideo isolates and suggested that the pistachio strain was responsible for four human salmonellosis cases in three successive years (2007, 2008, and 2009). The most recent common ancestor of the 6 isolates was inferred to have originated in October 2006 (95% HPD, July 2005 to June 2007). Without epidemiological data, we cannot conclusively link these human cases to consumption of pistachios, but it is conceivable that contaminated products entered the marketplace before March 2009 and caused sporadic cases, particularly as persistence of Salmonella in both primary production and processing facilities has been reported (3, 7, 21). Alternatively, a smaller clade within this group of isolates, which is also significantly supported (posterior probability of >0.95) and contains a single human isolate (FSL R8-4922) in addition to a pistachio isolate, may represent the pistachio clade, indicating identification of only a single additional human case that is both temporally and genetically closely linked to this contamination event. These scenarios illustrate how, in the future, epidemiological data, along with data on the time of the most common ancestor of isolates linked to a single outbreak, can be used to identify outbreak-associated cases and to predict the possible time when an outbreak or an associated contamination event started.
Control isolates show several previously undetected outbreak clusters.
Within the population of control isolates, we found at least three further clades that may represent previously undetected outbreaks. These clades include (i) a clade of five isolates obtained during the period of April to August 2004 at different geographically dispersed counties in New York State (putative outbreak clade 1 in Fig. 1), (ii) a clade of three isolates obtained between October and November 2008 from different counties surrounding the New York City area (Westchester, Nassau, and Suffolk counties; putative outbreak clade 2 in Fig. 1), and (iii) a clade of two isolates obtained in July and December 2005 from the greater New York City area (one isolate from the New York City Department of Health and Mental Health, and one isolate from Nassau county; putative outbreak clade 3 in Fig. 1). While the last two clades may represent minor regional clusters, the temporal pattern and geographic distribution of the 2004 isolates raise the possibility of a larger outbreak. These outbreaks may have gone unnoticed due to the inability to discriminate such clusters from the background population by means of standard subtyping techniques like PFGE.
We restricted putative outbreak clades to have no more than 2 years between the median age of the MRCA and the isolation date of the most recent isolate of the clade. This definition may exclude outbreaks that occurred over prolonged times; for example, one Salmonella outbreak linked to a single source occurred over 3 years (1). In the future, epidemiological data will need to be used in conjunction with genome sequence data representing both human cases and suspected food samples to identify likely outbreaks and to define outbreak durations. In particular, full genome sequence data may help to identify small outbreaks that may not be easily detected with lower-resolution subtyping approaches.
None of the putative clusters found in the SNP analysis showed distinct PFGE patterns, even when data for all three enzymes were analyzed simultaneously. The PFGE patterns obtained through XbaI and SpeI digestion were identical for all isolates (see Fig. S2 in the supplemental material), while the NotI PFGE patterns, though very variable, did not match any of the clades found in the phylogenetic analysis (Fig. 1). Our hypothesis is that the variability seen in the NotI PFGE patterns may be due to variable presence of mobile elements, such as plasmids and prophages, which were not included in the SNP analyses. The comparative analysis of PFGE and full genome SNP data clearly illustrates the advantages of an SNP-based approach when performing subtype analyses for highly clonal pathogens, particularly since SNP-based data can be analyzed to accurately identify phylogenetic relationships, which is not possible with PFGE data. The discrepancies between NotI restriction patterns and phylogenetically relevant SNP-based data illustrate the challenges of interpreting PFGE patterns, particularly for highly clonal pathogens.
SNP typing allows for rapid real-time (RT) PCR-based assay to discriminate between outbreak strains and the background population.
Three SNPs were found to be completely specific for the isolates associated with the pepper outbreak clade. Two of these SNPs are found in intergenic regions, while the third SNP causes a nonsynonymous mutation in envE, a gene found in Salmonella pathogenicity island 11 (18). TaqMan SNP genotyping assays targeting all three SNPs were developed; however, one of the three assays (AHN1G9M; see Table 2) did not give clear separation of fluorescent signal between the two alleles and was therefore used only to genotype a subset (20 isolates) of the panel. TaqMan SNP genotyping assays AHPAFFU and AHQJDL2 could successfully discriminate between isolates associated with the pepper outbreak clade and other Salmonella isolates, including isolates from the S. Montevideo background population with XbaI PFGE pattern JIXX01.0011. Only the isolates that were already known by sequencing to carry the pepper-associated outbreak SNPs were positive for the outbreak strain SNP alleles with these assays. None of the additional isolates (n = 65) in the panel carried these alleles. Therefore, the SNP genotyping assays are highly specific for isolates of the pepper-associated outbreak clade and could be used to discriminate this clade from other Salmonella isolates irrespective of their serovar or source.
Table 2.
Primers and probes used for SNP typing assays
| Assay | Forward primer sequence | Reverse primer sequence | Reporter 1 sequence | Reporter 2 sequence | SNP position in reference (AESM contig) | Description |
|---|---|---|---|---|---|---|
| AHN1G9 M | ACTAAAATTGCGCTGAGTCGGAAT | TTTACGCCAAGCCAGCTAACT | ATGTGGTTCGACAGAAGAA | ATGTGGTTTGACAGAAGAA | 27722 (AESM01000013.1) | SNP in intergenic region |
| AHPAFFU | GGTCAGCGGAGCGCAA | GGAGTTTGGCCAGCTTTTACGA | CTGACGCAAGTTAAAATG | CTGACGCAACTTAAAATG | 70295 (AESM01000017.1) | SNP causes nonsynonymous mutation in envE |
| AHQJDL2 | GCGGGAAAAAGCGGGAAATG | CAAGGCAAAGATGAACGTGATAGC | CATCACACCGACCTATTAGT | CACACCGGCCTATTAGT | 82461 (AESM01000030.1) | SNP in intergenic region |
Conclusion.
This study demonstrates that massively parallel sequencing can be used to assess the population structure of highly clonal, outbreak-associated pathogens at single-base resolution. This capability facilitates outbreak detection, assignment of isolates to a given outbreak, and development of specific molecular assays that allow for rapid, high-throughput screening of isolates. We envision an outbreak investigation workflow in which a draft genome assembly of an outbreak strain would first be generated using a sequencing platform that can sequence a bacterial genome in 2 to 3 days (17). This initial draft genome could then be used to characterize and predict phenotypic characteristics of the outbreak strain and serve as a reference genome for high-throughput resequencing and SNP discovery in additional related isolates. Outbreak strain-specific polymorphisms could then be targeted by rapid molecular assays that allow for large-scale rapid isolate screening to support the outbreak investigation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nellie Dumas of the New York State Department of Health for her collaboration and helpful comments on the manuscript. We thank Rebecca Gray for her help with implementing the ascertainment bias correction model in the BEAST analyses.
This project was partially supported by USDA special research grant numbers 2008-34459-19043 and 2009-34459-19750 (to M. Wiedmann) and a Life Technologies Collaborative Research Compact grant (to C. A. Cummings). Life Technologies Corporation partially funded this study by providing instruments and sequencing reagents and by compensating its employees (C. A. Cummings, L. Degoricija, R. Fang, and M. R. Furtado), who participated in study design, data collection and analysis, decision to publish, and preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 14 October 2011.
REFERENCES
- 1. Behravesh C. B., et al. 2010. Human Salmonella infections linked to contaminated dry dog and cat food, 2006-2008. Pediatrics 126:477–483 [DOI] [PubMed] [Google Scholar]
- 2. Bruen T. C., Philippe H., Bryant D. 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics 172:2665–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Centers for Disease Control and Prevention 2008. Investigation of outbreak of infections caused by Salmonella agona. CDC, Atlanta, GA: http://www.cdc.gov/salmonella/agona/ [Google Scholar]
- 4. Centers for Disease Control and Prevention 2009. Salmonella in pistachio nuts, 2009. CDC, Atlanta, GA: http://www.cdc.gov/salmonella/pistachios/update.html [Google Scholar]
- 5. Centers for Disease Control and Prevention 2010. Salmonella Montevideo infections associated with salami products made with contaminated imported black and red pepper—United States, July 2009-April 2010. MMWR Morb. Mortal. Wkly. Rep. 59:1647–1650 [PubMed] [Google Scholar]
- 6. Croucher N. J., et al. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331:430–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cummings K. J., et al. 2009. The duration of fecal Salmonella shedding following clinical disease among dairy cattle in the northeastern U.S.A. Prev. Vet. Med. 92:134–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Drummond A. J., Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Faustini A., et al. 1998. An outbreak of Salmonella hadar associated with food consumption at a building site canteen. Eur. J. Epidemiol. 14:99–106 [DOI] [PubMed] [Google Scholar]
- 10. Felsenstein J. 1985. Confidence-limits on phylogenies—an approach using the bootstrap. Evolution 39:783–791 [DOI] [PubMed] [Google Scholar]
- 11. Francis S., et al. 1989. An outbreak of paratyphoid fever in the UK associated with a fish-and-chip shop. Epidemiol. Infect. 103:445–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gray R. R., et al. 2011. Testing spatiotemporal hypothesis of bacterial evolution using methicillin-resistant Staphylococcus aureus ST239 genome-wide data within a Bayesian framework. Mol. Biol. Evol. 28:1593–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harris S. R., et al. 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327:469–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huson D. H., Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254–267 [DOI] [PubMed] [Google Scholar]
- 15. Kurtz S., et al. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lienau E. K., et al. 2011. Identification of a salmonellosis outbreak by means of molecular sequencing. N. Engl. J. Med. 364:981–982 [DOI] [PubMed] [Google Scholar]
- 17. Mellmann A., et al. 2011. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next-generation sequencing technology. PLoS One 6:e22751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morgan E. 2007. Salmonella pathogenicity islands, p. 67–88 In Rhen M., Maskell D. J., Mastroeni P., Threlfall J. (ed.), Salmonella. Molecular biology and pathogenesis. Horizon Bioscience, Norfolk, United Kingdom [Google Scholar]
- 19. Ribot E. M., et al. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67 [DOI] [PubMed] [Google Scholar]
- 20. Suchard M. A., Weiss R. E., Sinsheimer J. S. 2001. Bayesian selection of continuous-time Markov chain evolutionary models. Mol. Biol. Evol. 18:1001–1013 [DOI] [PubMed] [Google Scholar]
- 21. Uesugi A. R., Danyluk M. D., Mandrell R. E., Harris L. J. 2007. Isolation of Salmonella Enteritidis phage type 30 from a single almond orchard over a 5-year period. J. Food Prot. 70:1784–1789 [DOI] [PubMed] [Google Scholar]
- 22. Wilgenbusch J. C., Swofford D. 2003. Inferring evolutionary trees with PAUP*. Curr. Protoc. Bioinformatics 6:6.4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.