Abstract
Two isolates of enterohemorrhagic Escherichia coli (EHEC) O104:H4 were isolated in France in 2004 and 2009. Both were characterized and compared to the strain which caused the German outbreak in 2011 and to other O104:H4 strains. This suggests that different O104:H4 EHEC strains were present several years prior to the 2011 outbreak.
TEXT
From May to July 2011, a large-scale outbreak of enterohemorrhagic Escherichia coli (EHEC) was observed in several European countries, mainly affecting northern Germany. Altogether, about 4,000 cases of EHEC infections and nearly 50 fatalities were reported (8). This outbreak also led to massive economic losses for farmers and to widespread public concern, after various vegetables and sprouts as well as a possible bioterrorist attack were publicly discussed as possible sources of the infection. Recent epidemiological studies singled out imported fenugreek seeds, although this has not yet been substantiated by laboratory evidence (6).
In order to understand the evolution and possibly the origin of the recent outbreak strain, it is of great interest to identify related or ancestral isolates. Only a few cases of infections caused by E. coli O104:H4 were described earlier than 2011. One was a case of hemolytic-uremic syndrome (HUS) from South Korea (1), but the causative strain appeared not to be closely related to the current European outbreak strain, differing in toxin carriage, resistance properties, and pulsed-field gel electrophoresis profiles (10). Another one originated from Italy in 2009 (18). A recently described and fully sequenced strain, HUSEC 41 isolate 01-0991, originated from a HUS case in Germany in 2001 (4, 11).
Prior to the 2011 outbreak, E. coli O104:H4 was also detected in two isolated clinical cases in France. One patient was a 6-year-old male child from the Lyon area treated for HUS in 2009. He was hospitalized, treated with azithromycin, and cured. The other one was an adult male patient with hemorrhagic colitis from the town of Lille, in northern France, in 2004. The clinical course and outcome are not known. Both isolates were characterized by microarray analysis using a previously described system (9), and they yielded identical hybridization patterns. They were also compared to the current European outbreak strain, as represented by 13 identical isolates. These were three serial isolates from a young female patient from Dresden, Saxony, Germany, who was admitted with bloody diarrhea and HUS after traveling to northern Germany, and 10 isolates from clinical cases from northeastern Germany (provided by D. Bandt, Frankfurt/Oder, Germany).
Recently, the similarity of E. coli 55989, an enteroaggregative strain that was isolated in Central Africa in the late 1990s (13, 19), to the European outbreak strain (GenBank entries AFOB00000000 and GL989507.1) was noted based on genome sequence data (4, 5) and particularly identical multilocus sequence typing alleles (ST678) (https://github.com/ehec-outbreak-crowdsourced/BGI-data-analysis/wiki/MLST-and-serotyping-55989-in-silico and reference 4). Thus, this strain and the German HUSEC 41 isolate 01-0991 (4) were included in the comparison based on analyses of their genome sequences (GenBank numbers CU928145 and AFPS01000000, respectively).
Like E. coli 55989, the German HUSEC 41 isolate 01-0991, and the European outbreak strain, the two French O104:H4 isolates belonged to sequence type 678. All these strains carried wzx-O104 and fliC-H04 determinants as well as genes encoding a major fimbrial subunit (lpfA) and secreted serine proteases (pic, sepA, and sigA). Moreover, they were characterized by the absence of eae (intimin, locus of enterocyte effacement) and of Shiga-like toxin 1 (stx-A/B1).
The two French isolates carried Shiga-like toxin 2 (stx-A/B2) genes and the microcin operon (mchB, mchC, and mchF). These genes were also present in all tested isolates of the outbreak strain and in its published genome sequences, as well as in the German HUSEC 41 isolate 01-0991. They were absent from the E. coli 55989 genome sequence.
All strains carried aggR (encoding aggregative-adherence fimbriae; determined for the French isolates by PCR [14]), but there were differences with regard to the carriage of pAA plasmids. The two French isolates were positive for astA (enteroaggregative heat stable enterotoxin), which was absent from isolates of the European outbreak strain. It can be assumed that the French isolates carry a pAA plasmid similar or identical to those of E. coli 55989 (GenBank AF411067.1) and 01-0991 (4), which include astA as well as a type III (2) aggDCBA operon. In contrast, genome sequences of the outbreak strain indicate the presence of a type I operon (16, 17) and the lack of astA.
Further differences included the carriage of genes associated with resistance toward antibiotic compounds. The two French isolates carried none of the resistance genes covered by the array (9). The 2004 isolate was completely susceptible to nalidixic acid, whereas the 2009 isolate was resistant. The European outbreak strain harbored an extended-spectrum-β-lactamase gene, blaCTX-M-15 (4, 7), which is currently spreading worldwide across populations of different enterobacteria (15). This strain also carried an additional β-lactamase gene (blaTEM-1), genes encoding dihydrofolate reductase type 7 (dfrA7) and dihydropteroate synthetase types 1 and 2 (sul1 and sul2), and streptomycin resistance genes (strA [apha3] and strB [apha6]), as well as a gene for a tetracycline efflux protein (tetA). Analyses of the genome sequences showed that E. coli 55989 harbored the tetracycline resistance gene tetB, while 01-0991 was positive for strA, strB, sul2, and blaTEM-1.
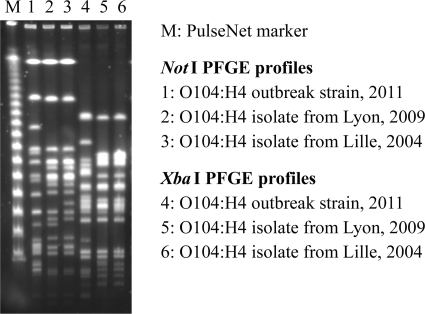
In conclusion, the two French isolates described herein are largely identical to the recently described HUSEC 41 isolate 01-0991 from Germany. It is tempting to regard these three isolates as some kind of intermediate form or missing link between the European outbreak strain and its putative ancestor, an E. coli 55989-like EAEC strain. However, there are differences in astA- and aggR-bearing plasmids, with the 2011 outbreak strain carrying another pAA than all other O104:H4 isolates, as well as in pulsed-field gel electrophoresis profiles (Fig. 1). It is also noteworthy that the French isolates lack blaTEM-1 and the other resistance genes detected in 01-09591, although they were isolated more recently. Therefore, it is unlikely that a direct line of ancestry leads from E. coli 55989 through 01-0991 and the French isolates to the recent outbreak strain. As proposed previously (12), it can be assumed that several O104:H4 EHEC lineages emerged from O104:H4 EAEC ancestors which differ most obviously in pAA and resistance gene carriage. Sporadic observations from France, Germany (4, 11), Italy (18), and Korea (10) indicate that several such strains might have persisted for several years without receiving much attention, apparently due to their rarity. In contrast to other EHEC strains, there are no reports of an O104:H4 EHEC strain isolated from cattle. This does not rule out a very rare occurrence, a presence in other animals, or a presence in cows from other parts of the world. However, because of the lack of such reports and because of their EAEC parentage (5), it should be investigated whether humans rather than cattle were the actual reservoir of O104:H4.
Fig. 1.
NotI (3) and XbaI pulsed-field gel electrophoresis patterns of the 2011 European outbreak strain, the isolate from Lyon (2009), and the isolate from Lille (2004).
Acknowledgments
We thank D. Bandt (Institute for Medical Diagnostics Oderland, Frankfurt/Oder, Germany) for clinical isolates from the recent outbreak as well as L. Geue (German National Reference Laboratory for verotoxin-forming Escherichia coli, Wusterhausen, Germany) for helpful discussions. We acknowledge E. Müller, I. Engelmann, G. Rößler, and J. Sachtschal (Alere Technologies GmbH, Germany) as well as N. Nguyen (Alere France) for excellent help and technical assistance.
Stefan Monecke, Peter Slickers, and Ralf Ehricht are employees of Alere Technologies GmbH.
There was no external funding for this study. Each institution granted the time needed to perform this study. Decisions on study design, data collection and analysis, publication and preparation of the manuscript were agreed upon by the authors involved without influence by the institutions.
Footnotes
Published ahead of print on 14 October 2011.
REFERENCES
- 1. Bae W. K., et al. 2006. A case of hemolytic uremic syndrome caused by Escherichia coli O104:H4. Yonsei Med. J. 47:437–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bernier C., Gounon P., Le Bouguenec C. 2002. Identification of an aggregative adhesion fimbria (AAF) type III-encoding operon in enteroaggregative Escherichia coli as a sensitive probe for detecting the AAF-encoding operon family. Infect. Immun. 70:4302–4311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bidet P., et al. 2005. Characterization of Escherichia coli O157:H7 isolates causing haemolytic uraemic syndrome in France. J. Med. Microbiol. 54:71–75 [DOI] [PubMed] [Google Scholar]
- 4. Bielaszewska M., et al. 2011. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect. Dis. 11:671–676 [DOI] [PubMed] [Google Scholar]
- 5. Brzuszkiewicz E., et al. 29 June 2011, posting date Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: entero-aggregative-haemorrhagic Escherichia coli (EAHEC). Arch. Microbiol. [Epub ahead of print.] doi:10.1007/s00203-011-0725-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bundesinstitut fur Risikobewertung 5 July 2011, release date EHEC O104:H4 outbreak event in Germany clarified: sprouts of fenugreek seeds imported from Egypt as underlying cause. Bundesinstitut fur Risikobewertung, Berlin, Germany: http://www.bfr.bund.de/en/press_information/2011/21/ehec_o104_h4_outbreak_event_in_germany_clarified__sprouts_of_fenugreek_seeds_imported_from_egypt_as_underlying_cause-83273.html [Google Scholar]
- 7. Dallenne C., Da Costa A., Decre D., Favier C., Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65:490–495 [DOI] [PubMed] [Google Scholar]
- 8. European Centre for Disease Prevention and Control 27 July 2011, posting date Shiga toxin-producing E. coli (STEC): update on outbreak in the EU (27 July 2011). ECDC. http://ecdc.europa.eu/en/activities/sciadvice/Lists/ECDC%20Reviews/ECDC_DispForm.aspx?List=512ff74f-77d4-4ad8-b6d6-bf0f23083f30&ID=1166&RootFolder=%2Fen%2Factivities%2Fsciadvice%2FLists%2FECDC%20Reviews
- 9. Geue L., et al. 2010. Rapid microarray-based genotyping of enterohemorrhagic Escherichia coli serotype O156:H25/H−/Hnt isolates from cattle and clonal relationship analysis. Appl. Environ. Microbiol. 76:5510–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim J., et al. 2011. Escherichia coli O104:H4 from 2011 European outbreak and strain from Republic of Korea. Emerg. Infect. Dis. 17:1755–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mellmann A., et al. 2008. Analysis of collection of hemolytic uremic syndrome-associated enterohemorrhagic Escherichia coli. Emerg. Infect. Dis. 14:1287–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mellmann A., et al. 2011. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One 6:e22751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mossoro C., et al. 2002. Chronic diarrhea, hemorrhagic colitis, and hemolytic-uremic syndrome associated with HEp-2 adherent Escherichia coli in adults infected with human immunodeficiency virus in Bangui, Central African Republic. J. Clin. Microbiol. 40:3086–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Muller D., et al. 2007. Identification of unconventional intestinal pathogenic Escherichia coli isolates expressing intermediate virulence factor profiles by using a novel single-step multiplex PCR. Appl. Environ. Microbiol. 73:3380–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Naseer U., Sundsfjord A. 2011. The CTX-M conundrum: dissemination of plasmids and Escherichia coli clones. Microb. Drug Resist. 17:83–97 [DOI] [PubMed] [Google Scholar]
- 16. Nataro J. P., et al. 1993. Aggregative adherence fimbria I expression in enteroaggregative Escherichia coli requires two unlinked plasmid regions. Infect. Immun. 61:1126–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Savarino S. J., Fox P., Deng Y., Nataro J. P. 1994. Identification and characterization of a gene cluster mediating enteroaggregative Escherichia coli aggregative adherence fimbria I biogenesis. J. Bacteriol. 176:4949–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Scavia G., Morabito S. T. R., Michelacci V., Marziano M., Minelli F. 2011. Similarity of Shiga toxin-producing Escherichia coli O104:H4 strains from Italy and Germany. Emerg. Infect. Dis. 17:1957–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Touchon M., et al. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5:e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]