Abstract
Lactic acid bacteria (LAB) (n = 152) in African pearl millet slurries and in the metagenomes of amylaceous fermented foods were investigated by screening 33 genes involved in probiotic and nutritional functions. All isolates belonged to six species of the genera Pediococcus and Lactobacillus, and Lactobacillus fermentum was the dominant species. We screened the isolates for the abilities to survive passage through the gastrointestinal tract and to synthesize folate and riboflavin. The isolates were also tested in vitro for their abilities to survive exposure to bile salts and to survive at pH 2. Because the ability to hydrolyze starch confers an ecological advantage on LAB that grow in starchy matrixes as well as improving the nutritional properties of the gruels, we screened for genes involved in starch metabolism. The results showed that genes with the potential ability to survive passage through the gastrointestinal tract were widely distributed among isolates and metagenomes, whereas in vitro tests showed that only a limited set of isolates, mainly those belonging to L. fermentum, could tolerate a low pH. In contrast, the wide distribution of genes associated with bile salt tolerance, in particular bsh, is consistent with the high frequency of tolerance to bile salts observed. Genetic screening revealed a potential for folate and riboflavin synthesis in both isolates and metagenomes, as well as high variability among genes related to starch metabolism. Genetic screening of isolates and metagenomes from fermented foods is thus a promising approach for assessing the functional potential of food microbiotas.
INTRODUCTION
In Africa, many amylaceous fermented foods made from cassava and cereals are used as gruels for the complementary feeding of children under five during weaning. In Burkina Faso and Ghana, ben-saalga and koko gruels prepared from fermented pearl millet (Pennisetum glaucum) slurries are frequently consumed by young children (36, 62). The microbiota of these fermented foods is dominated by lactic acid bacteria (LAB), which contribute to their nutritional and sanitary qualities (9, 47). We are currently studying pearl millet-based fermented slurries as a model (“ben-saalga model”) for investigating the microbiota of this type of cereal-based food with the aim of developing strategies to improve nutritional quality (26, 57, 62). Many studies have focused on the phenotypic diversity of the LAB that compose the microbiota of tropical fermented foods. However, their functional diversity in isolates and in metagenomes remains to be described.
Since niche-specific adaptation has played a central role in the evolution of LAB (41), the building of collections of bacteria from traditional fermented plant foods in tropical countries may enable the identification of a specific gene set that differs from those of LAB isolated from dairy or bakery products. LAB from microbiotas of plant origin that are found in traditional African foods may have probiotic characteristics (3, 28, 36). These bacteria first have to be selected for their ability to survive passage through the gastrointestinal tract. Probiotic functions may also be associated with other functions that are of interest for nutrition. This is of particular interest for at-risk populations such as pregnant women and young children in developing countries. For instance, the amylase activity of some LAB helps increase the energy content of gruels for the complementary feeding of young children through partial hydrolysis of starch in the food matrix (46, 57) but also helps sustain the growth of the microbiota of starchy foods (15, 62). Other functions—for example, folate and riboflavin synthesis—may improve the quality of the food matrix and may be beneficial for the host. Folate deficiency can lead to neural tube defects, early spontaneous abortion, and megaloblastic anemia, while riboflavin deficiencies can result in growth failure, inflammation of the skin, or vision deterioration (40, 52, 53). LAB capable of producing B vitamins could be used for fortification of cereal-based foods (27) and as probiotics (27, 54). In this way, bacteria that combine different functional characteristics could be useful for developing improved or new foods made from local raw materials that target specific nutritional needs and health issues. However, phenotypic analysis of probiotic and other functional traits is time-consuming, especially when a large number of bacteria have to be tested at the same time, and takes even longer when different functions (survival in the gastrointestinal tract, synthesis of compounds of interest, degradation of different factors, etc.) have to be taken into account. On the other hand, advances in our knowledge of the genetic diversity of LAB and the increasing number of sequenced LAB genomes mean that the functional properties of LAB strains can be studied more easily at the molecular level (31, 35, 63). From an ecological point of view, screening of the genomes of isolates in the same food niche and in food metagenomes will enable mapping of the distribution of genes related to specific functions and hence evaluation of the quality of the fermented food that may be linked to its microbiota.
In the present study, a collection of 152 LAB from ben-saalga and the metagenomes of starchy fermented foods were investigated by screening for genes involved in probiotic and nutritional functions. Among the many functional traits described in the literature, we selected genes for which at least one functional analysis had already been performed. The aim of screening was to detect genes involved in survival in the gastrointestinal tract, in starch metabolism, and in folate and riboflavin synthesis. In addition, we investigated the ability of selected isolates to survive in vitro exposure to low pHs and bile salts.
MATERIALS AND METHODS
Fermented samples, bacteria, and culture conditions.
Bacterial isolates (n = 178) from fermented pearl millet slurries sampled in 12 different traditional production units in Ouagadougou, Burkina Faso, were randomly isolated on De Man, Rogosa, and Sharpe (MRS) agar plates. Preliminary characterization showed that 152 of these isolates were Gram positive, catalase negative, and non-spore forming and that they produced lactic acid or an equimolar ratio of lactic acid and ethanol from glucose; these were assigned to the LAB group. The positive controls used for gene screening were Pediococcus pentosaceus ATCC 25745, Leuconostoc mesenteroides ATCC 8293, Lactobacillus plantarum A6 (LMG 18053), L. plantarum WCFS1, L. plantarum ATCC 14917, Lactobacillus fermentum IFO 3956, L. fermentum ATCC 14931, L. fermentum Ogi E1 (CNCM I-2028), L. fermentum MW2 (CNCM I-2029), Lactobacillus johnsonii NCC533, Lactobacillus reuteri ATCC 23272, and Lactobacillus brevis DSM1268. All LAB were routinely cultured at 30°C in MRS broth (Difco, Le Pont de Claix, France).
For metagenome analysis, samples of fermented pearl millet slurries were collected in five small production units in Ouagadougou, Burkina Faso, after 16 h of fermentation. In addition, a sample of wheat sourdough fermented for 20 h from a traditional bakery in Montpellier, France, and a sample of cassava fermented for 24 h (attiéké) from a local market at Abidjan, Ivory Coast, were screened. Samples were stored at −20°C until DNA extraction.
DNA extraction.
Because the food samples were very sticky, 3 g was diluted three times in 0.9% (wt/vol) NaCl, centrifuged twice for 10 min at 1,000 × g and 4°C to eliminate the starch, and then centrifuged for 10 min at 10,000 × g and 4°C to pellet the bacteria. The final pellet was then washed one more time in 0.9% (wt/vol) NaCl. DNA was extracted from the pellets of food samples and from the pellets of overnight pure cultures using the Wizard genomic DNA purification kit (Promega, Charbonnières, France) with an additional lysis step using an amalgamator with zirconium beads (VWR, Fontenay-sous-Bois, France). First, the cells were lysed with 0.1-mm-diameter zirconium beads for 30 s, followed by 1 h of incubation at 37°C with lysozyme (40 kU; Eurobio) and mutanolysin (10 U; Promega). According to the manufacturer's instructions, cell lysis was completed with the Nuclei lysis solution (Promega). RNA was removed with the RNase solution (Promega) and proteins with the Protein Precipitation solution (Promega). DNA was precipitated with isopropanol and washed with 70% ethanol. DNA quality was checked by separation on an agarose gel, followed by ethidium bromide staining.
Molecular identification of bacterial isolates.
For 16S rRNA gene sequencing, primers W001 (6) and 23S1 (GenBank accession no. J01695) (10, 25) were used to amplify the 16S rRNA gene, including the intergenic region located between 16S rRNA and 23S rRNA. PCR products were sequenced by Eurofins MWG GmbH (Ebersberg, Germany). Each sequence was identified by comparing it with sequences from the Ribosomal Database Project II (RDP2) (http://rdp.cme.msu.edu). Because of the high identity between the 16S rRNA gene sequences of L. plantarum, Lactobacillus paraplantarum, and Lactobacillus pentosus, we used the previously described multiplex PCR assay to precisely identify isolates belonging to the L. plantarum group (61).
Primer design.
Genetic screening was based on sets of genes involved in bile salt tolerance, pH survival, biogenic amine synthesis (which is involved in pH tolerance by releasing ammonium or by consuming protons in the intracellular media of the bacteria), riboflavin synthesis, folate synthesis, and starch metabolism. These genes are listed in Table 1. To detect their presence, the DNA extracted from the isolates or from the metagenomes was screened by PCR amplification. The primers for each PCR were designed by comparing sequences from functional analysis with a genomic and protein database (NCBI) using the BLASTn, BLASTp, and BLASTx algorithms (as of September 2010). This analysis was limited to the species identified during 16S rRNA gene sequencing. Once selected, nucleotide sequences were aligned using the ClustalW program (60) to generate a unique consensus sequence (19) for the design of the primers with Primer3 software (55). When no data on the gene sequences of the bacterial species in our collection were available, we used primers designed for other LAB species reported in the literature. All primers were synthesized by Eurogentec (Angers, France).
Table 1.
List of primers used to screen the bacterial collection and food metagenomes
| General function | Gene | Predicted function | Primer |
Melting temp (°C) | Expected amplicon size (bp) | Species used for primer design | Relevant reference(s) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Orientation | Sequence (5′to 3′) | Reference | ||||||||
| Survival | ||||||||||
| pH | hdc | Histidine decarboxylase | F | AGATGGTATTGTTTCTTATG | 12 | 52.0 | 367 | Lactobacillus sp. 30A, | 12 | |
| R | AGACCATACACCATAACCTT | Lactobacillus buchneri, Clostridium perfringens | ||||||||
| tdc | Tyrosine decarboxylase | F | CCACTGCTGCATCTGTTTG | 15, 16 | 50.0 | 370 | L. brevis | 12 | ||
| R | CCRTARTCNGGNATAGCRAARTCNGTRTG | |||||||||
| odc | Ornithine decarboxylase | F | TMTWCCAACHGATCGWAATGC | This study | 58.0 | 245 | L. salivarius, L. helveticus, | 2, 12 | ||
| R | CRCCCCAWGCACARTCRAA | L. johnsonii, L. gasseri, L. acidophilus, Lactobacillus delbrueckii, Lactobacillus casei | ||||||||
| aguA | Agmatine deiminase | F | GAACGACTAGCAGCTAGTTAT | 17 | 60.0 | 542 | L. brevis, P. pentosaceus | 39 | ||
| R | CCAATAGCCGATACTACCTTG | |||||||||
| groEL | Heat shock protein 60 | F | TTCCATGGCKTCAGCRATCA | This study | 58.0 | 168 | L. salivarius, Leuconostoc | 37, 38 | ||
| R | GCTAAYCCWGTTGGCATTCG | mesenteroides, L. casei, L. delbrueckii, P. pentosaceus, P. acidilactici | ||||||||
| LBA1272 | Cyclopropane FA synthase | F | GGCTTACCAATGGCCACCTT | This study | 57.5 | 210 | L. fermentum | 32, 49 | ||
| R | GATCAAAAAGCCGGTCACGA | |||||||||
| F | GGCCGGTGTTCCACTAGTCC | This study | 58 | 203 | L. plantarum | |||||
| R | ACGTTGGGTCGATTTGACGA | |||||||||
| F | AAGGACCCGGATTTTGACGA | This study | 58 | 151 | P. pentosaceus | |||||
| R | ACGTGGTTTGACCCAGTGCT | |||||||||
| dltD | d-Alanine transfer protein | F | TTCGCCTGTTCAAGCCACAT | This study | 58 | 283 | L. fermentum | 49 | ||
| R | ACGTGCCCTTCTTTGGTTCC | |||||||||
| La995 | Amino acid permease | F | AACGAAGGTCCCGACAAAGG | This study | 57.5 | 246 | L. fermentum | 2 | ||
| R | ACGACCTTCGGGCTGGTTAC | |||||||||
| La57 | Amino acid antiporter | F | GGTCGGGGGATCTGAAAAGA | This study | 58.0 | 274 | L. plantarum | 2 | ||
| R | GATTTGGGCAAGCACATTGG | |||||||||
| pH and bile salt | gtf | Glucan synthase | F | ACACGCAGGGCGTTATTTTG | This study | 58.0 | 374 | L. diolivorans, P. parvulus, | 58 | |
| R | GCCACCTTCAACGCTTCGTA | P. damnosus, L. suebicus | ||||||||
| clpL | ATPase | F | GCTGCCTTYAAAACATCATCTGG | This study | 56.0 | 158 | L. plantarum, L. salivarius, | 66 | ||
| R | AATACAATTTTGAARAACGCAGCTT | L. fermentum, P. pentosaceus, P. acidilactici | ||||||||
| lr1516 | Putative esterase | F | TRACCACTYTCWCCATTCAACAA | This study | 56.5 | 143 | L. plantarum, P. pentosaceus | 66 | ||
| R | CCACTAGCRATGACYAATACKGGTT | |||||||||
| Bile salt | bsh | Conjugated bile salt acid hydrolase | F | ATTGAAGGCGGAACSGGMTA | This study | 58.0 | 155 | P. pentosaceus, P. acidilactici | 14, 17, 34, 42 | |
| R | ATWACCGGWCGGAAAGCTG | |||||||||
| F | ATTCCWTGGWTWYTGGGACA | This study | 58.0 | 384 | L. plantarum, L. salivarius | |||||
| R | AAAAGCRGCTCTNACAAAWCKAGA | |||||||||
| F | GGTTGGTCGGCCAGTTCTTT | This study | 58.0 | 205 | L. fermentum | |||||
| R | CCAACATGCCCAAGTTCGAC | |||||||||
| lr0085 | Hypothetical protein | F | RCTTTGACCGRTGGGGCTRT | This study | 57.5 | 150 | L. reuteri, Lactobacillus | 67 | ||
| R | NNNATGGCCGCATGGAAA | vaginalis, Lactobacillus antri | ||||||||
| lr1584 | Major facilitator superfamily permease | F | TAYGCCRTTCGGWTGTTTGG | This study | 55.5 | 151 | L. plantarum, L. fermentum | 67 | ||
| R | TCAWRATGGCRGTCCCAATG | |||||||||
| LBA0552 | Major facilitator superfamily permease | F | GTGATTGCCCTAGCCCTGGT | This study | 58.0 | 180 | L. fermentum | 51 | ||
| R | GATCCCGATCACGATGCAAG | |||||||||
| LBA1429 | Major facilitator superfamily permease | F | AATTTCAGGATGCCCCGGTA | This study | 58.0 | 196 | P. pentosaceus | 50 | ||
| R | CCAAGCTCCCAACAATGCAC | |||||||||
| F | CTACAGCCCGCTGCTAACCA | This study | 58.0 | 174 | L. plantarum | |||||
| R | AGTTTGCATGGCAACCTGGA | |||||||||
| LBA1446 | Multidrug resistance protein | F | GCTGGAGCCACACCGATAAC | This study | 58.0 | 275 | L. plantarum, L. salivarius, P. acidilactici | 51 | ||
| R | CAACGGGATTATGATTCCCATTAGT | |||||||||
| LBA1679 | ABC transporter | F | ATGACAACGTCGTCGGGAGA | This study | 58.0 | 267 | L. fermentum | 51 | ||
| R | GCTCCTCGTTGTTGGGACCT | |||||||||
| F | GGBATVTACGGTGGVCTDGAA | This study | 58.0 | 101 | L. plantarum, L. salivarius, P. pentosaceus, P. acidilactici | |||||
| R | NGYTCCAGAAAGAATCTTGAACATYA | |||||||||
| LBA1432 | Hypothetical protein | F | TCCCATTCATCAYATGGAACAA | This study | 56.5 | 352 | P. pentosaceus, P. acidilactici | 50 | ||
| R | CTGGCCCACATATCCATWCC | |||||||||
| apf | Aggregation-promoting factors | F | YAGCAACACGTTCTTGGTTAGCA | This study | 53.0 | 112 | L. plantarum, L. salivarius, | 22 | ||
| R | GAATCTGGTGGTTCATAYWCAGC | L. fermentum, P. pentosaceus, P. acidilactici | ||||||||
| Synthesis of B vitamins | ||||||||||
| Folate synthesis | folP | Dihydropteroate synthase/dihydropteroate pyrophosphorylase | F | CCASGRCSGCTTGCATGAC | This study | 59.5 | 261 | L. plantarum, L. fermentum | 13 | |
| R | TKACGCCGGACTCCTTTTWY | |||||||||
| folK | 2-Amino-4-hydroxy-6- | F | CCATTTCCAGGTGGGGAATC | This study | 59.5 | 214 | L. plantarum, L. | 13, 59 | ||
| hydroxymethyldihydropteridine diphosphokinase | R | GGGGTGGTCCAAGCAAACTT | fermentum | |||||||
| Riboflavin synthesis | ribH | 6,7-Dimethy-8-ribityllumazine synthase | F | AGGGCGAAACCGACCACTAC | This study | 60.0 | 179 | L. fermentum | 7 | |
| R | CGATTGGGCAGTCATCGAAC | |||||||||
| ribB | Riboflavin synthase subunit alpha | F | AGTAAACGGAACGGGCAAGC | This study | 60.0 | 235 | L. fermentum | 7 | ||
| R | GTTGACCAGGGCACCAACTG | |||||||||
| ribA | 3,4-Dihydroxy-2-butanone 4-phosphate synthase/ | F | TTTACGGGCGATGTTTTAGG | This study | 60.0 | 121 | L. fermentum, P. | 7 | ||
| GTP cyclohydrolase II | R | CGACCCTCTTGCCGTAAATA | pentosaceus, L. plantarum | |||||||
| ribG | Diaminohydroxyphosphoribosylaminopyrimidine deaminase | F | TGGKAAGACGCCKCCKTGT | This study | 56.0 | 351 | P. pentosaceus, L. plantarum | 7 | ||
| R | TTCACCAAYCARAATYGCTTGA | |||||||||
| Starch metabolism | glgP | Glycogen phosphorylase | F | GCGGGTGTTCAAAGTATCGT | This study | 60.0 | 229 | L. plantarum | 30 | |
| R | TCTCGAGGGCCTCTTGTAAA | |||||||||
| malL | Oligo-1,6-glucosidase | F | TTGCCTAACAACTGGGGTTC | This study | 60.0 | 177 | L. plantarum | 30 | ||
| R | ATCAACGCCTTTGTTCAACC | |||||||||
| agl | α-Glucosidase | F | GCSAAAATGCTAGCGACYMT | This study | 59.5 | 236 | L. plantarum | 30 | ||
| R | CCACTGCATYGGYGTACGY | |||||||||
| F | AACCTGGTGAAATGGCAGAC | This study | 60.0 | 206 | L. fermentum | |||||
| R | TTGGTCATTCCCAGTTCCTC | |||||||||
| α-amy | α-Amylase | F | AGATCAGGCGCAAGTTCAGT | This study | 60.0 | 220 | L. plantarum | 16, 30 | ||
| R | TTTTATGGGCACACCACTCA | |||||||||
| dexC | Neopullulanase | F | CCAGACGAGCAAGAACAACA | This study | 60.0 | 212 | L. plantarum | 30 | ||
| R | ATTGGCGATACGCCACTTAC | |||||||||
| F | ACTTTTCTGCAGCCTGGTGT | This study | 60.0 | 249 | L. fermentum | |||||
| R | ACGGCCATTAAACTGTCGTC | |||||||||
| malP | Maltose phosphorylase | F | TGCCAYAAYGARTGGGARAT | This study | 60.0 | 161 | L. plantarum, L. | 30 | ||
| R | ACSCKATCWGCCCARAAAC | fermentum, P. pentosaceus | ||||||||
PCR amplification for the detection of genes of interest.
Each PCR mixture (20 μl) contained a reaction cocktail of 200 μM each deoxynucleoside triphosphate, 0.5 μM each primer, 3.5 mM MgCl2, 0.5 U of Taq DNA polymerase (Promega), 10× Taq buffers, and 150 ng of the DNA template. The PCR conditions were as follows: 1 cycle at 95°C for 5 min; 40 cycles of 95°C for 30 s, an annealing temperature depending on the primer for 10 s (Table 1), and 72°C for 15 s; and 1 cycle at 72°C for 5 min. An Applied Biosystems Veriti thermal cycler (VWR, Strasbourg, France) was used. The PCR products were separated on an agarose gel, followed by ethidium bromide staining to check for the presence of a unique amplicon. When a gene from a particular species was amplified using a primer initially designed for a different species, the corresponding amplicon was purified and sequenced by Eurofins MWG Operon (Germany). The primers used for the sequence reaction were the same as those used for gene screening.
In vitro determination of acid and bile tolerance.
Acid and bile salt tolerance was determined according to the method described by Valdez and Taranto (64). Isolates (n = 38) that differed in the gene set related to their abilities to survive low pHs and exposure to bile salts were selected. LAB were grown in MRS broth at 37°C for 16 h and were centrifuged for 10 min at 10,000 × g. The pellets were washed twice with phosphate buffer (0.1 M; pH 7.0) and were resuspended in 10 ml of phosphate buffer. For acid resistance tests, artificial gastric juice (0.2% [wt/vol] NaCl, 0.32% [wt/vol] pepsin; Sigma, Saint-Quentin-Fallavier, France), adjusted with 3 M HCl to pH 2 and pH 7 (positive control) and sterilized by filtration (pore diameter, 0.20 μm; Terumo), was inoculated with 2% washed bacteria, and the mixture was incubated at 37°C for 4 h. Viable cells were counted by plating on MRS agar, and the results are expressed as the percentage of the viable count that was living after 1, 2, 3, and 4 h of exposure. For bile salt tolerance tests, MRS broth and MRS broth with 0.3% (wt/vol) oxgall (Sigma) (MRSO) were inoculated with 0.5% washed bacteria and were incubated at 37°C. The absorbance at 560 nm (A560) was measured at hourly intervals for 8 h. The results are expressed as the time difference in growth in the two culture media, measured by a 0.3-U increase in A560 during the early-exponential-growth stage, as described by Gilliland et al. (21). All experiments were performed in triplicate.
Nucleotide sequence accession numbers.
The sequences of the 16S rRNA gene amplicons have been deposited in the GenBank database under accession numbers FR873843 to FR873994. The GenBank accession numbers for the sequences of amplicons corresponding to the other genes are FR874135 to FR874204.
RESULTS
Identification of LAB isolates.
The 152 LAB isolates identified by sequencing of the 16S rRNA gene are listed in Table 2. All the bacteria from our collection were shown to be members of the family Lactobacillaceae, and identification at the species level showed 99% similarity or higher with the corresponding species in the RDP2 database. Forty-five percent of the sequences corresponded to L. fermentum, 26% to P. pentosaceus, 10% to Pediococcus acidilactici, 18% to the L. plantarum group, and 1% to Lactobacillus salivarius. The L. plantarum group comprised 14% L. plantarum and 4% L. paraplantarum, but no L. pentosus was found.
Table 2.
Identification of the bacteria isolated from 12 traditional production units by sequencing of the 16S rRNA coding gene
| Species | No. of isolates | Designation(s) of isolate(s)a (n = 152) |
|---|---|---|
| L. fermentum | 70 | 1.1, 1.10, 1.2, 1.3, 1.4, 1.5.1, 1.5.2, 1.6, 1.7.1, 1.7.2, 1.8, 1.9, 10.4, 11.1, 11.11.1, 11.11.2, 11.4, 11.5.1, 11.7, 2.10, 2.17.1, 2.17.2, 2.3, 2.5, 2.7.1, 2.7.2, 2.8, 2.9, 3.1, 3.10.1, 3.10.2, 3.2, 3.3, 3.4, 3.5, 3.6, 3.7, 3.8, 3.9.1, 3.9.2, 4.10, 4.11.1, 4.11.2, 4.2, 4.5, 4.7.1, 4.7.2, 4.8.1, 4.8.2, 4.9, 5.1, 5.10, 5.11, 5.3.1, 5.4.2, 5.7, 6.10.1, 6.3, 6.4.1, 6.4.2, 6.5.1, 6.5.2, 6.6.1, 6.6.2, 6.7, 6.9, 7.4, 7.9.1, 8.2, 8.5.2 |
| L. paraplantarum | 6 | 4.1, 2.2, 7.3.1, 4.4, 7.8.1, 7.8.2 |
| L. plantarum | 20 | 2.1, 6.2, 2.4.1, 2.13, 5.8, 11.3, 11.10, 5.9, 2.4.2, 11.6.2, 2.11.1, 11.5.2, 11.2, 2.6, 11.6.1, 5.2.2, 8.4, 6.1, 2.11.2, 2.12 |
| L. salivarius | 1 | 4.6 |
| P. acidilactici | 16 | 10.3.1, 10.3.2, 12.1, 12.11, 12.12, 12.2, 12.4.1, 12.4.2, 12.6, 12.7, 12.8.1, 12.8.2, 12.9, 9.12, 9.7, 9.8 |
| P. pentosaceus | 39 | 10.1, 10.5.1, 10.5.2, 10.6.2, 10.7, 11.8, 11.9, 12.5.1, 2.16.1, 2.16.2, 5.5.2, 5.6.2, 7.1, 7.10, 7.11, 7.2, 7.5.1, 7.5.2, 7.6, 7.7, 8.1.1, 8.10.1, 8.10.2, 8.12, 8.3, 8.5.1, 8.6, 8.7, 8.8, 8.9, 9.1, 9.10, 9.11, 9.2, 9.3.2, 9.4, 9.5.1, 9.5.2, 9.6 |
A total of 152 isolates were identified. Isolates are coded as follows. The first number corresponds to the unit production number (1 to 12), while the second number corresponds to the numbering of isolates. All isolates were checked repeatedly for purity. For some isolates, additional purification steps were necessary, and a third number was assigned to distinguish between isolates with different cell and colony morphologies.
Primer design.
In 58% of the 33 genes selected for which at least one functional analysis had already been performed, no conserved regions among Lactobacillaceae were identified, so primers were designed at the species level (Table 1). Since 15% of the genes in the collection shared conserved regions, designing the primers was easy, except for the primers targeting the hdc, tdc, and aguA genes, for which we used those reported in the literature (Table 1). All primers produced amplicons of the desired size with a single band on the agarose gel. Positive controls were made by testing the primers on the DNA from reference strains containing the targeted genes (Table 3).
Table 3.
Strains of lactic acid bacteria used as positive controls in PCR assays
| Strain(s) used as positive control | Targeted gene |
|---|---|
| Lactobacillus sp. 30A | hdc |
| L. brevis DSM1268 | tdc |
| L. johnsonii NCC533 | odc |
| P. pentosaceus ATCC 25745 | aguA |
| L. plantarum WCFS1, L. fermentum IFO3956, P. pentosaceus ATCC 25745, Leuconostoc mesenteroides ATCC 8293 | groEL |
| L. plantarum WCFS1, L. fermentum IFO3956, P. pentosaceus ATCC 25745, Leuconostoc mesenteroides ATCC 8293 | LBA1272 |
| L. plantarum WCFS1, L. fermentum IFO3956, P. pentosaceus ATCC 25745, Leuconostoc mesenteroides ATCC 8293 | dltD |
| L. plantarum WCFS1, L. fermentum IFO3956, P. pentosaceus ATCC 25745, Leuconostoc mesenteroides ATCC 8293 | La995 |
| L. plantarum WCFS1, L. fermentum IFO3956, P. pentosaceus ATCC 25745, Leuconostoc mesenteroides ATCC 8293 | La57 |
| L. plantarum WCFS1, L. fermentum IFO3956, P. pentosaceus ATCC 25745, Leuconostoc mesenteroides ATCC 8293 | clpL |
| L. plantarum WCFS1, L. fermentum IFO3956, P. pentosaceus ATCC 25745 | lr1516 |
| L. plantarum WCFS1, L. fermentum IFO3956, P. pentosaceus ATCC 25745 | bsh |
| L. reuteri ATCC 23272 | lr0085 |
| L. plantarum WCFS1, L. fermentum IFO3956, P. pentosaceus ATCC 25745 | lr1584 |
| L. plantarum WCFS1, L. fermentum IFO3956, P. pentosaceus ATCC 25745, Leuconostoc mesenteroides ATCC 8293 | LBA0552 |
| L. plantarum WCFS1, L. fermentum IFO3956, P. pentosaceus ATCC 25745, Leuconostoc mesenteroides ATCC 8293 | LBA1429 |
| L. plantarum WCFS1, L. fermentum IFO3956, P. pentosaceus ATCC 25745, Leuconostoc mesenteroides ATCC 8293 | LBA1446 |
| L. plantarum WCFS1, P. pentosaceus ATCC 25745, Leuconostoc mesenteroides ATCC 8293 | LBA1679 |
| L. plantarum WCFS1, L. fermentum IFO3956, P. pentosaceus ATCC 25745, Leuconostoc mesenteroides ATCC 8293 | LBA1432 |
| L. plantarum WCFS1, L. fermentum IFO3956, P. pentosaceus ATCC 25745, Leuconostoc mesenteroides ATCC 8293 | apf |
| L. plantarum WCFS1, L. fermentum IFO3956 | folP |
| L. plantarum WCFS1, L. fermentum IFO3956 | folK |
| L. plantarum WCFS1, L. fermentum IFO3956, P. pentosaceus ATCC 25745, Leuconostoc mesenteroides ATCC 8293 | ribH |
| L. plantarum ATCC 14917, L. fermentum IFO3956, P. pentosaceus ATCC 25745, Leuconostoc mesenteroides ATCC 8293 | ribB |
| L. plantarum WCFS1, L. fermentum IFO3956, P. pentosaceus ATCC 25745, Leuconostoc mesenteroides ATCC 8293 | ribA |
| L. plantarum ATCC 14917, L. fermentum IFO3956, P. pentosaceus ATCC 25745, Leuconostoc mesenteroides ATCC 8293 | ribG |
| L. plantarum WCFS1, L. plantarum ATCC 14917 | glgP |
| L. plantarum ATCC 14917, P. pentosaceus ATCC 25745 | malL |
| L. plantarum WCFS1, L. fermentum IFO3956, P. pentosaceus ATCC 25745, Leuconostoc mesenteroides ATCC 8293 | agl |
| L. plantarum WCFS1, L. plantarum A6, L. fermentum Ogi E1, L. fermentum MW2 | α-amy |
| L. plantarum WCFS1, L. plantarum ATCC 14917, L. fermentum ATCC 14931 | dexC |
| L. plantarum WCFS1, L. fermentum IFO3956, P. pentosaceus ATCC 25745, Leuconostoc mesenteroides ATCC 8293 | malP |
Database analysis revealed that some genes were differently distributed among species and strains. For example, at the strain level, the ribG gene, involved in riboflavin synthesis, was absent in the genome of L. plantarum WCFS1 but present in the genomes of L. plantarum JDM1 and L. plantarum ATCC 14917. This was also the case for the LBA1679 gene, which codes for an ABC transporter permease found in the genome of L. fermentum 28-3-CHN but not in the genomes of L. fermentum strains IFO 3956, ATCC 14931, and CECT 5716. Some genes were present in different copy numbers with quite different sequences depending on the species and strain. For example, the bsh gene was present in four copies in the genome of L. plantarum WCFS1 but only in one copy in P. pentosaceus ATCC 25745. In those cases, we decided to focus on the bsh (lp_3536) sequence, whose role in bile salt survival had already been demonstrated (34).
Cluster analysis of isolates and food metagenomes.
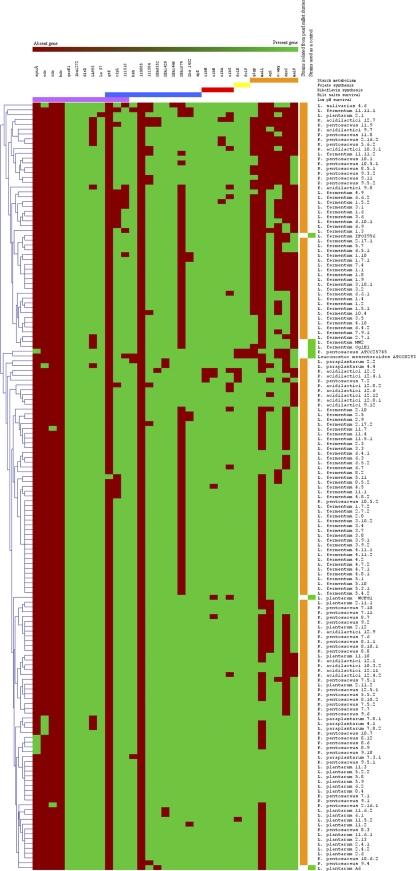
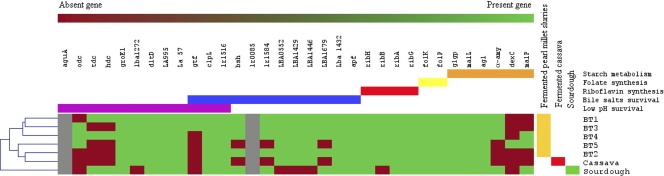
The 33 genes detected with the primers listed in Table 1 were used for cluster analysis of isolates and metagenomes from amylaceous foods. The dendrogram in Fig. 1 shows intraspecies diversity relative to the set of genes investigated. In some cases, isolates grouped at the same level, indicating that they shared the same gene profile. These groups were either homogeneous (composed of isolates belonging to the same species) or heterogeneous (composed of isolates belonging to different species) (Fig. 1). Cluster analysis of gene distribution in food metagenomes showed that the isolates from fermented pearl millet slurries and attiéké (fermented cassava) clustered separately from the isolates from wheat sourdough (Fig. 2). The pearl millet samples were distributed into two subclusters.
Fig. 1.
Distribution of genes involved in survival in the gastrointestinal tract, in folate and riboflavin synthesis, and in starch metabolism in a collection of 152 LAB isolates from fermented pearl millet slurries (indicated by orange bars on the right) and of strains used as positive controls (indicated by green squares on the right). Genes are given at the top, and their roles are indicated by the following color code: purple, low-pH survival; blue, bile salt resistance; red, riboflavin synthesis; yellow, folate synthesis; orange, starch metabolism. In the dendrogram, red indicates the absence of a gene, while green indicates its presence. The dendrogram shows estimated relationships among the strains and was constructed by average-linkage hierarchical analysis using Mev software, version 4.4 (56).
Fig. 2.
Distribution of genes involved in survival in the gastrointestinal tract, in folate and riboflavin synthesis, and in starch metabolism in metagenomes extracted from different starch-rich fermented foods. Genes are given at the top, and their roles are indicated by the following color code: purple, low-pH survival; blue, bile salt resistance; red, riboflavin synthesis; yellow, folate synthesis; orange, starch metabolism. In the dendrogram, red indicates the absence of a gene, while green indicates its presence. The aguA and lr0085 genes are indicated in gray, because the primer used did not allow the amplification of a single band. The dendrogram shows estimated relationships among the strains and was constructed by average-linkage hierarchical analysis using Mev software, version 4.4 (56).
Detection of genes involved in survival in the gastrointestinal tract.
The results of gene detection are presented in Fig. 1. As expected, the housekeeping gene groEL, which is also involved in survival at low pHs, was found in all isolates. Some of the other genes involved in low-pH survival (clpL, lr1516, LBA1272, dltD, La995, La57) were found in 91% to 100% of the bacteria from the collection, while others (gtf, aguA, odc, tdc, hdc) were found in 0% to 4% of the isolates. For each gene screened for low-pH survival, one amplicon obtained from PCR amplification of DNA extracted from one isolate for each species in the collection was sequenced. At least 91% similarity was found with the corresponding gene in the reference strains L. plantarum WCFS1, L. plantarum ST-III, L. fermentum IFO 3956, P. pentosaceus ATCC 25745, Lactobacillus helveticus DPC 4571, Lactobacillus curvatus HSCC1736, and Lactobacillus suebicus CUPV221.
Most of the genes involved in bile salt tolerance (bsh, lr1584, LBA0552, LBA1679, LBA1429, LBA1446, LBA1432, apf) were detected in almost all isolates at a frequency of 92% to 100% (Fig. 1). In contrast, the LBA1679 gene was detected in only 52% of the isolates, but never in those belonging to the L. fermentum species. Because no other data were available, primers designed on the basis of species absent from our bacterial collection were used to search for the lr0085 gene (Table 1; Fig. 1). No amplification of this gene was obtained on any DNA extracted from our isolates. The sequencing of 35 amplicons from DNA from different species showed 93% to 99% similarity with the corresponding genes in the reference strains of L. plantarum WCFS1, L. fermentum IFO 3956, and P. pentosaceus ATCC 25745. Most of the bacteria isolated from the pearl millet slurries exhibited a genetic profile favorable to their survival in the gastrointestinal tract, and the distribution of the survival-related genes was mostly not species specific but was distributed equally among all the isolates of the six species that make up the collection.
Survival at low pHs and in the presence of bile salts.
On the basis of the results of the molecular screening, 38 isolates belonging to different species and/or differing in their genetic equipment were selected for evaluation of their survival at low pHs (Table 4) and in the presence of bile salts (Table 5). Resistance to low pHs was estimated by incubating the isolates in a pH 2 gastric juice solution to mimic stomach conditions. As a control, we checked that all the isolates survived for at least 4 h in the artificial gastric juice at pH 7. Among the 38 isolates tested, 55% survived for at least 1 h at pH 2, 34% survived for 2 h, 18% for 3 h, and 11% for 4 h (Table 4). It is noteworthy that 91% and 33% of the L. fermentum isolates survived for 2 and 4 h, respectively, whereas only 8% of the isolates from other species survived for 2 h in the synthetic gastric juice at pH 2.
Table 4.
Results of the pH survival assay
| Species and straina | Initial count (CFU/ml) | % of living bacteria after incubation at pH 2b for the following time: |
Presence or absence of the following genec: |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 h | 2 h | 3 h | 4 h | clpL | lr1516 | LBA1272 | dltD | La995 | La57 | gtf | aguA | odc | tdc | hdc | groEL | ||
| L. fermentum | |||||||||||||||||
| 1.1 | (2.0 ± 0.9) × 107 | 16.1 ± 6.5 | 0 | 0 | 0 | + | + | + | + | + | + | − | − | − | − | − | + |
| 1.3 | (4.0 ± 4.8) × 107 | 35.4 ± 20.0 | 38.6 ± 22.7 | 27.5 ± 13.9 | 16.7 ± 10.0 | + | − | + | + | + | + | − | − | − | − | − | + |
| 1.6 | (1.1 ± 0.9) × 107 | 46.5 ± 17.3 | 30.1 ± 18.1 | 26.3 ± 14.5 | 24.5 ± 12.3 | − | + | + | + | + | + | − | − | − | − | − | + |
| 2.10 | (4.5 ± 1.3) × 107 | 6.7 ± 3.0 | 0.7 ± 0.7 | 0 | 12.6 ± 12.6 | + | + | + | + | + | + | − | − | − | − | − | + |
| 3.1 | (3.5 ± 4.4) × 107 | 36.5 ± 30.0 | 31.4 ± 31.4 | 31.1 ± 31.1 | 0 | − | − | + | + | + | + | − | − | − | − | − | + |
| 3.3 | (2.5 ± 0.7) × 107 | 37.0 ± 20.4 | 13.1 ± 13.1 | 2.6 ± 2.6 | 0 | + | + | + | + | + | + | − | − | − | − | − | + |
| 3.9.2 | (5.6 ± 1.2) × 107 | 32.6 ± 18.9 | 24.9 ± 23.4 | 8.9 ± 8.6 | 0.2 ± 0.2 | + | + | + | + | + | + | − | − | − | − | − | + |
| 4.9 | (4.0 ± 3.2) × 107 | 25.8 ± 25.1 | 6.9 ± 6.9 | 0 | 0 | − | − | + | + | + | + | − | − | − | − | − | + |
| 6.4.2 | (8.0 ± 2.2) × 107 | 42.8 ± 19.5 | 4.9 ± 2.5 | 0.6 ± 0.6 | 0 | + | + | + | + | + | + | − | − | − | − | − | + |
| 11.1 | (5.2 ± 2.6) × 107 | 4.0 ± 1.0 | 1.5 ± 1.5 | 0 | 0 | − | + | + | + | + | + | − | − | − | − | − | + |
| 11.11.1 | (4.6 ± 4.0) × 107 | 7.5 ± 5.4 | 1.0 ± 1.0 | 0 | 0 | − | + | + | + | + | + | − | − | − | − | − | + |
| IFO 3956* | (9.2 ± 0.9) × 106 | 83.8 ± 5.8 | 11.5 ± 3.2 | 6.3 ± 6.3 | 0 | + | + | + | + | + | + | − | − | − | − | − | + |
| P. pentosaceus | |||||||||||||||||
| 5.6.2 | (1.5 ± 1.8) × 107 | 0 | 0 | 0 | 0 | + | + | + | + | − | + | − | − | − | − | − | + |
| 8.6 | (6.4 ± 0.9) × 107 | 0 | 0 | 0 | 0 | + | + | + | + | + | + | − | + | − | − | − | + |
| 8.9 | (5.5 ± 0.5) × 107 | 4.3 ± 1.0 | 0.6 ± 0.6 | 0 | 0 | + | + | + | + | + | + | − | + | − | − | − | + |
| 8.12 | (4.7 ± 2.7) × 107 | 6.6 ± 1.6 | 0 | 0 | 0 | + | + | + | + | + | + | − | + | − | − | − | + |
| 9.1 | (2.5 ± 1.5) × 107 | 0 | 0 | 0 | 0 | + | + | + | + | + | + | − | − | − | − | − | + |
| 9.3.2 | (1.2 ± 1.7) × 107 | 0 | 0 | 0 | 0 | + | + | + | + | + | + | − | − | − | − | − | + |
| 9.10 | (2.3 ± 1.8) × 107 | 0 | 0 | 0 | 0 | + | + | + | + | + | + | − | + | − | − | − | + |
| 10.6.2 | (8.2 ± 0.8) × 106 | 34.6 ± 34.6 | 0 | 0 | 0 | + | + | + | + | + | + | − | − | − | − | − | + |
| 10.7 | (1.6 ± 0.2) × 107 | 0 | 0 | 0 | 0 | + | + | + | + | + | + | − | − | + | − | − | + |
| 11.8 | (3.5 ± 1.0) × 107 | 0 | 0 | 0 | 0 | + | + | + | + | − | + | − | − | − | − | − | + |
| 11.9 | (7.2 ± 0.4) × 106 | 29.0 ± 29.0 | 12.9 ± 12.9 | 0 | 0 | + | + | + | + | − | + | − | − | − | − | − | + |
| ATCC 25745* | (2.8 ± 5.0) × 106 | 0 | 0 | 0 | 0 | + | + | + | + | + | + | − | + | − | − | − | + |
| L. plantarum | |||||||||||||||||
| 2.1 | (2.7 ± 2.6) × 107 | 0 | 0 | 0 | 0 | + | + | + | + | + | − | − | − | − | − | − | + |
| 11.3 | (2.8 ± 0.4) × 107 | 0 | 0 | 0 | 0 | + | + | + | + | + | + | − | − | − | − | − | + |
| 11.5.2 | (5.9 ± 0.6) × 107 | 0 | 0 | 0 | 0 | + | + | + | + | + | + | − | − | − | − | − | + |
| 11.6.2 | (4.2 ± 0.8) × 107 | 0 | 0 | 0 | 0 | + | + | + | + | + | + | − | − | − | − | − | + |
| WCFS1* | (1.1 ± 0.3) × 107 | 5.3 ± 5.3 | 0 | 0 | 0 | + | + | + | + | + | + | − | − | − | − | − | + |
| P. acidilactici | |||||||||||||||||
| 12.6 | (3.4 ± 0.9) × 107 | 0 | 0 | 0 | 0 | + | + | + | + | + | + | − | − | − | − | − | + |
| 12.8.2 | (3.8 ± 1.1) × 107 | 3.5 ± 2.2 | 0 | 0 | 0 | + | + | + | + | + | + | − | − | − | − | − | + |
| 12.9 | (8.2 ± 9.8) × 107 | 0.3 ± 0.3 | 0 | 0 | 0 | + | + | + | + | + | + | − | − | − | − | − | + |
| L. paraplantarum | |||||||||||||||||
| 4.1 | (5.3 ± 0.3) × 107 | 4.0 ± 2.2 | 0 | 0 | 0 | + | + | + | + | − | + | − | − | + | − | − | + |
| 4.4 | (4.1 ± 0.8) × 107 | 2.9 ± 2.9 | 0 | 0 | 0 | + | + | + | + | − | + | − | − | + | − | − | + |
| 7.8.1 | (4.0 ± 0.9) × 107 | 0 | 0 | 0 | 0 | + | + | + | + | − | + | − | − | + | − | − | + |
| 7.8.2 | (2.7 ± 0.9) × 107 | 0 | 0 | 0 | 0 | + | + | + | + | − | + | − | − | − | − | − | + |
| L. salivarius 4.6 | (2.1 ± 1.0) × 107 | 0 | 0 | 0 | 0 | + | − | + | + | + | + | − | − | + | − | − | + |
| Leuconostoc mesenteroides ATCC 8293* | (2.7 ± 0.2) × 106 | 0 | 0 | 0 | 0 | + | − | + | + | + | + | − | − | − | − | − | + |
Asterisks indicate reference strains used as positive controls in the PCR assays.
The quantity of bacteria living after incubation for 1, 2, 3, or 4 h in a gastric juice at pH 2 is expressed as a percentage of the initial count. Where living bacteria represent less than 0.0001% of the initial count, a value of zero is given.
Genes involved in survival at low pH. +, presence; −, absence.
Table 5.
Results of the bile salt tolerance assay, expressed as the difference in growth in MRS versus MRSO mediuma
| Species and isolate or strainb | Growth delay (min) | Sensitivity | Presence or absence of the following genec: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| bsh | clpL | lr1516 | lr0085 | lr1584 | LBA0552 | LBA1429 | LBA1446 | LBA1679 | LBA1432 | LBA0493 | gtf | |||
| L. fermentum | ||||||||||||||
| 1.1 | 15 ± 3 | Tolerant | + | + | + | − | + | + | + | + | − | + | + | − |
| 1.3 | 29 ± 8 | Tolerant | + | + | − | − | + | + | + | + | − | + | + | − |
| 1.6 | 24 ± 6 | Tolerant | + | + | + | − | + | + | + | + | − | + | + | − |
| 2.10 | 43 ± 18 | Low tolerant | + | + | + | − | + | + | + | + | − | + | + | − |
| 3.1 | 10 ± 8.0 | Resistant | + | + | − | − | + | + | + | + | − | + | + | − |
| 3.3 | 35 ± 1 | Tolerant | + | + | + | − | + | + | + | + | − | + | + | − |
| 3.9.2 | 33 ± 21 | Tolerant | + | + | + | − | + | + | + | + | − | + | + | − |
| 4.9 | 37.5 ± 2 | Tolerant | + | + | − | − | + | + | + | + | − | + | + | − |
| 6.4.2 | 99 ± 38 | Sensitive | + | + | + | − | + | + | + | + | − | + | + | − |
| 11.1 | 27 ± 12 | Tolerant | + | + | + | − | + | + | + | + | − | + | + | − |
| 11.11.1 | 91 ± 27 | Sensitive | − | − | + | − | − | + | + | + | − | − | + | − |
| IFO 3956* | 14 ± 10.5 | Resistant | + | + | + | − | + | + | + | + | − | + | + | − |
| L. paraplantarum | ||||||||||||||
| 4.1 | 9 ± 13 | Resistant | + | + | + | − | + | + | + | + | + | + | + | − |
| 4.4 | 31 ± 11 | Tolerant | + | + | + | − | + | + | − | + | + | + | + | − |
| 7.8.1 | 6 ± 8 | Resistant | + | + | + | − | + | + | + | + | + | + | + | − |
| 7.8.2 | 7 ± 2 | Resistant | + | + | + | − | + | + | + | + | + | + | + | − |
| L. plantarum | ||||||||||||||
| 2.1 | 4 ± 4 | Resistant | + | + | + | − | + | + | + | + | + | − | + | − |
| 11.3 | 22 ± 2 | Tolerant | + | + | + | − | + | + | + | + | + | + | + | − |
| 11.5.2 | 16 ± 19 | Tolerant | + | + | + | − | + | + | + | + | + | + | + | − |
| 11.6.2 | 24 ± 17 | Tolerant | + | + | + | − | + | + | − | + | + | + | + | − |
| WCFS1* | 29 ± 5 | Tolerant | + | + | + | − | + | + | + | + | + | + | + | − |
| L. salivarius 4.6 | 34 ± 2 | Tolerant | + | + | − | − | − | + | + | − | − | + | + | − |
| Leuconostoc mesenteroides ATCC 8293* | 78 ± 19 | Sensitive | − | + | − | − | + | + | + | + | + | + | + | − |
| P. acidilactici | ||||||||||||||
| 12.6 | 9 ± 10 | Resistant | + | + | + | − | + | + | + | + | + | + | + | − |
| 12.8.2 | 11 ± 9 | Resistant | + | − | + | − | − | + | + | + | + | + | + | − |
| 12.9 | 3 ± 4 | Resistant | + | + | + | − | + | + | + | + | + | + | + | − |
| P. pentosaceus | ||||||||||||||
| 5.6.2 | 6 ± 2 | Resistant | + | + | + | − | + | + | + | + | + | + | + | − |
| 8.6 | 7 ± 6 | Resistant | + | + | + | − | + | + | + | + | + | + | + | − |
| 8.9 | 14 ± 12 | Resistant | + | + | + | − | + | + | + | + | + | + | + | − |
| 8.12 | 12 ± 17 | Resistant | + | + | + | − | + | + | + | + | + | + | + | − |
| 9.1 | 15 ± 4 | Tolerant | + | + | + | − | + | + | + | + | + | + | + | − |
| 9.3.2 | 22 ± 2 | Tolerant | + | + | + | − | + | + | + | + | + | + | + | − |
| 9.10 | 3 ± 2 | Resistant | + | + | + | − | + | + | + | + | + | + | + | − |
| 10.6.2 | 1 ± 10 | Resistant | + | + | + | − | + | + | + | + | + | + | + | − |
| 10.7 | 55 ± 27 | Low tolerant | + | + | + | − | + | + | + | + | + | + | + | − |
| 11.8 | 20 ± 9 | Tolerant | + | + | + | − | + | + | + | + | + | + | + | − |
| 11.9 | 23 ± 11 | Tolerant | + | − | + | − | − | − | + | + | + | + | + | − |
| ATCC 25745* | 7 ± 4 | Resistant | + | + | + | − | + | + | + | + | + | + | + | − |
Measured by an increase of 0.3 U in A560 during the early-exponential-growth stage.
Asterisks indicate reference strains used as positive controls in the PCR assays.
Genes involved in tolerance to bile salt. +, presence; −, absence.
Resistance to bile salts was evaluated by the ability of the same 38 isolates to grow in MRSO medium. Most of the isolates were resistant or tolerant to bile salts (Table 5). Only three L. fermentum isolates, one P. pentosaceus isolate, and Leuconostoc mesenteroides ATCC 8293 were sensitive or displayed low tolerance. Unlike that of low pH resistance, the distribution of resistance or sensitivity to bile salts was not linked to a particular species. We can thus consider that the majority of the isolates have the potential to survive these conditions.
Detection of genes coding for enzymes involved in the biosynthesis of riboflavin and folate.
The potential of LAB isolates to synthesize folate was assessed by screening for the presence or absence of two signature genes, folP and folK (13), encoding dihydropteroate synthase and 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase, respectively (Table 1). Almost all the bacteria in the collection were potentially able to produce folate: 100% and 98% of the isolates showed good amplification at the expected size for folP and folK, respectively (Fig. 1). Only two P. acidilactici isolates failed to display folK amplification.
LAB that were putatively able to synthesize riboflavin were screened using the four genes involved in its production: ribA, ribG, ribB, and ribH (Table 1; Fig. 1). These genes were found in 92% to 98% of the isolates. In results similar to those of the screening for genes involved in folate synthesis, P. acidilactici had the lowest frequency of rib genes, with only half of the isolates harboring ribG, while 94% to 100% of the isolates belonging to the other species harbored this gene. The distribution of the ribA, ribB, and ribH genes generally followed the same pattern.
The sequences of the amplicons obtained with primers whose design was based on the genes of other bacterial species (Table 1) showed high similarity (80% to 100%) to the corresponding genes in the reference strains L. fermentum IFO 3956, L. plantarum JDM1, L. plantarum WCFS1, and P. pentosaceus ATCC 25745.
Detection of genes involved in starch metabolism.
Six genes were screened to evaluate the potential of the isolates to produce enzymes involved in starch metabolism, from hydrolysis of the macromolecules amylose and amylopectin (e.g., α-amylase, neopullulanase) to hydrolysis of starch degradation products such as maltose, enabling their entry into the glycolytic pathway (maltose phosphorylase, α-glucosidase). The isolates differed considerably with respect to the presence or absence of each gene (Fig. 1). Distribution of genes among all the isolates were as follows: 79% for agl, 76% for glgP, 75% for α-amy, 66% for malP, 54% for dexC, and 19% for malL. Only 8% of the isolates tested were positive for all the genes, while 3% were negative for all the genes. Sequencing of PCR amplicons revealed at least 74% similarity with the corresponding fragment in the L. plantarum JDM1 and L. plantarum WCFS1 genomes.
Screening of metagenomes from different fermented starchy foods.
The results of gene detection in the metagenomes extracted from samples of different fermented starchy foods are presented in Fig. 2. Almost all the PCRs led to the amplification of a single band, reflecting the high specificity of the primers. The only exceptions were aguA and lr0085, which displayed several bands, reflecting nonspecific amplification. Among the 33 genes screened, 16, including most of the genes involved in riboflavin and folate synthesis, were found in all the metagenomes. The main differences observed between the food samples concerned genes related to survival at low pHs, survival after exposure to bile salts, and starch metabolism. Although all the samples of the fermented pearl millet slurries tested (BT1 to BT5) were produced using the same traditional process, there were differences in their genetic profiles. For example, the metagenome of sample BT1 harbored 90% of the genes related to survival at low pHs and after exposure to bile salts, while BT5 harbored only 66% of them. All the foods chosen in this study were rich in starch, and their metagenomes shared a common set of genes related to starch metabolism (glgP, malL, agl), but the genes encoding α-amylase, neopullulanase, and maltose phosphorylase (α-amy, dexC, and malP, respectively) were not systematically detected, reflecting different capacities or different possible pathways for starch metabolism.
DISCUSSION
Very few studies have dealt with molecular screening in bacterial genomes and metagenomes for genes involved in functions of interest for different food and health applications. Five wine-related genes in 120 bacterial isolates belonging to seven Lactobacillus species were recently screened (45). Kaushik et al. (2009) used a PCR-based test to detect three genes involved in probiotic functions in two L. plantarum strains (31). We performed molecular screening of a larger set of genes involved in probiotic functions and in nutrition in order to identify their distribution in 152 isolates and in the metagenomes of amylaceous fermented foods. Microbial diversity was limited to five species, among which L. fermentum was dominant, followed by P. pentosaceus and L. plantarum. Based on the PCR detection profiles of the 33 genes, cluster analysis showed that in general, isolates belonging to the same species were grouped together but in several separate clusters (e.g., L. fermentum, L. plantarum), pointing to the relative specificity of gene distribution and associated functions.
Species-level identification of the isolates enabled us to reduce in silico analysis by designing species-targeted primers. The 21 genes that were chosen because they are involved in gastrointestinal survival were found in several probiotic strains whose genomes have already been sequenced (Lactobacillus acidophilus NCFM, Lactobacillus gasseri ATCC 33323, L. johnsonii NCC533, L. plantarum WCFS1, and L. salivarius UCC118). We found that 82% of our bacterial collection harbored 14 of the 21 genes, and 63% of the metagenomes from fermented foods harbored at least 12 of these genes. Only four of the genes were detected in all isolates: one was the housekeeping gene groEL, but the others were nonessential: LBA1272, dltD, and LBA0493 (20). The existence of a conserved domain in the DltD_M protein (pfam04918), involved in the biosynthesis of d-alanyl-lipoteichoic acid, and in the lysophospholipid acyltransferases (cd07989), encoded by dltD and LBA1272, respectively, from several Lactobacillaceae species could explain why these genes were present in the entire bacterial collection. No such large conserved domains were identified in the aggregation-promoting protein LBA0493, but the wide distribution of the corresponding gene among Lactobacillus species and the existence of a small conserved sequence confined mostly to the C-terminal region of the protein (22) could explain the detection of this gene in all the bacteria. Less frequently detected genes were gtf, aguA, odc, tdc, hdc, and lr0085, which were found in 0% to 4% of the 152 isolates. The gtf gene was detected only in L. fermentum 5.11 and in two metagenomes. Since no other data were available, the primers used to detect this gene were designed on the basis of sequences from Lactobacillus diolivorans, Pediococcus parvulus, Pediococcus damnosus, and L. suebicus. This gene codes for glycosyltransferase, an enzyme that, in addition to its role in survival, is also involved in beta-glucan production. This exopolysaccharide may modify the organoleptic properties of the food and has also been reported to have many health-promoting properties (58).
Among the genes screened for survival at low pHs, it is noteworthy that there were two clearly defined groups (Fig. 1): one group that was detected in most of the isolates (clpL, lr1516, LBA1272, dltD, La995, La57) and a second group that was less frequently detected (gtf, aguA, odc, tdc, hdc). However, despite the presence of genes belonging to the first group in most of the 152 isolates, nearly half the isolates did not survive for 1 h at pH 2. All L. fermentum isolates tested in vitro presented remarkable resistance to pH 2 despite the absence of genes from the second set, in contrast with the results of functional analysis, which showed that the odc and aguA genes were involved in survival at low pHs (2, 65). However, only a few LAB harboring one of these genes had variable but low survival abilities at low pHs, ranging from 0 to 2 h.
Despite the wide distribution of the first set of genes and the occasional occurrence of some of the second set, no relationship was found between the presence of these genes and low-pH-survival capacity. This screening failed to preselect isolates that were able to survive at low pHs, underlining the difficulty of selecting markers that would enable an appropriate molecular strategy to be designed. Nevertheless, the detection of genes involved in other functions could be useful for future metabolic investigations. For example, the aguA, odc, tdc, and hdc genes are also involved in putrescine, histamine, and tyrosine formation and have been studied mainly for their toxicological effects (24). As a result, their low frequency should be considered a positive characteristic given the potential deleterious effect on health of biogenic amines in foods. The genes reported to be related to bile salt resistance were widely distributed in the collection. Resistance to bile salts was also high in the 38 isolates tested. The important role of bsh genes in several LAB species has been extensively discussed, but it is still difficult to assess their role in bile salt survival (14, 17, 34, 42). In our study, L. fermentum 11.11.1 and the collection strain Leuconostoc mesenteroides ATCC 8293, lacking bsh, were shown to be sensitive to bile salts. The lrl0085 and gtf genes did not appear to play a functional role in bile salt resistance, since their absence in all isolates and reference strains did not affect the tolerance of these isolates to bile salts. The same was true for the LBA1679 gene, which was absent in all L. fermentum isolates tested and in L. salivarius 4.6. However, the low bile salt tolerance of wild isolates and of the reference strain concomitant with the absence of bsh, the conserved regions in bsh genes from different LAB species, and the reported role of the corresponding hydrolases in the deconjugation of bile salts by LAB belonging to different species (29, 34) make this marker the best available target, at least enabling the exclusion of isolates without this gene.
Analysis of LAB isolates from the fermented pearl millet slurries showed that despite the expected high potential for survival at low pHs revealed by molecular data, isolates were more frequently tolerant of exposure to bile salts than of low pHs. When the metagenomes of the five traditional fermented pearl millet samples were examined, the distribution of genes for pH resistance was shown to be similar to that in the isolates, and four samples out of five were positive for bsh. In addition, three samples were positive for odc. Variability among pearl millet slurries indicates that the same type of food produced in different production units located in the same geographical area (Ouagadougou, Burkina Faso) will not necessarily have the same functional potential.
Studying survival in the gastrointestinal tract using genetic screening is complex due to its multifactorial character, which combines different metabolic functions, making the choice of a pertinent marker difficult. On the other hand, this approach is simpler when more-focused and well-defined metabolic pathways are targeted, for example, biogenic amine synthesis or other characteristics of interest, such as vitamin B synthesis and starch degradation. Vitamin B synthesis by LAB offers interesting perspectives for both food fortification and probiotic applications (7, 27, 43, 54). But despite some evidence for higher vitamin B contents in fermented cereal foods, information on the capacity of LAB from cereal-based fermented foods to produce B vitamins is surprisingly scarce (9). Both in our LAB collection and in the metagenomes of the starchy fermented foods, genetic screening revealed a high potential to produce folate and riboflavin, whereas the genomes of the reference strains lack some of the genes coding for the synthesis of these vitamins (7, 11, 33, 41, 44). Genes involved in folate and riboflavin synthesis were well distributed among the six Lactobacillus and Pediococcus species. Lactobacillus species are usually auxotrophic for B vitamins (5, 48). However, compared to other raw materials of plant origin, processed cereals are poor sources of B vitamins, and this phenomenon probably represents a selection pressure for LAB with the ability to produce B vitamins. Nevertheless, it should be borne in mind that even though yeasts are minor components of the microbiota of such foods (62), they can also be a source of B vitamins. The potential of the isolates and the metagenomes revealed by our study opens the way for future research on the production of B vitamins by LAB in fermented cereals or other plant foods.
The ecological flexibility of certain LAB species, such as L. plantarum, or their specificity to certain environments is due to their capacity to metabolize and/or transport different types of carbohydrates and/or to modulate their metabolic pathways (4). Given that starch is a carbon and energy source, only a few LAB are able to use this substrate. Nonetheless, amylolytic LAB (ALAB) are common members of the microbiota of amylaceous fermented foods (23). Screening revealed high genetic variability related to starch metabolism and led to a clearer picture than that for the other groups of genes, probably reflecting complex interactions with the food matrix and between the LAB species in their particular food niche. The majority of our isolates displayed the potential for starch hydrolysis, mainly through the presence of α-amy (the α-amylase gene) and, to a lesser extent, dexC (the neopullulanase gene). Partial starch hydrolysis by α-amylase to liquefy starch is a potentially useful characteristic that could be exploited to improve the energy density of gruels used for the complementary feeding of young children (46). Furthermore, the presence of dexC suggests that pullulanases may also play a role in starch hydrolysis by ALAB. The contribution of pullulanases to modifying the rheology of fermented gruels has not yet been investigated and deserves further attention. The high potential for α-amylase synthesis in many isolates belonging to the L. plantarum and L. fermentum species is consistent with the fact that amylolytic isolates from these species have frequently been recovered from starchy fermented tropical foods (23). As expected from their reported α-amylase activity (1, 18), the ALAB strains used as positive controls, L. plantarum A6, L. fermentum Ogi E1, and L. fermentum MW2, were positive for α-amy, and α-amy was often associated with genes coding for enzymes that enable the use of maltose, i.e., malP (encoding maltose phosphorylase) and/or agl (encoding α-glucosidase). Among isolates that do not have α-amy or dexC, mainly a few P. pentosaceus and L. fermentum isolates, some nevertheless have the potential to use maltose (malP, agl), suggesting that they could establish a trophic relation with ALAB. In addition, α-glucosidase activity was detected in L. fermentum Ogi E1 (8), whereas the strain showed no maltose phosphorylase activity, consistent with the presence of agl and the absence of malP. However, both genes were present in some L. fermentum isolates, suggesting some flexibility in maltose metabolism within the same species.
Analysis of the metagenomes of the starchy foods gave a rather narrow view of amylolytic potential compared to the presence of the other groups of genes tested, except for those encoding amino acid decarboxylases. Unexpectedly, two out of five metagenomes from fermented pearl millet slurries were not positive for the presence of α-amy and dexC, pointing to possible alternative metabolisms or carbon sources for energy generation and growth. It is interesting that these two metagenomes clustered separately from those of the other pearl millet samples and shared characteristics with the attiéké metagenome, including the absence of genes encoding amino acid decarboxylases. Even though our main purpose here was to examine the feasibility of using molecular screening by applying the method to a limited number of food metagenomes in different categories, we observed that French wheat sourdough differed from its tropical counterparts. This may be only an occasional phenomenon, and in order to draw conclusions, molecular screening would have to include a larger number of samples from different locations in a dedicated study. In addition, the metagenomic approach did not differentiate between prokaryotic and eukaryotic DNAs (mainly from yeasts, which share the same ecological food niche with LAB). Therefore, some useful functions may also be shared with yeasts, even if these are not dominant in the food microbiota.
In conclusion, genetic screening is a promising way to assess the potential of microbiotas for specific functions and to orient strategies for ecological studies. Indeed, this approach revealed that traditional African cereal-based fermented foods, as in the “ben-saalga model,” have the genetic potential for functions of interest in both probiotics and nutrition. In particular, thanks to their ability to survive the conditions prevailing in the gastrointestinal tract, L. fermentum isolates merit further investigations to assess their probiotic potential.
ACKNOWLEDGMENTS
We thank Michiel Kleerebezem and Stéphane Duboux for supplying reference strains and Claire Mouquet for slurry samples.
Footnotes
Published ahead of print on 14 October 2011.
REFERENCES
- 1. Agati V., Guyot J. P., Morlon-Guyot J., Talamond P., Hounhouigan D. J. 1998. Isolation and characterization of new amylolytic strains of Lactobacillus fermentum from fermented maize doughs (mawè and ogi) from Benin. J. Appl. Microbiol. 85:512–520 [Google Scholar]
- 2. Azcarate-Peril M. A., Altermann E., Hoover-Fitzula R. L., Cano R. J., Klaenhammer T. R. 2004. Identification and inactivation of genetic loci involved with Lactobacillus acidophilus acid tolerance. Appl. Environ. Microbiol. 70:5315–5322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blandino A., Al-Aseeri M. E., Pandiella S. S., Cantero D., Webb C. 2003. Cereal-based fermented foods and beverages. Food Res. Int. 36:527–543 [Google Scholar]
- 4. Boekhorst J., et al. 2004. The complete genomes of Lactobacillus plantarum and Lactobacillus johnsonii reveal extensive differences in chromosome organization and gene content. Microbiology 150:3601–3611 [DOI] [PubMed] [Google Scholar]
- 5. Bowman W. C., DeMoll E. 1993. Biosynthesis of biotin from dethiobiotin by the biotin auxotroph Lactobacillus plantarum. J. Bacteriol. 175:7702–7704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brosius J., Palmer M. L., Kennedy P. J., Noller H. F. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 75:4801–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burgess C., O'Connell-Motherway M., Sybesma W., Hugenholtz J., van Sinderen D. 2004. Riboflavin production in Lactococcus lactis: potential for in situ production of vitamin-enriched foods. Appl. Environ. Microbiol. 70:5769–5777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Calderon M., Loiseau G., Guyot J. P. 2003. Fermentation by Lactobacillus fermentum Ogi E1 of different combinations of carbohydrates occurring naturally in cereals: consequences on growth energetics and alpha-amylase production. Int. J. Food Microbiol. 80:161–169 [DOI] [PubMed] [Google Scholar]
- 9. Chavan J. K., Kadam S. S. 1989. Nutritional improvement of cereals by fermentation. Crit. Rev. Food Sci. Nutr. 28:349–400 [DOI] [PubMed] [Google Scholar]
- 10. Cibik R., Lepage E., Talliez P. 2000. Molecular diversity of Leuconostoc mesenteroides and Leuconostoc citreum isolated from traditional French cheeses as revealed by RAPD fingerprinting, 16S rDNA sequencing and 16S rDNA fragment amplification. Syst. Appl. Microbiol. 23:267–278 [DOI] [PubMed] [Google Scholar]
- 11. Claesson M. J., et al. 2006. Multireplicon genome architecture of Lactobacillus salivarius. Proc. Natl. Acad. Sci. U. S. A. 103:6718–6723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Costantini A., Cersosimo M., Del Prete V., Garcia-Moruno E. 2006. Production of biogenic amines by lactic acid bacteria: screening by PCR, thin-layer chromatography, and high-performance liquid chromatography of strains isolated from wine and must. J. Food Prot. 69:391–396 [DOI] [PubMed] [Google Scholar]
- 13. de Crecy-Lagard V., El Yacoubi B., de la Garza R. D., Noiriel A., Hanson A. D. 2007. Comparative genomics of bacterial and plant folate synthesis and salvage: predictions and validations. BMC Genomics 8:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Denou E., et al. 2008. Identification of genes associated with the long-gut-persistence phenotype of the probiotic Lactobacillus johnsonii strain NCC533 using a combination of genomics and transcriptome analysis. J. Bacteriol. 190:3161–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Diaz-Ruiz G., Guyot J. P., Ruiz-Teran F., Morlon-Guyot J., Wacher C. 2003. Microbial and physiological characterization of weakly amylolytic but fast-growing lactic acid bacteria: a functional role in supporting microbial diversity in pozol, a Mexican fermented maize beverage. Appl. Environ. Microbiol. 69:4367–4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eom H. J., Moon J. S., Seo E. Y., Han N. S. 2009. Heterologous expression and secretion of Lactobacillus amylovorus alpha-amylase in Leuconostoc citreum. Biotechnol. Lett. 31:1783–1788 [DOI] [PubMed] [Google Scholar]
- 17. Fang F., et al. 2009. Allelic variation of bile salt hydrolase genes in Lactobacillus salivarius does not determine bile resistance levels. J. Bacteriol. 191:5743–5757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Florencio J. A., et al. 2000. Lactobacillus plantarum amylase acting on crude starch granules. Native isoforms and activity changes after limited proteolysis. Appl. Biochem. Biotechnol. 84–86:721–730 [DOI] [PubMed] [Google Scholar]
- 19. Giegerich R., Meyer F., Schleiermacher C. 1996. GeneFisher: software support for the detection of postulated genes. Proc. Int. Conf. Intell. Syst. Mol. Biol. 4:68–77 [PubMed] [Google Scholar]
- 20. Gil R., Silva F. J., Pereto J., Moya A. 2004. Determination of the core of a minimal bacterial gene set. Microbiol. Mol. Biol. Rev. 68:518–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gilliland S. E., Staley T. E., Bush L. J. 1984. Importance of bile tolerance of Lactobacillus acidophilus used as a dietary adjunct. J. Dairy Sci. 67:3045–3051 [DOI] [PubMed] [Google Scholar]
- 22. Goh Y. J., Klaenhammer T. R. 2010. Functional roles of aggregation-promoting-like factor in stress tolerance and adherence of Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 76:5005–5012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guyot J.-P. 2010. Fermented cereal products, p. 247–261 In Tamang J. P., Kailasapthy K. (ed.), Fermented foods and beverages of the world. CRC Press, Boca Raton, FL [Google Scholar]
- 24. Halász A., Baráth Á., Simon-Sarkadi L., Holzapfel W. 1994. Biogenic-amines and their production by microorganisms in food. Trends Food Sci. Technol. 5:42–49 [Google Scholar]
- 25. Humblot C., et al. 2005. 1H nuclear magnetic resonance spectroscopy-based studies of the metabolism of food-borne carcinogen 2-amino-3-methylimidazo[4,5-f]quinoline by human intestinal microbiota. Appl. Environ. Microbiol. 71:5116–5123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Humblot C., Guyot J. P. 2009. Pyrosequencing of tagged 16S rRNA gene amplicons for rapid deciphering of the microbiomes of fermented foods such as pearl millet slurries. Appl. Environ. Microbiol. 75:4354–4361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Iyer R., Tomar S. K. 2009. Folate: a functional food constituent. J. Food Sci. 74:R114–R122 [DOI] [PubMed] [Google Scholar]
- 28. Jacobsen C. N., et al. 1999. Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl. Environ. Microbiol. 65:4949–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jiang J., et al. 2010. Diversity of bile salt hydrolase activities in different lactobacilli toward human bile salts. Ann. Microbiol. 60:81–88 [Google Scholar]
- 30. Kanehisa M., Goto S. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kaushik J. K., et al. 2009. Functional and probiotic attributes of an indigenous isolate of Lactobacillus plantarum. PLoS One 4:e8099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Klaenhammer T. R., Barrangou R., Buck B. L., Azcarate-Peril M. A., Altermann E. 2005. Genomic features of lactic acid bacteria affecting bioprocessing and health. FEMS Microbiol. Rev. 29:393–409 [DOI] [PubMed] [Google Scholar]
- 33. Kleerebezem M., et al. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. U. S. A. 100:1990–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lambert J. M., Bongers R. S., de Vos W. M., Kleerebezem M. 2008. Functional analysis of four bile salt hydrolase and penicillin acylase family members in Lactobacillus plantarum WCFS1. Appl. Environ. Microbiol. 74:4719–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lebeer S., Vanderleyden J., De Keersmaecker S. C. 2008. Genes and molecules of lactobacilli supporting probiotic action. Microbiol. Mol. Biol. Rev. 72:728–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lei V., Jakobsen M. 2004. Microbiological characterization and probiotic potential of koko and koko sour water, African spontaneously fermented millet porridge and drink. J. Appl. Microbiol. 96:384–397 [DOI] [PubMed] [Google Scholar]
- 37. Lim E. M., Ehrlich S. D., Maguin E. 2000. Identification of stress-inducible proteins in Lactobacillus delbrueckii subsp. bulgaricus. Electrophoresis 21:2557–2561 [DOI] [PubMed] [Google Scholar]
- 38. Lorca G. L., Font de Valdez G., Ljungh A. 2002. Characterization of the protein-synthesis dependent adaptive acid tolerance response in Lactobacillus acidophilus. J. Mol. Microbiol. Biotechnol. 4:525–532 [PubMed] [Google Scholar]
- 39. Lucas P. M., et al. 2007. Agmatine deiminase pathway genes in Lactobacillus brevis are linked to the tyrosine decarboxylation operon in a putative acid resistance locus. Microbiology 153:2221–2230 [DOI] [PubMed] [Google Scholar]
- 40. Lucock M. 2000. Folic acid: nutritional biochemistry, molecular biology, and role in disease processes. Mol. Genet. Metab. 71:121–138 [DOI] [PubMed] [Google Scholar]
- 41. Makarova K., et al. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U. S. A. 103:15611–15616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McAuliffe O., Cano R. J., Klaenhammer T. R. 2005. Genetic analysis of two bile salt hydrolase activities in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 71:4925–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mohammad M. A., Molloy A., Scott J., Hussein L. 2006. Plasma cobalamin and folate and their metabolic markers methylmalonic acid and total homocysteine among Egyptian children before and after nutritional supplementation with the probiotic bacteria Lactobacillus acidophilus in yoghurt matrix. Int. J. Food Sci. Nutr. 57:470–480 [DOI] [PubMed] [Google Scholar]
- 44. Morita H., et al. 2008. Comparative genome analysis of Lactobacillus reuteri and Lactobacillus fermentum reveal a genomic island for reuterin and cobalamin production. DNA Res. 15:151–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mtshali P. S., Divol B., van Rensburg P., du Toit M. 2010. Genetic screening of wine-related enzymes in Lactobacillus species isolated from South African wines. J. Appl. Microbiol. 108:1389–1397 [DOI] [PubMed] [Google Scholar]
- 46. Nguyen T. T. T., et al. 2007. Effect of fermentation by amylolytic lactic acid bacteria, in process combinations, on characteristics of rice/soybean slurries: a new method for preparing high energy density complementary foods for young children. Food Chem. 100:623–631 [Google Scholar]
- 47. Nout M. J. R., Motarjemi Y. 1997. Assessment of fermentation as a household technology for improving food safety: a joint FAO/WHO workshop. Food Control 8:221–226 [PMC free article] [PubMed] [Google Scholar]
- 48. Ouwehand A. C., Kirjavainen P. V., Shortt C., Salminen S. 1999. Probiotics: mechanisms and established effects. Int. Dairy J. 9:43–52 [Google Scholar]
- 49. Perea Velez M., et al. 2007. Functional analysis of d-alanylation of lipoteichoic acid in the probiotic strain Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 73:3595–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pfeiler E. A., Azcarate-Peril M. A., Klaenhammer T. R. 2007. Characterization of a novel bile-inducible operon encoding a two-component regulatory system in Lactobacillus acidophilus. J. Bacteriol. 189:4624–4634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pfeiler E. A., Klaenhammer T. R. 2009. Role of transporter proteins in bile tolerance of Lactobacillus acidophilus. Appl. Environ. Microbiol. 75:6013–6016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Powers H. J. 2003. Riboflavin (vitamin B-2) and health. Am. J. Clin. Nutr. 77:1352–1360 [DOI] [PubMed] [Google Scholar]
- 53. Rohner F., Zimmermann M. B., Wegmueller R., Tschannen A. B., Hurrell R. F. 2007. Mild riboflavin deficiency is highly prevalent in school-age children but does not increase risk for anaemia in Cote d'Ivoire. Br. J. Nutr. 97:970–976 [DOI] [PubMed] [Google Scholar]
- 54. Rossi M., Amaretti A., Raimondi S. 2011. Folate production by probiotic bacteria. Nutrients 3:118–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rozen S., Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365–386 [DOI] [PubMed] [Google Scholar]
- 56. Saeed A. I., et al. 2006. TM4 microarray software suite. Methods Enzymol. 411:134–193 [DOI] [PubMed] [Google Scholar]
- 57. Songre-Ouattara L. T., et al. 2008. Enzyme activities of lactic acid bacteria from a pearl millet fermented gruel (ben-saalga) of functional interest in nutrition. Int. J. Food Microbiol. 128:395–400 [DOI] [PubMed] [Google Scholar]
- 58. Stack H. M., Kearney N., Stanton C., Fitzgerald G. F., Ross R. P. 2010. Association of beta-glucan endogenous production with increased stress tolerance of intestinal lactobacilli. Appl. Environ. Microbiol. 76:500–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sybesma W., et al. 2003. Increased production of folate by metabolic engineering of Lactococcus lactis. Appl. Environ. Microbiol. 69:3069–3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Torriani S., Felis G. E., Dellaglio F. 2001. Differentiation of Lactobacillus plantarum, L. pentosus, and L. paraplantarum by recA gene sequence analysis and multiplex PCR assay with recA gene-derived primers. Appl. Environ. Microbiol. 67:3450–3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tou E. H., et al. 2006. Study through surveys and fermentation kinetics of the traditional processing of pearl millet (Pennisetum glaucum) into ben-saalga, a fermented gruel from Burkina Faso. Int. J. Food Microbiol. 106:52–60 [DOI] [PubMed] [Google Scholar]
- 63. Turpin W., Humblot C., Thomas M., Guyot J. P. 2010. Lactobacilli as multifaceted probiotics with poorly disclosed molecular mechanisms. Int. J. Food Microbiol. 143:87–102 [DOI] [PubMed] [Google Scholar]
- 64. Valdez G. F., Taranto M. P. 2000. Probiotic properties of lactobacilli. Methods Biotechnol. 14:173–181 [Google Scholar]
- 65. Vrancken G., Rimaux T., Weckx S., De Vuyst L., Leroy F. 2009. Environmental pH determines citrulline and ornithine release through the arginine deiminase pathway in Lactobacillus fermentum IMDO 130101. Int. J. Food Microbiol. 135:216–222 [DOI] [PubMed] [Google Scholar]
- 66. Wall T., et al. 2007. The early response to acid shock in Lactobacillus reuteri involves the ClpL chaperone and a putative cell wall-altering esterase. Appl. Environ. Microbiol. 73:3924–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Whitehead K., Versalovic J., Roos S., Britton R. A. 2008. Genomic and genetic characterization of the bile stress response of probiotic Lactobacillus reuteri ATCC 55730. Appl. Environ. Microbiol. 74:1812–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]