Abstract
The aims of this study were to determine if marine bacteria from Danish coastal waters produce antifouling compounds and if antifouling bacteria could be ascribed to specific niches or seasons. We further assess if antibacterial effect is a good proxy for antifouling activity. We isolated 110 bacteria with anti-Vibrio activity from different sample types and locations during a 1-year sampling from Danish coastal waters. The strains were identified as Pseudoalteromonas, Phaeobacter, and Vibrionaceae based on phenotypic tests and partial 16S rRNA gene sequence similarity. The numbers of bioactive bacteria were significantly higher in warmer than in colder months. While some species were isolated at all sampling locations, others were niche specific. We repeatedly isolated Phaeobacter gallaeciensis at surfaces from one site and Pseudoalteromonas tunicata at two others. Twenty-two strains, representing the major taxonomic groups, different seasons, and isolation strategies, were tested for antiadhesive effect against the marine biofilm-forming bacterium Pseudoalteromonas sp. strain S91 and zoospores of the green alga Ulva australis. The antiadhesive effects were assessed by quantifying the number of strain S91 or Ulva spores attaching to a preformed biofilm of each of the 22 strains. The strongest antifouling activity was found in Pseudoalteromonas strains. Biofilms of Pseudoalteromonas piscicida, Pseudoalteromonas tunicata, and Pseudoalteromonas ulvae prevented Pseudoalteromonas S91 from attaching to steel surfaces. P. piscicida killed S91 bacteria in the suspension cultures, whereas P. tunicata and P. ulvae did not; however, they did prevent adhesion by nonbactericidal mechanism(s). Seven Pseudoalteromonas species, including P. piscicida and P. tunicata, reduced the number of settling Ulva zoospores to less than 10% of the number settling on control surfaces. The antifouling alpP gene was detected only in P. tunicata strains (with purple and yellow pigmentation), so other compounds/mechanisms must be present in the other Pseudoalteromonas strains with antifouling activity.
INTRODUCTION
The bacterial colonization of marine surfaces and subsequent macrofouling is a ubiquitous phenomenon (20), and mature marine biofouling communities are complex and highly dynamic ecosystems which are difficult to eradicate once established (22). The biofouling of ship hulls (or other immersed structures) greatly increases the hydrodynamic drag and thereby leads to high fuel consumption (40) and CO2 emissions. Costly mechanical processes coupled with toxic heavy metal-based paint containing, e.g., tin and copper, traditionally have been used to combat marine biofouling. Due to the nonspecific effects of heavy metal leaching, such paints are environmentally hazardous. An example is the very effective toxic compound tributyl tin, which is believed to contribute to the development of antimicrobial tolerance in marine organisms (35) and to cause imposex in some invertebrates (1, 16). Antifouling paints containing tin have been banned since 2003 and gradually removed from shipping fleets (27). Consequently, the search for antifouling compounds or principles that reduce or eliminate the attachment of marine organisms is intense (37). The key focus of the present study and other studies (7, 36) is to find environmentally friendly molecules or organisms (10) to replace the toxic/biocidal compounds being used to day. All macroorganisms in the marine environment have developed natural antifouling strategies, and our hypothesis is that any chemical components developed as part of a natural antifouling defense likely would be environmentally compatible.
Macroorganisms produce antifouling compounds, such as the halogenated furanones produced by the red algae Delisea pulchra (8), but they also rely on epiphytic bacteria as producers of antifouling compounds (11, 24, 36). From a biotechnological perspective, microorganisms are an exploitable source of antifouling compounds (37), hence we focused our search on marine bacteria. Some marine bacteria, primarily belonging to the Pseudoalteromonas genus, the Vibrionaceae family, and the Roseobacter clade, excrete compounds that can reduce bacterial biofilm formation and settlement by larger microorganisms on surfaces (6, 12, 38), suggesting their suitability as antifouling bacteria. However, a large-scale search for bacteria producing antifouling compounds is hampered by the fact that a high-throughput assay for screening large numbers of bacteria for antifouling capacity is not available. Typically, studies have focused on bacteria already known to inhibit the growth of other bacteria, such as the well-studied Pseudoalteromonas tunicata strain D2 (23, 25), or on a small subcollection of bacteria from a very specific niche (12, 15).
The purpose of the present study was to probe the antifouling potential of marine bacteria on a broader scale. We based our primary screen on the search for antibacterial activity, as this can be done in a high-throughput manner. Although we assume that bacteria producing antibacterial compounds also were likely to display antifouling capacity, it has never been determined that these characteristics are linked, i.e., if the search for antibacterial activity is a good proxy for antifouling activity. We deliberately took samples from different sources (water and surfaces), locations, and seasons to determine if bacteria with antibacterial and antifouling activity were associated with specific niches or seasons. We focused our sampling on bacteria isolated from live or inert marine surfaces, as other studies have indicated that marine bacteria associated with surfaces are more likely to display antibacterial activity than planktonic bacteria isolated from open waters (31, 33, 47).
MATERIALS AND METHODS
Sample collection.
Seawater, swabs from marine surfaces (an area of between 1 and 10 cm2 was swabbed), and samples such as seaweed, barnacles, and mussels were collected from 11 coastal sites in Denmark, representing different salinity and temperature conditions (Table 1). A total of 467 samples were analyzed and were derived from approximately two seawater and eight surface samples from each of the 11 samplings sites during different seasons (April, June, August, and November 2009 and February 2010). Water temperature and salinity were measured in situ with a handheld Professional Plus instrument (YS6050000; YSI, Yellow Springs, OH).
Table 1.
Chemical and microbiological parameters in seawater collected during a 1-year survey from 11 coastal sites in Denmark
| Sample site and collection time | Chemical parameter |
Microbiological parametera |
||
|---|---|---|---|---|
| Temp (°C) | Salinity (%) | Total cell count (log cells/ml) | Culturable counts (log CFU/ml) | |
| Jyllinge Habour | ||||
| April 2009 | 12.2 ± 0.1 | 1.4 ± 0.0 | 6.60 ± 0.03 | 2.98 ± 0.02 |
| June 2009 | 16.0 ± 0.0 | 1.4 ± 0.0 | 6.32 ± 0.06 | 3.74 ± 0.06 |
| August 2009 | 18.8 ± 0.0 | 1.4 ± 0.0 | 6.74 ± 0.05 | 2.90 ± 0.00 |
| November 2009 | 7.4 ± 0.1 | 1.5 ± 0.0 | 6.43 ± 0.07 | 2.87 ± 0.08 |
| February 2010 | NDb | ND | 5.85 ± 0.02 | 2.65 ± 0.10 |
| Gilleje | ||||
| April 2009 | 12.0 ± 0.4 | 1.7 ± 0.0 | 6.26 ± 0.03 | 3.38 ± 0.24 |
| June 2009 | 14.2 ± 0.0 | 1.5 ± 0.1 | 6.54 ± 0.02 | 4.72 ± 0.05 |
| August 2009 | 21.3 ± 0.3 | 1.7 ± 0.3 | 6.57 ± 0.03 | 3.00 ± 0.00 |
| November 2009 | 8.6 ± 0.0 | 1.8 ± 0.1 | 5.47 ± 0.17 | 4.35 ± 0.04 |
| February 2010 | −1.0 ± 0.1 | ND | 5.87 ± 0.07 | 2.04 ± 0.06 |
| Bellevue | ||||
| April 2009 | 13.7 ± 0.1 | 0.8 ± 0.1 | 6.41 ± 0.11 | 3.48 ± 0.06 |
| June 2009 | 13.7 ± 0.0 | 1.0 ± 0.0 | 6.60 ± 0.06 | 3.65 ± 0.23 |
| August 2009 | 18.8 ± 0.1 | 1.0 ± 0.0 | 6.69 ± 0.03 | 4.02 ± 0.08 |
| November 2009 | 6.1 ± 0.0 | 1.0 ± 0.0 | 6.55 ± 0.04 | 3.98 ± 0.02 |
| February 2010 | −0.2 ± 0.1 | 1.0 ± 0.0 | 6.21 ± 0.02 | 2.81 ± 0.05 |
| Limfjorden | ||||
| April 2009 | 9.5 ± 0.1 | 2.2 ± 0.3 | 6.87 ± 0.06 | 4.58 ± 0.23 |
| June 2009 | 16.7 ± 0.1 | 2.3 ± 0.2 | 6.55 ± 0.06 | 4.45 ± 0.03 |
| August 2009 | 17.4 ± 0.1 | 2.6 ± 0.0 | 6.80 ± 0.04 | 3.93 ± 0.00 |
| November 2009 | 7.5 ± 0.0 | 2.0 ± 0.0 | 6.48 ± 0.03 | 5.22 ± 0.18 |
| February 2010 | ND | ND | 4.96 ± 0.26* | 3.72 ± 0.07* |
| North Sea (north Agger) | ||||
| April 2009 | 8.2 ± 0.0 | 3.3 ± 0.0 | 7.00 ± 0.04 | 4.77 ± 0.22 |
| June 2009 | 12.3 ± 0.0 | 3.5 ± 0.0 | 6.24 ± 0.11 | 3.32 ± 0.13 |
| August 2009 | 17.7 ± 0.0 | 2.9 ± 0.1 | 6.04 ± 0.06 | 3.77 ± 0.23 |
| November 2009 | 9.5 ± 0.0 | 3.0 ± 0.3 | 5.67 ± 0.07 | 3.01 ± 0.00 |
| February 2010 | −0.9 ± 0.0 | 3.4 ± 0.0 | 5.76 ± 0.09 | 2.74 ± 0.04 |
| North Sea (halfway Hvide sande) | ||||
| April 2009 | 11.2 ± 0.0 | 2.2 ± 0.0 | 6.92 ± 0.06 | 4.89 ± 0.02 |
| June 2009 | 13.2 ± 0.0 | 2.1 ± 0.1 | 6.55 ± 0.03 | 4.67 ± 0.42 |
| August 2009 | 16.7 ± 0.1 | 2.0 ± 0.1 | 6.68 ± 0.06 | 4.11 ± 0.13 |
| November 2009 | 8.2 ± 0.0 | 2.0 ± 0.1 | 6.26 ± 0.05 | 3.06 ± 0.02 |
| February 2010 | 0.0 | 2.4 ± 0.0 | 5.59 ± 0.04 | 2.72 ± 0.14 |
| North Sea (south Blåvand) | ||||
| April 2009 | 14.9 ± 0.1 | 2.7 ± 0.3 | 6.85 ± 0.07 | 4.90 ± 0.02 |
| June 2009 | 14.6 ± 0.1 | 2.7 ± 0.1 | 6.47 ± 0.05 | 4.09 ± 0.04 |
| August 2009 | 17.8 ± 0.1 | 3.2 ± 0.0 | 6.40 ± 0.03 | 3.59 ± 0.24 |
| November 2009 | 8.3 ± 0.0 | 3.1 ± 0.0 | 6.26 ± 0.06 | 3.08 ± 0.07 |
| February 2010 | −0.3 ± 0.0 | 3.0 ± 0.0 | 6.07 ± 0.04 | 3.38 ± 0.04 |
| Middelfart | ||||
| April 2009 | 11.9 ± 0.4 | 2.1 ± 0.1 | 6.82 ± 0.05 | 4.62 ± 0.08 |
| June 2009 | 15.3 ± 0.1 | 2.1 ± 0.0 | 6.70 ± 0.08 | 4.54 ± 0.06 |
| August 2009 | 17.9 ± 0.1 | 2.3 ± 0.0 | 6.82 ± 0.05 | 4.21 ± 0.07 |
| November 2009 | 8.9 ± 0.1 | 1.8 ± 0.0 | 6.53 ± 0.10 | 5.70 ± 0.31 |
| February 2010 | 0.9 ± 0.0 | 1.1 ± 0.3 | 6.19 ± 0.02 | 3.87 ± 0.02 |
| Korsør | ||||
| April 2009 | 13.7 ± 1.2 | 1.5 ± 0.0 | 6.19 ± 0.05 | 4.83 ± 0.10 |
| June 2009 | 15.3 ± 0.1 | 1.6 ± 0.0 | 6.32 ± 0.11 | 3.65 ± 0.21 |
| August 2009 | 18.9 ± 0.0 | 1.4 ± 0.0 | 6.62 ± 0.03 | 4.61 ± 0.20 |
| November 2009 | 7.1 ± 0.1 | 1.2 ± 0.0 | 6.65 ± 0.07 | 3.04 ± 0.16 |
| February 2010 | 0.6 ± 0.0 | 1.2 ± 0.0 | 6.24 ± 0.05 | 2.86 ± 0.26 |
| Kalvehave Møn | ||||
| April 2009 | 15.3 ± 0.8 | 0.9 ± 0.0 | 6.63 ± 0.09 | 2.98 ± 0.34 |
| June 2009 | 12.7 ± 0.0 | 0.9 ± 0.0 | 5.64 ± 0.21 | 5.29 ± 1.01 |
| August 2009 | 20.4 ± 0.0 | 1.0 ± 0.0 | 6.72 ± 0.05 | 4.35 ± 0.21 |
| November 2009 | 6.3 ± 0.4 | 1.0 ± 0.0 | 6.65 ± 0.03 | 4.07 ± 0.40 |
| February 2010 | ND | ND | ND | 5.01 ± 0.00* |
| Stensballe Horsens | ||||
| April 2009 | 15.1 ± 0.4 | 2.1 ± 0.0 | 6.48 ± 0.15 | 5.42 ± 0.09 |
| June 2009 | 19.5 ± 0.3 | 2.4 ± 0.1 | 6.19 ± 0.07 | 4.49 ± 0.22 |
| August 2009 | 17.7 ± 0.0 | 2.3 ± 0.1 | 6.62 ± 0.06 | 5.26 ± 0.13 |
| November 2009 | 7.7 ± 0.1 | 1.9 ± 0.0 | 5.10 ± 0.26 | 4.07 ± 0.40 |
| February 2010 | ND | ND | ND | 5.26 ± 0.00* |
A 0.02-μm filter was used. *, measured on frozen water.
ND, not determined.
Total bacterial cell density in seawater.
Total bacterial densities were determined using epifluorescence counts of SYBR gold-stained bacteria on 5-μm polycarbonate filters (no. K50BP02500; GE Water & Process Technologies) and 0.02-μm anodisc filters (no. 6809-6002; Whatman) as previously described (19). Cells were counted using an Olympus BX51 microscope with 460- to 490-nm excitation and >510-nm emission filters. Five fields were counted per sample.
Plate counts of culturable bacteria for all samples.
Bacteria were extracted from solid samples of seaweed, algae, etc., and 10-fold serially diluted in 3% Instant Ocean (IO; Aquarium Systems Inc., Sarrebourg, France). Samples were divided into two groups according to their texture. Soft tissues, such as seaweeds, were homogenized using an Ultraturrax T25, whereas hard samples, such as small stones, barnacles, and mussel shells, were mixed by vortexing at maximum speed for 30 s. Seawater, swabs, and diluted samples also were 10-fold serially diluted in 3% IO and plated on 50% marine agar (MA; diluted with 1.5% IO; 212185; Difco). Plates were incubated at 20°C for 4 to 5 days, and colonies were counted to determine CFU (CFU/ml, CFU/g, or CFU/swab).
Enrichment of bacteria.
From all water samples and from 10-fold-diluted surface/swab samples, 0.1 ml was mixed with 5 ml marine broth (279110; Difco) and incubated at 20°C for 2 weeks. In all of these enrichment samples, a biofilm formed on the glass at the air-water interface, and bacteria from these biofilm rings were streaked on MA and incubated at 20°C. Subsequently the bacteria were tested for antibacterial activity by replica plating against V. anguillarum strain 90-11-287 (41) as described below.
Selection of pigmented bacteria.
Approximately 10 pigmented colonies were isolated from each sampling site at each sampling time after culturable plate count. Colonies were streaked on MA and tested for antibacterial activity against V. anguillarum strain 90-11-287 in a replica assay.
Antibacterial bioassay.
Plate counts, colonies from enriched samples, and pigmented strains were tested for inhibitory activity against V. anguillarum strain 90-11-287 in agar-based assays in which V. anguillarum was incorporated into a 1.2% agar with 3% (wt/vol) IO salts, 0.3% (wt/vol) Bacto Casamino acids (product number 223050; BD, MD), and 0.4% (wt/vol) glucose as previously described (21). V. anguillarum strain 90-11-287 was selected as an indicator strain for antibacterial activity, as it is very sensitive to inhibitory compounds produced by other marine bacteria (19). Colonies causing a clearing zone in the turbid V. anguillarum layer were isolated from the original plate, restreaked, and stored at −80°C.
Identification of bacteria.
Identification to the genus level was based on a combination of phenotypic characteristics (19) and 16S rRNA gene sequence analysis. Chromosomal DNA was purified from the marine bacteria grown for 3 days at 25°C in marine broth using the NucleoSpin tissue kit from Macherey-Nagel (M740952). The 16S rRNA genes were amplified using the universal primers 27F (AGAGTTTGATCMTGGCTCAG) and 1492R (TACGGYTACCTTGTTACGACTT) (19). Following gel electrophoresis, the PCR products were purified using the GFX PCR DNA and gel band purification kit from GE Healthcare (Buckinghamshire, United Kingdom). Sequencing was carried out by DNA Technology, Århus, Denmark, with 518F (CCAGCAGCCGCGGTAATACG) and 800R (TACCAGGGTATCTAATCC) as primers. Identification to the genus level was done using the BLAST-based program leBIBI (9) and NCBI databases. The inhibitory activities of all strains were retested in agar diffusion assays against V. anguillarum strain 90-11-287 after being frozen at −80°C for at least 3 months.
Presence of a known antifouling gene.
All Pseudoalteromonas strains were tested for the presence of the alpP gene by PCR with primers previously described (42). Aliquots (2 μl) of purified DNA were applied to the following to give a 25-μl PCR mixture: 13 μl Brilliant II quantitative PCR master mix (2×) (Agilent Technology), 1 μl (12.5 μM) of primer alpP-1F and primer alpP-2R, and 8 μl sterile MilliQ water. The PCR amplification (15 min at 95°C and then 40 cycles of three steps consisting of 30 s at 92°C, 60 s at 55°C, and 30 s at 72°C, with a final extension of 5 min at 72°C) was performed with a 9800 Fast Thermal Cycler (Applied Biosystems). The size of the final PCR product was determined by electrophoresis in a 1% agarose gel.
Collection and sporulation of Ulva australis.
U. australis cells were transported (for approximately 20 min) on ice after collection and then frozen at −20°C for half an hour. The plants were thawed and rinsed gently in sterile filtered seawater, placed in a beaker with sterile filtered seawater, and positioned close to a light source (desk lamp) to induce sporulation (13). The phototactic response of the zoospores was used to remove other unicellular organisms associated with the algal spores and to select spores with higher chances of survival (13). Spores were added to one side of a glass tray (20 cm long) containing sterile filtered seawater, and spores were allowed to migrate toward a light source at the other end. After 10 to 15 min, spores that reached the other side of the tray were collected.
Streptomycin resistance.
Twenty-two isolates representing Pseudoalteromonas, Vibrio, Photobacterium, and Phaeobacter were tested for antiadhesive properties. Apart from representing the major taxonomic groups, the isolates also represented different seasons and isolation strategies (plate count, enrichment, and pigmentation). The strains were tested for their ability to prevent the attachment of a marine biofilm-forming bacterium, Pseudoalteromonas strain S91 (45). Strain S91 is streptomycin resistant, and the 22 strains were tested for streptomycin resistance to determine if streptomycin-containing agar medium would allow us to differentiate between S91 and the potential antifouling bacteria. The strains were plated on marine agar containing 25, 50, 100, and 200 μg/ml of streptomycin and incubated at 25°C for up to 1 week.
Antiadhesive activity and growth inhibition.
Adhesion to and prevention of adhesion to inert surfaces was tested using stainless steel coupons (5 by 5 by 1 mm). The reporter strain S91 was chosen as a marker for antiadhesive effect against marine bacteria, as it is a nonantagonistic marine environmental strain which attaches to both abiotic and marine biotic surfaces and produces extended biofilms (44). The strain also is streptomycin resistant, which allowed us to distinguish between S91 and the potential antifouling marine strains. We quantified bacteria on the steel coupons with removal by sonication, followed by plate count on marine agar with and without streptomycin. We have previously demonstrated in pure cultures that the bacterial numbers determined by sonication removal and subsequent plating agrees well with numbers determined by the direct staining of attached bacteria (3). In situ quantification by staining with different probes followed by fluorescence microscopy also could have been used but would require strain-specific probes and that the number of bacteria on the surface was quantifiable by microscopy.
Prior to use, the coupons were cleaned by soaking overnight in a 15% Deconex solution, rinsed, degreased with acetone, and sterilized by autoclaving. The coupons were tilted and placed individually in wells of a microtiter plate (no. 167008; Nunc). The 22 marine bacteria were cultured in marine broth for 3 days at 25°C and diluted by a factor of 1,000 in marine broth, and 200 μl was transferred to each well containing the coupons. The bacteria were allowed to grow and attach to the stainless steel surface for 3 days at 25°C before the coupons were washed twice in 2 ml of 3% IO, transferred to a new microtiter plate, and exposed to a marine fouling bacterium. The amount of biofilms formed on the stainless steel coupons prior to exposure to S91 was determined by sonication and plate counts as described below. The spontaneous streptomycin-resistant mutant S91 of the marine fouling bacteria Pseudoalteromonas sp. strain S9 (45) was grown for 3 days in marine broth and diluted by a factor of 10,000 in 3% sea salt (S9883; Sigma-Aldrich). Two hundred μl was transferred to each well in the microtiter plate containing the biofilm-coated coupons, and the bacterial adhesion of strain S91 was allowed to take place on both sites of the coupons. Samples of the suspension and of the steel plates were taken after 1, 4, and 24 h. The stainless steel coupons (with attached bacteria) were immersed in polystyrene tubes (Sterikin LDT; Bibby Sterin LDT, Stones, United Kingdom) containing 2 ml sterile 3% sea salt (Sigma). Bacteria were removed from the surface by sonication (4) and vortexed at maximum speed for 15 s to further facilitate removal. The samples were serially diluted in sterile 3% sea salt (Sigma), and colony counts of the total number of adhered bacteria and the number of adhered S91 were determined by plating on 50% marine agar and 50% marine agar containing 400 μg/ml streptomycin, respectively. The efficiency of the detachment procedure was verified by the SYBR gold staining of the steel coupons followed by fluorescence microscopy (3). Bacterial counts in the suspensions also were done. All adhesion assays were carried out in triplicate. Phaeobacter strain 27-4 (21) and Pseudoalteromonas tunicata D2 (23) were included as positive (antifouling) controls, and marine broth was used as a negative control.
Ulva zoospore adhesion and germination assay.
The effect of bacteria on algal zoospore settlement and germination was studied using the marine alga U. australis. It was collected prior to sporulation from rock surfaces located at Shark Point, Clovelly, Sydney, Australia. Algal spores were exposed directly to monospecies bacterial biofilms of the 22 strains. The marine bacteria were cultured in marine broth for 3 days at 25°C and diluted by a factor of 100, and 1 ml was transferred to a 24-multiwell culture plate (Sigma). The bacteria were allowed to grow and form biofilms on the plastic surfaces for 3 days at 25°C before the growth medium was discarded. The wells were washed twice with sterile filtered (0.22-μm pore size; Millex; Millipore, Carrigtwohill, Ireland) seawater prior to adding the spores. The amount of bacterial biofilms was assessed by the crystal violet staining of biofilms grown under conditions identical to those used for the spore settlement assay (29). One ml of the Ulva spore suspension (described above) was added to each well in the 24 multiwells coated with bacteria, and the plates were placed in darkness for 2 h to allow the even settlement of spores. The plates then were incubated at room temperature under natural light for 24 h (for spore settlement), 1 ml of fresh sterile seawater was added, and the incubation was extended for six more days (for germination).
The quantification of the number of settled spores and germination was done by counting 10 fields of vision under 40× magnification using an inverted light microscope (Zeiss). Treatments were compared to controls consisting of noncoated and marine broth-coated wells.
Statistical analysis.
The comparison of the prevalence of inhibitory colonies isolated in the different sampling months (April, June, August, November, and February) was done using Pearson's chi-squared test. The comparison of antifouling activity against strain S91 was done by a t test comparison of log-transformed cell densities (CFU/ml or CFU/cm2).
RESULTS
Sample collection.
A total of 467 samples were analyzed from the 11 coastal sites in April, June, August, and November 2009 and February 2010. The average temperatures varied during the year, being just below 0°C in February and above 18°C in August (Table 1). Variation was seen between the sampling sites, and as expected the water temperatures in the fjords compared to coastal waters were higher in the summer months and lower in the winter months (Table 1). We were only able to measure temperature and salinity at 6 of 11 sampling sites in February due to snow and sea ice. The average salinity level from April to November for each sampling site varied from below 10 practical salinity units (psu) at the southeast cost of Denmark to above 30 psu at the northwest coast (Table 1). The variation between salinity measurements at the same sampling site over time was low (Table 1).
Total bacterial cell density and culturable bacterial counts.
The total cell counts of the seawater were between 5.1 and 7.0 log cells per ml, of which approximately 0.4% were culturable on 50% marine agar. Large variations were observed in the culturable counts between the different locations at each sampling month (Table 1).
Seawater samples (105 in total) contained 3.9 ± 0.3 log CFU/ml of culturable bacteria, whereas numbers of culturable organisms were higher in swab samples (165 in total) and whole-surface samples (197 in total), 6.8 ± 1.3 and 6.9 ± 1.2 log CFU/g, respectively. Approximately 124,000 bacterial colonies, of which 19,000 were from water samples, 43,000 from swab samples, and 61,000 from whole surface samples, were replica plated and tested for inhibitory activity against V. anguillarum strain 90-11-287. Of these 300 colonies (1.6%) from seawater, 515 (1.2%) from swab samples and 505 (0.8%) from whole-surface samples were inhibitory against V. anguillarum strain 90-11-287 in the primary replica plating. Forty-one of the 1,320 inhibitory colonies were restreaked and identified as described below. These were chosen to represent the different isolation times, places, and sample types (surface and water samples).
Enrichment of bacteria.
Enrichment in marine broth for 2 weeks resulted in the development of a biofilm at the air-liquid interface. Streaking from 229 biofilms on 50% marine agar and replica plating resulted in 48 strains inhibiting V. anguillarum (Table 2). The inhibitory strains isolated after enrichment all were nonpigmented or had only a slight pigmentation (light yellow or orange). All of these 48 strains were included in further analyses.
Table 2.
Influence of isolation strategy on marine bacteria from Danish coastal waters capable of inhibiting Vibrio anguillarum strain 90-11-287
| Identification based on 16S rRNA gene sequence | No. of isolated Vibrio anguillarum inhibitory strains based on isolation method |
|||
|---|---|---|---|---|
| Colony count | Pigmented colonies | Enrichment in MB | Total | |
| Pseudoalteromonas | 18 | 17 | 19 | 54 |
| Vibrio | 4 | 2 | 18 | 24 |
| Phaeobacter | 10 | 0 | 0 | 10 |
| Photobacterium | 0 | 0 | 8 | 8 |
| Shewanella | 0 | 1 | 1 | 2 |
| Marinomonas | 5 | 0 | 0 | 5 |
| Other | 4 | 1 | 2 | 7 |
| Total | 41 | 21 | 48 | 110 |
Selection of pigmented bacteria.
Since several pigmented marine bacteria display either antibacterial or antifouling activity (6), we also randomly isolated highly pigmented colonies from the 50% MA plates. A total of 294 strains were isolated, and 21 (7.1%) were inhibitory against V. anguillarum, and they all were isolated from surface samples. Seventeen of the 21 inhibitory strains were identified as Pseudoalteromonas and were white, yellow, orange, purple, brown, or black.
Identification of marine bacteria with antibacterial activity.
One hundred ten strains inhibiting V. anguillarum were selected based on the size of inhibition zones in the replica plates against V. anguillarum, different colony morphology, and different pigmentation (see Table S1 in the supplemental material). Also, we ensured that strains were from different sample types, times of sampling, and sampling places. The 110 strains consisted of 41 strains isolated from plate counts, 48 after selective enrichment and 21 selected based on pigmentation (Table 2). All antagonistic isolates were Gram-negative rods with positive oxidase and catalase reactions. Forty-five strains were intensely pigmented, being yellow, purple, orange, or black (21 of these were directly isolated due to their pigmentation). The similarity of 16S rRNA gene sequences identified the majority as Pseudoalteromonas (54 strains), Vibrio (24 strains), Photobacterium (8 strains), and Phaeobacter (10 strains). Eighty-seven strains retained inhibition upon retesting against V. anguillarum. It was mainly the nonpigmented Pseudoalteromonas strains and Vibrio strains isolated in November and February that lost their activity (data not shown).
Species identification of Pseudoalteromonas strains.
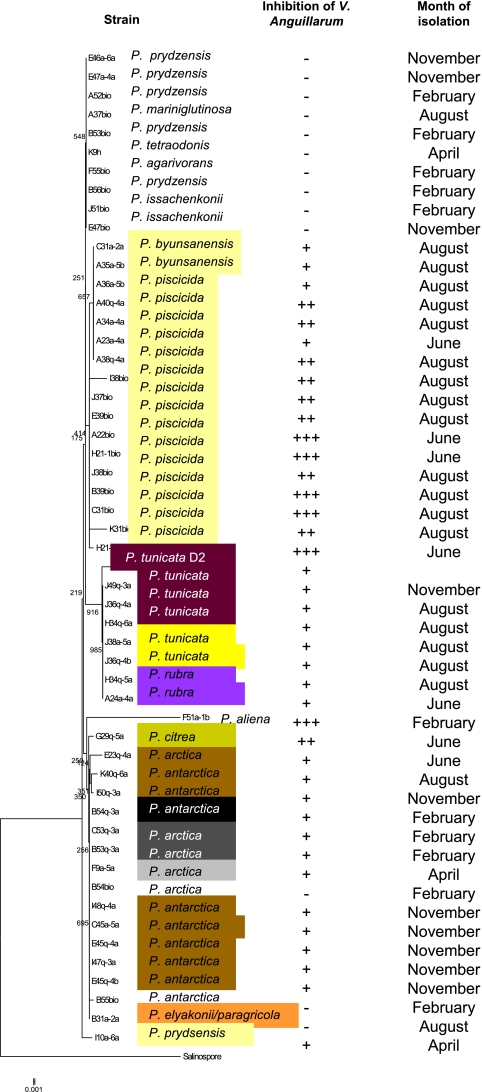
Our subsequent findings (see below) indicated that some of the isolated Pseudoalteromonas strains had pronounced antifouling activity, and as stated in the introduction, this has been described previously for P. tunicata. The initial BLAST searches querying the 16S rRNA gene sequences of our Pseudoalteromonas strains were ambiguous (i.e., gave large numbers of hits with identical scores). A BLAST (http://blast.ncbi.nlm.nih.gov) search of our isolated Pseudoalteromonas strains against a compilation of Pseudoalteromonas type strain sequences retrieved from GenBank (the list of type strains was obtained from http://www.bacterio.cict.fr) therefore was done to identify isolates to the species level. The best type strain BLAST match identified 15 strains as P. piscicida, 5 strains as P. tunicata (being either dark purple or yellow on marine agar), and 2 strains as P. ulvae (Fig. 1). To group the bacteria according to 16S rRNA gene sequence similarity, a phylogenetic tree based on 52 of our isolated Pseudoalteromonas strains (for the last two of our strains the 16S rRNA samples were short, therefore these strains were left out of the analysis) and P. tunicate D2 (23) was done using Salinispora arenicola strain CNS-205 as an outgroup as previously described (47). The clustering of the phylogenetic analysis reflected both species and pigmentation (Fig. 1). The presence of pigmentation was indicative of a stable antibacterial activity against V. anguillarum strains except strain F51a-1 (identified as P. aliena), which was nonpigmented and had strong inhibitory activity (Fig. 1).
Fig. 1.
Phylogenetic tree based on 16S-rRNA gene sequences of Pseudoalteromonas collected in Danish coastal waters. Sequences were aligned by MAFFT (default options), and the resulting alignment was used to generate a neighbor-joining tree in the MEGA4 software package (default settings; 1,000 bootstrap replicates). Salinispora arenicola CNS-205 (GenBank accession number CP000850; GeneID 5705939) was used as the outgroup. The suffix bio indicates that bacteria were isolated after enrichment in marine broth for 2 weeks. Zone diameters in well diffusion assays with V. anguillarum strain 90-11-287 were designated with the following symbols: ≤6 mm, −; 6.1 to 10 mm, +; 10.1 to 15 mm, ++; and >15 mm, +++. The well itself has a diameter of 6 mm. No inhibition of supernatants occurred.
The alpP gene was detected in all strains that were identified by 16S rRNA gene similarity as being P. tunicata independently of their pigmentation (blackish or yellow).
Effect of spatial and seasonal variation on isolation of bioactive bacterial strains.
The prevalence of inhibitory culturable bacteria varied by season (temperature), with the lowest prevalence being seen in the colder months. Of the approximately 124,000 bacterial colonies tested for inhibitory activity against V. anguillarum, 35,000 were isolated in April, 27,000 in June, 19,000 in August, 24,000 in November, and 18,000 in February. The inhibitory colonies constituted 1.5 (April), 1.2 (June), 1.6 (August), 0.5 (November), and 0.03% (February) of the populations. This monthly difference between the prevalence of inhibitory bacteria was highly significant in the Pearson's chi-squared test (P < 0.001). Antibacterial Pseudoalteromonas and Vibrio strains were isolated year-round, whereas antibacterial Phaeobacter and Photobacterium were detected only in August and November (Table 3). The Pseudoalteromonas species isolated varied according to season/water temperature (Fig. 1). P. piscicida was frequently isolated in June and August, whereas P. tunicata was isolated in August and November (Fig. 1). Pseudoalteromonas antarctica and P. arctica were isolated mainly in November and February (Fig. 1).
Table 3.
Influence of season on marine bacteria from Danish coastal waters capable of inhibiting Vibrio anguillarum strain 90-11-287
| Identification based on 16S rRNA gene sequence | No. of Vibrio anguillarum inhibitory strains by time pointa |
|||||
|---|---|---|---|---|---|---|
| April (12.5 ± 2.3) | June (14.8 ± 2.0) | August (18.5 ± 1.3) | November (7.8 ± 1.0) | February (−0.1 ± 0.7) | Total | |
| Pseudoalteromonas | 3 | 8 | 21 | 11 | 11 | 54 |
| Vibrio | 1 | 5 | 12 | 5 | 1 | 24 |
| Phaeobacter | 0 | 0 | 6 | 4 | 0 | 10 |
| Photobacterium | 0 | 1 | 5 | 2 | 0 | 8 |
| Shewanella | 0 | 1 | 0 | 0 | 1 | 2 |
| Marinomonas | 0 | 0 | 0 | 5 | 0 | 5 |
| Other | 1 | 1 | 2 | 2 | 3 | 7 |
| Total | 5 | 16 | 46 | 30 | 16 | 110 |
Numbers in parentheses indicate temperatures (average ± standard deviation).
There were geographical patterns in the isolation of antibacterial bacteria strains, since some genera were detected only in certain areas (Table 4). All 10 Phaeobacter strains were isolated from samples from Jyllinge harbor, whereas all except one of the P. tunicata strains were isolated from Møn. Both sites are characterized by waters with low turbulence and NaCl concentrations of 1.4 and 1.0%, respectively (Table 4).
Table 4.
Influence of site of isolation on marine bacteria from Danish coastal waters capable of inhibiting Vibrio anguillarum strain 90-11-287
| Identification based on 16S rRNA gene sequence | No. of Vibrio anguillarum inhibitory strains from locationa: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | K | Total | |
| Pseudoalteromonas | 11 | 8 | 4 | 0 | 7 | 3 | 1 | 4 | 5 | 7 | 4 | 54 |
| Vibrio | 2 | 2 | 3 | 1 | 4 | 4 | 4 | 1 | 2 | 1 | 0 | 24 |
| Phaeobacter | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 |
| Photobacterium | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 2 | 1 | 8 |
| Shewanella | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 2 |
| Marinomonas | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 5 |
| Other | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 2 | 2 | 7 |
| Total | 25 | 12 | 9 | 1 | 12 | 9 | 5 | 9 | 8 | 13 | 7 | 110 |
Locations: A, Jyllinge harbor; B, Gilleje; C, Bellevue; D, Thisted; E, Agger; F, Hvide Sande; G, Blåvand; H, Middelfart; I, Korsør; J, Kalvehave at Møn; and K, Horsens.
Influence of isolation method on phylogenetic distribution of bioactive strains.
Bacterial strains producing bioactive compounds were isolated with three different isolation strategies, (i) plate count, (ii) selection for pigmented strains, and (iii) enrichment for 2 weeks in marine broth, all followed by replica plating against V. anguillarum. These different isolation strategies resulted in different bacterial genera (Table 2). Enrichment in marine broth favored the growth of bacteria belonging to the Vibrionaceae, since 18 of 24 inhibitory Vibrio strains and all eight Photobacterium strains were isolated after enrichment (Table 2). The opposite was seen for Phaeobacter and Marinomonas, where all strains were found after direct plate counting (Table 2). Bacteria belonging to the Pseudoalteromonas group were isolated by all procedures; however, the vast majority of nonpigmented strains in this genus were isolated after enrichment (Fig. 1). Ten of 15 P. piscicida strains were isolated after enrichment (Fig. 1), indicating a strong ability of this species to outcompete other marine bacteria under these culturing conditions.
Antiadhesive activity of antibacterial marine bacteria.
Twenty-two strains were chosen (based on the criteria described in Materials and Methods) for antiadhesive studies against the marine fouling bacteria Pseudoalteromonas S91 and against the settlement and germination of Ulva australis zoospores. From the Pseudoalteromonas group we included different species and chose three P. piscicida strains, since this species repeatedly caused large and clear inhibition zones when replica plated against V. anguillarum, and it represented different degrees of pigmentation (yellow and very light yellow). We also included three strains of P. tunicata (two yellow and a dark purple strain), since pigmentation and antifouling properties have previously been linked in P. tunicata strain D2 (14). Twenty-one of the 22 marine strains with inhibitory activity against V. anguillarum attached to and formed a biofilm on the stainless steel coupons after growth for 3 days in marine broth with cell densities above 6.2 log CFU/cm2. Vibrio strain B37bio formed a thin layer, with only 4.8 log CFU/cm2. The highest cell density in the 3-day-old biofilms was observed for all P. tunicata strains independently of color and for Phaeobacter strains (Table 5). When marine broth-coated steel plates were submerged in a suspension of the biofouler Pseudoalteromonas S91, it attached at a level of 5.9 log CFU/cm2 after 24 h.
Table 5.
Biofilm formation by marine antibacterial strains and their killing and antiadhesive effect against Pseudoalteromonas S91 and antisettlement effect against Ulva australis
| Species and strain | Pigmentation | Biofilm formation on stainless steela | Biofilm formation on plasticb | Reduced no. of adhered S91 cellsc | Reduced no. of S91 cells in suspensiond | Reduced no. of settled Ulva zoosporese |
|---|---|---|---|---|---|---|
| P. piscicida | ||||||
| A23a-4a | Yellow | ++ | + | +++ | +++ | +++ |
| A38q-4a | Yellow | ++ | + | +++ | +++ | +++ |
| B39bio | Yellow | ++ | +++ | +++ | +++ | ++ |
| P. tunicata | ||||||
| J49q-3a | Purple | +++ | ++ | +++ | + | +++ |
| J36q-4a | Yellow | +++ | + | +++ | ++ | +++ |
| J38a-5a | Yellow | +++ | + | +++ | + | +++ |
| P. ulvae | ||||||
| H34q-5a | Purple | +++ | +++ | +++ | + | ++ |
| A24a-4a | Purple | ++ | +++ | +++ | + | ++ |
| P. agarivorans | ||||||
| F55bio | Beige | ++ | ++ | + | + | +++ |
| P. aliena | ||||||
| F51a-1b | White | ++ | ++ | +++ | + | ++ |
| P. antarctica | ||||||
| B54q-3a | Black | ++ | + | + | + | +++ |
| E45q-4a | Brown | ++ | + | + | + | − |
| Vibrio sp. | ||||||
| A31bio | White | ++ | +++ | − | + | − |
| A32bio | White | ++ | ++ | − | − | + |
| B37bio | White | + | + | − | − | − |
| Photobacterium sp. | ||||||
| K29bio | Light yellow | ++ | + | − | + | − |
| J46bio | White | ++ | + | − | − | − |
| H38bio | White | ++ | + | + | + | +++ |
| I46bio | White | ++ | + | ++ | + | − |
| Phaeobacter sp. | ||||||
| A36a-5a | Brown | +++ | +++ | + | ++ | − |
| A49a-4a | Brown | ++ | +++ | + | ++ | − |
| A40a-4a | Brown | ++ | +++ | + | ++ | − |
| Controls | ||||||
| P. tunicata | ||||||
| D2 | Purple | +++ | ++ | +++ | + | +++ |
| Phaeobacter | ||||||
| 27-4 | Brown | +++ | +++ | + | ++ | − |
The amount of bacterial cells was quantified by plate count on marine agar after removal by ultrasonication. +, <6 log CFU/cm2; ++, between 6 and 7 log CFU/cm2; +++, >7 log CFU/cm2.
The amount of biofilm after crystal violet staining, measured at the OD550. +, OD550 of <0.5; ++, OD550 between 0.5 and 1.5; +++, OD550 of >1.5.
Log reduction (CFU/cm2) of adhered Pseudoalteromonas S91 after 4 h of incubation compared to that on marine broth-coated surfaces. −, <0.5 log reduction; +, between 0.5 and 1.5 log reduction; ++, between 1.5 and 3 log reduction; +++, >3 log reduction.
Killing of Pseudoalteromonas S91 after 4 h of incubation. −, <0.5 log reduction; +, between 0.5 and 1.5 log reduction; ++, between 1.5 and 3.0 log reduction; +++, >3 log reduction.
Settlement of Ulva australis spores on bacterial biofilm compared to that on marine broth-coated surfaces. −, >90% settlement; +, between 50 and 90% settlement; ++, between 10 and 50% settlement; +++, <10% settlement.
In particular, strains of the four Pseudoalteromonas species P. piscicida, P. tunicate, P. ulvae, and P. aliena had a strong antiadhesive effect. All strains of these species significantly reduced the numbers of attaching Pseudoalteromonas strain S91 cells to below or equal to the detection limit (1.6 log CFU/cm2) (Fig. 2a). Two different patterns were observed for the antiadhesive effect. The P. piscicida strains were bactericidal, since no S91 bacteria could be detected in the suspension surrounding the coupons after 1 (data not shown), 4 (Table 5), or 24 h (Fig. 2b). The P. tunicate, P. ulvae, and P. aliena strains only reduced the number of S91 in the suspension with 1 to 2 log CFU/ml independently of the pigmentation of the strains (Table 5). This level of growth inhibition equals the inhibition observed for the other strains.
Fig. 2.
Number of Pseudoalteromonas S91 organisms (a) attached to stainless steel (CFU/cm2) precoated with different marine bacteria and (b) in the surrounding suspension (CFU/ml) after 24 h. *, **, and *** indicate that the number of Pseudoalteromonas S91 cells adhered to stainless steel and in the suspension is significantly different from that of the control (marine broth) at 5, 1, and 0.1% levels, respectively.
The Phaeobacter strains, including the control Phaeobacter strain 27-4, reduced the number of adhered S91 cells by approximately 1 log unit, and it may be linked to a bactericidal effect, since numbers of S91 were reduced with approximately 2 log CFU/ml in the suspension surrounding the coupons.
The Photobacterium strain I46bio reduced the number of adhered S91 cells by almost 2 log units, which differed from the other strains in the Vibrionaceae group that did not have any or had only a slight reducing effect (Table 5). The bactericidal or bacteriostatic effect of the Vibrionaceae against S91 was less than 1.5 log CFU/ml.
Anti-Ulva spore settlement and germination activity of marine antibacterial bacteria.
Anti-Ulva spore activity was tested by exposing spores directly to monoculture biofilms. All 22 bacteria formed biofilms on the surface of the microtiter wells as measured by crystal violet staining (Table 5).
Twelve of the 22 strains were able to reduce the number of settling Ulva spores to below 10% of the level on control surfaces coated with marine broth (Table 5). All except one of these strains belonged to the Pseudoalteromonas group (Table 5). Of the 12 Pseudoalteromonas strains tested, only one (identified as P. antarctica) was unable to inhibit the settlement of Ulva spores, indicating a strong effect on zoospore attachment in this genus. The inhibitory effect against Ulva spores varied between strains belonging to the Vibrionaceae group (Table 5). One Vibrio strain reduced the number of settled spores slightly; one Photobacterium strain reduced the number of spores to 8%, whereas three strains enhanced the settlement of spores. None of three Phaeobacter strains inhibited Ulva spore settlement, and two of the strains did in fact mildly stimulate the settlement (data not shown).
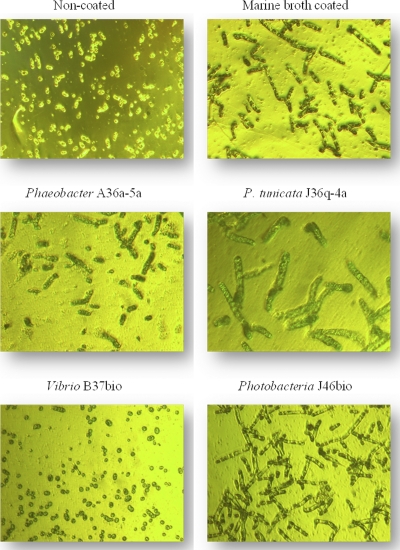
The inhibitory effect on the germination of Ulva spores by bacterial biofilms was tested after incubation for 7 days. There was no correlation between the number of attached Ulva spores and the number of these that actually germinated (Fig. 3). The presence of marine broth on the surface did not change the number of spores that settled (data not shown) but enhanced the number of spores germinating (Fig. 3). The germination of the settled spores depended on the bacterial biofilm. Spores that attached to a biofilm of Pseudoalteromonas elongated and had a wide diameter compared to that of attached spores on a Vibrio biofilm, which did not germinate within 7 days of incubation (Fig. 3).
Fig. 3.
Germination of Ulva on bacterial biofilms (40× magnification).
DISCUSSION
Bacteria from marine environments represent a relatively untapped resource of bioactive compounds (19, 31, 33). In this study, we isolated bacteria with antibacterial and antifouling activity (summarized as bioactive bacteria) within a relatively small geographic area (the coastline of Denmark). Bioactive bacteria predominantly belonged to the Vibrionaceae family, Pseudoalteromonas, and the Roseobacter clade, and the isolation strategy (Table 2) influenced the type of genera being isolated. The screening for bioactive bacteria during this 1-year study at 11 coastal sites of Denmark demonstrated that numbers and types of bioactive bacteria varied with both season and niche. This indicates that both temporal and spatial screening is important if different bioactive bacteria are to be isolated.
Our prime purpose was not the identification of antibacterial activity per se but the detection of additional antifouling activity expressed by these bacteria. Antibacterial activity could not, however, directly serve as a proxy for broader antifouling effect as measured against bacterial or algal spore attachment. Twenty-two of the marine bacteria that inhibited the growth of V. anguillarum were tested for their ability to prevent the adhesion of Pseudoalteromonas strain S91, and Pseudoalteromonas species were particularly effective in preventing bacterial adhesion. P. piscicida, P. tunicata, P. ulvae, and P. aliena prevented any adhesion of Pseudoalteromonas S91 by two distinct mechanisms. While P. piscicida had strong bactericidal activity, P. tunicata (including strain D2), P. ulvae, and P. aliena were not bactericidal against S91 but prevented the adhesion. In the agar-screening (replica) plating, P. piscicida strains consistently caused the largest clearing zones, which could indicate that they produce an antibacterial compound more potent and diffusible than those of the other Pseudoalteromonas species, thus explaining the rapid killing of S91 in suspension. We speculate that the non-piscicida Pseudoalteromonas strains produce a specific antiadhesive molecule(s). Such a compound(s) has been found in other Pseudoalteromonas strains. In strain 3J6, a proteinaceous substance reduced attachment by other bacteria (10). Further, the strains may produce exopolysaccharides (EPS) containing galactosamine, which reduce the adhesion of marine bacteria which use the autoinducer type-2 signaling system (30). Several Pseudoalteromonas species contain galactosamine in their EPS layer (18), and the target strain may use the type 2 signaling system, as previously shown for other Pseudoalteromonas strains (5). The antifouling Pseudoalteromonas also may produce quorum-sensing inhibitors or biosurfactants (10). Further studies clearly are required to elucidate the mechanism behind the nonbactericidal antifouling effect.
Pseudoalteromonas strains caused more pronounced inhibitory effects in the algal spore settlement assay than strains belonging to the Vibrionaceae and Phaeobacter. This parallels a study (34) in which spores of the green algae Enteromorpha were more efficiently repelled by Pseudoalteromonas strains than Vibrio spp. We did not observe any correlation between bactericidal effect and the prevention of Ulva zoospore settlement.
The presence of pigmentation in Pseudoalteromonas species has been linked to their antifouling capacity, as expressed by their ability to reduce algal spore and larval settlement (13, 14, 22, 26). Nonpigmented mutants of P. tunicata D2 (14) or of the red Pseudoalteromonas strain sf57 (26) lost the antisettlement activity seen in the purple/red pigmented wild types, and Holmström et al. (22) also suggested that pigmentation and the antisettlement effect are correlated, as they isolated two white Pseudoalteromonas strains without anti-Ulva activity and eight pigmented strains with activity. However, our results indicate that mechanisms other than pigmentation can be responsible for antisettlement activity in Pseudoalteromonas, since our two white/beige strains (P. aliena and P. agarivorans) reduced the settlement of Ulva spores, whereas the brown P. antarctica did not.
The antibacterial protein AlpP has been suggested to be the mediator for the anti-adhesive activity and bactericidal effect, particularly in P. tunicata D2 (28, 32, 38, 39), and may be a useful proxy in screening studies. All four P. tunicata strains harbored the gene coding for AlpP. However, the alpP gene was not detected in other Pseudoalteromonas strains with antifouling ability (42), indicating that either another compound(s) is responsible for the observed antifouling effect or that the alpP primers are too specific, as they are constructed from only one known P. tunicata sequence (42). One could speculate that a similar mechanism is used by all three bacteria, since they are genetically similar at least on the 16S rRNA gene sequence level (Fig. 1) and since we could not link the antifouling activity to pigmentation as previously suggested (14). P. aliena is nonpigmented, whereas two P. tunicata strains were bright yellow and one P. tunicata strain and the P. ulvae strains were purple.
Another relevant feature of antifouling activity is the ability of a bacterial biofilm to prevent the germination and growth of the zoospores already attached. A study using supernatants from a Pseudomonas sp. and different Bacillus spp. suggested that the anti-settlement effect is not necessarily related to the prevention of germination and growth of Ulva zoospores. A Pseudomonas strain was able to prevent both settlement and germination, whereas Bacillus licheniformis only reduced settlement (7). The germination of already settled zoospores on bacterial biofilms was, in the present study, related to the bacterial genus, as all Pseudoalteromonas and Phaeobacter strains resulted in large germinated spores, whereas the zoospores attached to Vibrio biofilm did not germinate at all independently of the number of spores settled (Fig. 3). Part of the difference in the germination may be linked to different amounts of nutrient left for the zoospores, since we observed a lack of germination on noncoated surfaces, whereas marine broth-coated surfaces resulted in germination to slime algae. The germination on the marine broth-coated surface also may be a result of a bacterial biofilm being developed over time, since zoospores are not sterilized prior to use. It may, however, also be due to bioactive compounds being produced by the marine bacteria, and further studies are required to verify this hypothesis.
The Phaeobacter strains inhibited the growth of both V. anguillarum and Pseudoalteromonas S91, but interestingly they facilitated spore settlement to their bacterial biofilms. We tested the Phaeobacter strains in a monitor assay for acylated homoserine lactones (AHLs) (Agrobacterium tumefaciens) and all were positive (data not shown), indicating that AHL production could be the cue in promoting algal settlement by these strains as well. Rao et al. (38) similarly observed that high cell densities of Phaeobacter sp. strain 2.10 attracted Ulva sp. spores (38), and the production of the density-sensing signal molecules of acyl homoserine lactones (AHLs) has been suggested previously to act as a settlement cue for Ulva spores in general (44).
We observed a significant difference in the number of bioactive bacteria isolated in different months, with the largest numbers being observed in August and November. This may be an artifact of the isolation method used (incubation at 20°C), but we continuously observed large variations within the Roseobacter clade bacteria and the Pseudoalteromonas genus during the year in particular, indicating a season-specific marine microbiota. This was most profound for bacteria belonging to the Roseobacter clade, which all were isolated from one location and only when the water was visually green and turbid, indicating phytoplankton blooms. Roseobacter clade bacteria often are associated with algal blooms, where they can constitute more than 20% of the prokaryotic DNA (17); however, we did not isolate Phaeobacter from the alga-containing water samples but did isolate them from heavily fouled surfaces in Jyllinge harbor, indicating that they used algal components but were specifically associated with the fouling macroalgal community. Cooccurrence between such fouling and bacteria within the Roseobacter clade has been described and suggested to be related to the need for cell-surface interaction for some lineage members in this clade (43).
Bioactive Pseudoalteromonas species were isolated year-round; however, there was a seasonal pattern in the Pseudoalteromonas species isolation, with P. arctica, P. antarctica, and P. prydzensis occurring predominantly in the colder months. It is well known that several pigmented Pseudoalteromonas species are antibacterial (6), hence it is not surprising that these were isolated in our sampling because we deliberately included pigmented colonies when testing for antibacterial activity against V. anguillarum. In contrast to a recent global sampling for antibacterial bacteria, we did not isolate P. luteoviolacea and P. rubra (19, 47), which is likely explained by their preference for tropical waters (47). This indicates that temperature is an important factor influencing the species composition of the antibacterial Pseudoalteromonas population. However, local adaptation to specific econiches also was found. P. tunicata was isolated from only 2 of 11 locations, and both were heavily fouled with algae and sea grass. The P. tunicata strains all were surface associated, indicating a preference of this bacterium to colonize surfaces, which corresponds to results from previous isolation sites (15, 23) and to genome analysis revealing properties of a surface-associated lifestyle (46).
The battle against fouling is ongoing, and the search for antifouling compounds or principles that can reduce or eliminate the attachment of marine organisms is intense (2, 37). While several studies have demonstrated that specific marine bacteria produce compounds with antifouling activity against a range of marine micro- and macroorganisms (15, 48), our study is the first to elucidate the antifouling potential of a range of antibacterial bacteria from different niches and seasons. We demonstrated that several groups of antibacterial bacteria could be isolated even within a relatively small geographic region, but that both season and local parameters influenced the genus or species of bacteria isolated. Our study further indicated that antifouling activity against both a marine fouling bacteria and against Ulva zoospores were predominantly observed within the antibacterial Pseudoalteromonas group. Interestingly, two different mechanisms caused the antibacterial fouling effect of Pseudoalteromonas. One was likely caused by the bactericidal activity of P. piscicida, whereas the other was not dependent on bactericidal activity. This observation is relevant if marine bacteria or their bioactive compounds are to be used as antifouling agents, since the antifouling effect caused by bactericidal activity may result in the build-up of microbial resistance. Hence, elucidating the antifouling principle of P. tunicata, P. ulvae, and P. aliena may result in the identification of novel antifouling compounds which can prevent fouling without killing marine microorganisms.
Supplementary Material
ACKNOWLEDGMENTS
This work was financed by the Danish Directorate for Food, Fisheries, and Agribusiness (3304-FVFP-08-M-15-01). We acknowledge financial support from the Otto Mønsteds Fond, Oticon Fonden, and Christian og Ottilia Brorsons Rejselegat for Yngre Videnskabsmænd og-Kvinder to N.B. for her visit to the University of New South Wales, Australia.
Strain S91 was kindly donated by Amanda Goodman and Marina Delpin, Flinders University, Adelaide, Australia.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 14 October 2011.
REFERENCES
- 1. Alzieu C. 2000. Impact of tributyltin on marine invertebrates. Ecotoxicology 9:71–76 [Google Scholar]
- 2. Banerjee I., Pangule R. C., Kane R. S. 2011. Antifouling coatings: recent developments in the design of surfaces that prevent fouling by proteins, bacteria, and marine organisms. Adv. Mater. 23:690–718 [DOI] [PubMed] [Google Scholar]
- 3. Bernbom N., et al. 2006. Bacterial adhesion to stainless steel is reduced by aqueous fish extract coatings. Biofilms 3:25–36 [Google Scholar]
- 4. Bernbom N., et al. 2009. Adhesion of food-borne bacteria to stainless steel is reduced by food conditioning films. J. Appl. Microbiol. 106:1268–1279 [DOI] [PubMed] [Google Scholar]
- 5. Bodor A., Elxnat B., Thiel V., Schulz S., Wagner-Döbler I. 2008. Potential for luxS related signalling in marine bacteria and production of autoinducer-2 in the genus Shewanella. BMC Microbiol. 8:13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bowman J. P. 2007. Bioactive compounds synthetic capacity and ecological significance of marine bacterial genus Pseudoalteromonas. Mar. Drugs 5:220–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burgess J. G., et al. 2003. The development of a marine natural product-based antifouling paint. Biofouling 19(Suppl.):197–205 [DOI] [PubMed] [Google Scholar]
- 8. De Nys R., Givskov M., Kumar N., Kjelleberg S., Steinberg P. D. 2006. Furanones. Prog. Mol. Subcell. Biol. 42:55–86 [DOI] [PubMed] [Google Scholar]
- 9. Devulder G., Perriere G., Baty F., Flandrois J. P. 2003. BIBI, a bioinformatics bacterial identification tool. J. Clin. Microbiol. 41:1785–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dheilly A., et al. 2010. Antibiofilm activity of the marine bacterium Pseudoalteromonas sp. strain 3J6. Appl. Environ. Microbiol. 76:3452–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dobretsov S. 2009. Inhibition and induction of marine biofouling by biofilms, p. 293–313 In Flemming H.-C., Murthy P. S., Venkatesan R., Cooksey K. (ed.), Marine and industrial biofouling. Springer-Verlag, Berlin, Germany [Google Scholar]
- 12. Dobretsov S., Qian P. Y. 2004. The role of epibotic bacteria from the surface of the soft coral Dendronephthya sp. in the inhibition of larval settlement. J. Exp. Mar. Biol. Ecol. 299:35–50 [Google Scholar]
- 13. Egan S., James S., Holmström C., Kjelleberg S. 2001. Inhibition of algal spore germination by the marine bacterium Pseudoalteromonas tunicata. FEMS Microbiol. Ecol. 35:67–73 [DOI] [PubMed] [Google Scholar]
- 14. Egan S., James S., Holmström C., Kjelleberg S. 2002. Correlation between pigmentation and antifouling compounds produced by Pseudoalteromonas tunicata. Environ. Microbiol. 4:433–442 [DOI] [PubMed] [Google Scholar]
- 15. Egan S., Thomas T., Holmström C., Kjelleberg S. 2000. Phylogenetic relationship and antifouling activity of bacterial epiphytes from the marine alga Ulva lactuca. Environ. Microbiol. 2:343–347 [DOI] [PubMed] [Google Scholar]
- 16. Evans S. M., Kerrigan E., Palmer N. 2000. Causes of imposex in the dogwhelk Nucella lapillus (L.) and its use as a biological indicator of tributyltin contamination. Mar. Pollut. Bull. 40:212–219 [Google Scholar]
- 17. González J. M., et al. 2000. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 66:4237–4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gorshkova N. M., Gorshkova R. P., Ivanova E. P., Nazarenko E. L., Zubkov V. A. 2001. Diversity in the monosaccharide composition of antigenic polysaccharides from proteobacteria Pseudoalteromonas and Marinomonas genera. Mikrobiologia 70:651–655 [PubMed] [Google Scholar]
- 19. Gram L., Melchiorsen J., Bruhn J. B. 2010. Antibacterial activity of marine culturable bacteria collected from a global sampling of ocean surface waters and surface swabs of marine organisms. Mar. Biotechnol. 12:439–451 [DOI] [PubMed] [Google Scholar]
- 20. Harder T. 2009. Marine epibiosis: concepts, ecological, consequences and host defence, p. 219–231 In Flemming H.-C., Murthy P. S., Venkatesan R., Cooksey K. (ed.), Marine and industrial biofouling. Springer-Verlag, Berlin, Germany [Google Scholar]
- 21. Hjelm M., et al. 2004. Selection and identification of autochthonous potential probiotic bacteria from turbot larvae (Scophthalmus maximus) rearing units. Syst. Appl. Microbiol. 27:360–371 [DOI] [PubMed] [Google Scholar]
- 22. Holmström C., Egan S., Franks A., McCloy S., Kjelleberg 2002. Antifouling activities expressed by marine surface associated Pseudoalteromonas species. FEMS Microbiol. Ecol. 41:47–58 [DOI] [PubMed] [Google Scholar]
- 23. Holmström C., James S., Neilan B. A., White D. C., Kjelleberg S. 1998. Pseudoalteromonas tunicata sp. nov., a bacterium that produces antifouling agents. Int. J. Syst. Bacteriol. 48:1205–1212 [DOI] [PubMed] [Google Scholar]
- 24. Holmström C., Kjelleberg S. 1999. Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents. FEMS Microbiol. Ecol. 30:285–293 [DOI] [PubMed] [Google Scholar]
- 25. Holmström C., Rittschof D., Kjelleberg S. 1992. Inhibition of settlement by larvae of Balanus amphitrite and Ciona intestinalis by a surface-colonizing marine bacterium. Appl. Environ. Microbiol. 58:2111–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang Y. L., Li M., Yu Z., Qian P. Y. 2011. Correlation between pigmentation and larval settlement deterrence by Pseudoalteromonas sp. sf57. Biofouling 27:287–293 [DOI] [PubMed] [Google Scholar]
- 27. International Maritime Organisation 2007. Harmful ships' paints systems to be outlawed as international convention meets entry into force criteria. IMO News 4:6 [Google Scholar]
- 28. James S. G., Holmström C., Kjelleberg S. 1996. Purification and characterization of a novel antibacterial protein from the marine bacterium D2. Appl. Environ. Microbiol. 62:2783–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jensen A., Larsen M. H., Ingmer H., Vogel B. F., Gram L. 2007. Sodium chloride enhances adherence and aggregation and strain variation influences invasiveness of Listeria monocytogenes strains. J. Food Prot. 70:592–599 [DOI] [PubMed] [Google Scholar]
- 30. Kim M., et al. 2011. The antifouling potentiality of galactosamine characterized from Vibrio vulnificus exopolysaccharide. Biofouling 27:851–857 [DOI] [PubMed] [Google Scholar]
- 31. Long R. A., Azam F. 2001. Antagonistic interactions among marine pelagic bacteria. Appl. Environ. Microbiol. 67:4975–4983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mai-Prochnow A., et al. 2004. Biofilm development and cell death in the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 70:3232–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nair S., Simidu U. 1987. Distribution and significance of heterotrophic marine bacteria with antibacterial activity. Appl. Environ. Microbiol. 53:2957–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Patel P., Callow M. E., Joint I., Callow J. A. 2003. Specificity in the settlement modifying response of bacterial biofilms towards zoospores of the marine alga Enteromorpha. Environ. Microbiol. 5:338–349 [DOI] [PubMed] [Google Scholar]
- 35. Petersen S., Gustavson K. 2000. Direct toxic effects of TBT on natural enclosed phytoplankton at ambient TBT concentrations of coastal waters. Ecotoxicology 9:273–285 [Google Scholar]
- 36. Qian P. Y., Lau S. C., Dahms H. U., Dobretsov S., Harder T. 2007. Marine biofilms as mediators of colonization by marine macroorganisms: implications for antifouling and aquaculture. Mar. Biotechnol. 9:399–410 [DOI] [PubMed] [Google Scholar]
- 37. Qian P. Y., Xu Y., Fusetani N. 2010. Natural products as antifouling compounds: recent progress and future perspectives. Biofouling 26:223–234 [DOI] [PubMed] [Google Scholar]
- 38. Rao D., et al. 2007. Low densities of epiphytic bacteria from the marine alga Ulva australis inhibit settlement of fouling organisms. Appl. Environ. Microbiol. 73:7844–7852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rao D., Webb J. S., Kjelleberg S. 2005. Competitive interactions in mixed-species biofilms containing the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 71:1729–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schultz M. P. 2007. Effects of coating roughness and biofouling on ship resistance and powering. Biofouling 23:331–341 [DOI] [PubMed] [Google Scholar]
- 41. Skov M. N., Pedersen K., Larsen J. L. 1995. Comparison of pulsed-field gel electrophoresis, ribotyping, and plasmid profiling for typing of Vibrio anguillarum serovar O1. Appl. Environ. Microbiol. 61:1540–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Skovhus T. L., Holmström C., Kjelleberg S., Dahllof I. 2007. Molecular investigation of the distribution, abundance and diversity of the genus Pseudoalteromonas in marine samples. FEMS Microbiol. Ecol. 61:348–361 [DOI] [PubMed] [Google Scholar]
- 43. Slightom R. N., Buchan A. 2009. Surface colonization by marine roseobacters: integrating genotype and phenotype. Appl. Environ. Microbiol. 75:6027–6037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tait K., et al. 2005. Disruption of quorum sensing in seawater abolishes attraction of zoospores of the green alga Ulva to bacterial biofilms. Environ. Microbiol. 7:229–240 [DOI] [PubMed] [Google Scholar]
- 45. Techkarnjanaruk S., Pongpattanakitshote S., Goodman A. E. 1997. Use of a promoterless lacZ gene insertion to investigate chitinase gene expression in the marine bacterium Pseudoalteromonas sp. strain S9. Appl. Environ. Microbiol. 63:2989–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thomas T., et al. 2008. Analysis of the Pseudoalteromonas tunicata genome reveals properties of a surface-associated life style in the marine environment. PLoS One 3:e3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vynne N. G., Månsson M., Nielsen K. F., Gram L. 2011. Bioactivity, chemical profiling, and 16S rRNA-based phylogeny of Pseudoalteromonas strains collected on a global research cruise. Mar. Biotechnol. [Epub ahead of print.] doi:10.1007/s10126-011-9369-4 [DOI] [PubMed] [Google Scholar]
- 48. Xu Y., Li H., Li X., Xiao X., Qian P. Y. 2009. Inhibitory effects of a branched-chain fatty acid on larval settlement of the polychaete Hydroides elegans. Mar. Biotechnol. 11:495–504 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.