Abstract
Fusion of herpesviruses with their target cells requires a minimum of three glycoproteins, namely, gB and a complex of gH and gL. Epstein-Barr virus (EBV) fusion with an epithelial cell requires no additional virus glycoproteins, and we have shown previously that it can be initiated by an interaction between integrin αvβ6 or αvβ8 and gHgL. We now report that integrin αvβ5 can also bind to gHgL and trigger fusion. Binding of gHgL to integrins is a two-step reaction. The first step, analyzed by surface plasmon resonance, was fast, with high association and low dissociation rate constants. The second step, detected by fluorescence spectroscopy of gHgL labeled at cysteine 153 at the domain I-domain II interface with the environmentally sensitive probes acrylodan and IANBD, involved a slower conformational change. Interaction of gHgL with neutralizing monoclonal antibodies or Fab′ fragments was also consistent with a two-step reaction involving fast high-affinity binding and a subsequent slower conformational change. None of the antibodies bound to the same epitope, and none completely inhibited integrin binding. However, binding of each decreased the rate of conformational change induced by integrin binding, suggesting that neutralization might involve a conformational change that precludes fusion. Overall, the data are consistent with the interaction of gHgL with an integrin inducing a functionally important rearrangement at the domain I-domain II interface.
INTRODUCTION
Epstein-Barr virus (EBV) is an orally transmitted human gammaherpesvirus that is carried by the majority of the adult population worldwide. Many primary infections are asymptomatic, but the virus is an etiologic agent of infectious mononucleosis and is also associated with and implicated in development of both lymphoid and epithelial malignancies (reviewed in reference 38). B lymphocytes and epithelial cells are its primary targets.
Like all herpesviruses, EBV enters its target cells by fusion. Fusion with a B cell first requires endocytosis, is sensitive to chlorpromazine, and occurs at acidic pH. Fusion with an epithelial cell is not sensitive to chlorpromazine and occurs at neutral pH, possibly at the cell surface (28). In both cases, it requires the activities of trimers of the glycoprotein gB and heterodimers of the glycoproteins gH and gL (18). Homologs of these three glycoproteins are thought to be responsible for fusion of all herpesviruses and are referred to as the core fusion machinery (45). Glycoprotein B has been posited to be the protein that is proximal to the fusion event, in large part because of its structural similarity to the vesicular stomatitis virus fusion protein G (3, 16, 39), but full fusion activity is achieved only in the presence of gHgL.
One of the major differences in entry of individual herpesviruses into various cell types is the way in which the core fusion machinery is initially activated. Several herpesviruses, including herpes simplex virus (HSV) and EBV, can require a fourth protein for activation of fusion. Herpes simplex virus requires glycoprotein gD and the interaction of gD with one of four known unique cell surface molecules (44). Current models for HSV propose that engagement of gD induces a conformational change in the glycoprotein and transmission of an activating signal to gB and gHgL (21). This is followed by insertion of putative fusion loops in gB into the membrane (15) and a physical association of gB with gHgL, which somehow leads to full fusion (1). Fusion of EBV with a B cell requires glycoprotein gp42 in addition to gB and gHgL, though in contrast to HSV gD, which exists as a separate structure, gp42 constitutively forms a trimer with gHgL. Interaction of gp42 with HLA class II molecules is thought to be the initiator of fusion, and the crystal structures of gp42 in the presence or absence of HLA class II molecules suggest that, in parallel with gD, binding leads to a conformational change in gp42 (20, 31). Events subsequent to this have not been studied as well as they have for HSV, but they might be assumed to be broadly similar.
Fusion of EBV with an epithelial cell is initiated differently. Instead of using gp42 as an intermediary, a direct interaction of gHgL with integrin αvβ6 or αvβ8 has been shown to trigger the event (8). Fusion of epithelial cells lacking human integrins and transfected with EBV gHgL and gB can be induced directly by addition of soluble forms of αvβ6 or αvβ8. The presence of gp42 in a trimeric complex with gHgL prevents triggering by this mechanism, and to accommodate this, the EBV virion carries not only trimeric complexes but also dimeric complexes of gHgL that lack gp42 (48). Modulation of the ratio of the two complexes in virion particles influences the preferred tropism of the virus, and switching such tropism is accomplished by the cellular environment in which the virus replicates. In B cells, the levels of trimeric complexes available for incorporation into the virion are reduced as a result of interaction with HLA class II molecules and trafficking with these molecules, presumably to the peptide loading compartment, which is rich in proteases. In HLA class II-negative cells, such as epithelial cells, they are not (4).
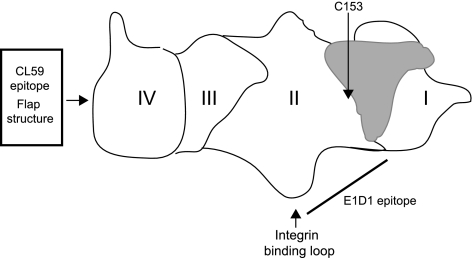
The ability to trigger fusion in the absence of an intermediary virus protein such as gp42 or gD facilitates analysis of any conformational changes that might be induced in gHgL as a result of integrin engagement. Although HSV gH contains an integrin-binding RGD motif, mutation of the sequence has shown that the motif plays no role in virus penetration (12). The recently completed crystal structures of EBV and the homologous HSV and pseudorabies virus (PRV) gHgL complexes have as yet provided no clear picture of how they function, in that none resembles any known viral fusogen (2, 9, 27). However, some potentially relevant features of the complexes were inferred following comparison of a core fragment of the four-domain structure of PRV and EBV gHgL (Fig. 1) with that of HSV gHgL, in which domain boundaries have been interpreted differently (2). A “long crossover segment of polypeptide chain, the flap” (2) is found in the carboxy-terminal domain IV of the complex in all three viruses, adjacent to the virus membrane, and has been postulated to represent a flexible structure that, if moved, would expose a conserved, potentially membrane-interactive hydrophobic surface. Mutations in this flap affect fusion supported by EBV gHgL, and a monoclonal antibody (MAb) (CL59) which maps to the flap can neutralize infection of an epithelial cell (51, 52). What may be a “syntaxin-like bundle” has been identified in domain II (2), and the integrin binding site of EBV gHgL is also in a prominent loop in domain II (27). The monoclonal antibody E1D1 reduces binding to integrins (5), binds only to gH complexed with gL, and has a reduced ability to recognize gH mutated at residue 65, located in the domain I-domain II linker helix (34). The antibody thus presumably binds surfaces at the junctional region between domain I, which comprises the amino-terminal 65 residues of gH and the entirety of gL, and domain II. The putative binding site for gp42 is also probably nearby, given the ability of gp42 to block integrin binding (5). Mutations that map to the domain I-domain II interface affect fusion (34), and it has been suggested that a domain I-domain II conformational change could be part of the mechanism for triggering membrane fusion (27).
Fig. 1.
Cartoon of the structure of gHgL. The four domains of gHgL are indicated, as are the positions of the flap structure, the CL59 epitope, and the region of the protein thought to contain the E1D1 epitope. The position of C153 is shown, and gL is depicted in gray.
The crystal structure of EBV gHgL indicates that there is a single unpaired cysteine residue close to the domain I-domain II interface. We took advantage of this to label the residue with thiol-reactive, environmentally sensitive fluorescent probes. We report that binding of gHgL to integrins is a two-step reaction. The first, detected by surface plasmon resonance (SPR), is fast, with high association and low dissociation rate constants. The second, detected by fluorescence spectroscopy, involves a slower conformational change. Interaction between a truncated form of gH together with gL (gHtgL) and monoclonal antibodies or Fab′ fragments was also consistent with a two-step reaction involving fast, high-affinity binding and a subsequent, slower conformational change involving the domain I-domain II interface. Antibodies decreased the rate of conformational change induced by integrin binding, suggesting that neutralization might involve a premature or nonproductive conformational change. We also report that integrin αvβ5, in addition to integrins αvβ6 and αvβ8, can bind to gHgL and trigger fusion.
MATERIALS AND METHODS
Cells and virus.
AGS cells, which are human gastric carcinoma cells (American Type Culture Collection) that have been cured of parainfluenza virus type 5 infection (54) by treatment with ribavirin, and CHO-K1 cells were grown in Ham's F-12 medium (Sigma). SVKCR2, a simian virus 40 (SV40)-transformed keratinocyte cell line stably transfected with a plasmid expressing CD21 (24), was grown in Joklik's modified Eagle's medium supplemented with 10 ng/ml cholera toxin (Sigma). Hybridoma cells, EBV-negative Akata Burkitt lymphoma-derived cells (42), and Akata-GFP cells, carrying a recombinant EBV in which the thymidine kinase gene is interrupted with a double cassette expressing neomycin resistance and green fluorescent protein (GFP) (29), were grown in RPMI 1640 (Sigma). Akata-GFP medium was also supplemented with 500 μg active G418 (Mediatech). 293-B6 AVAP, 293-B3 AVAP (13), and 293-B8 AVAP cells (30) (a gift of Stephen Nishimura, University of California at San Francisco), which secrete truncated αvβ8, αvβ6, and αvβ3, respectively, conjugated to alkaline phosphatase (AP), were grown in Dulbecco's modified Eagle's medium (DMEM; Sigma) supplemented with 1% nonessential amino acids. Media for all mammalian cells except for SVKCR2 cells were also supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco). SVKCR2 medium was supplemented with heat-inactivated HyClone Cosmic calf serum (Sigma). Insect Sf9 cells were grown in Sf900 II medium (Invitrogen) and infected at a multiplicity of 3 with baculovirus expressing gHtgL (37) (a gift of Andrew Morgan, University of Bristol, England) or at a multiplicity of 2 with equal ratios of baculoviruses expressing truncated forms of αv and β5 cloned in frame with fos and jun dimerization domains, respectively (26) (a gift of Glen Nemerow, Scripps Research Institute). Spent supernatant was harvested at 72 h and clarified by low-speed centrifugation. Akata-GFP virus was collected from clarified culture medium of cells that had been induced by treatment with anti-human immunoglobulin (22).
Antibodies.
Antibodies used were MAbs E1D1 to gHgL (23), CL40 and CL59 to gH (29), F-2-1 to gp42 (23), CL55 to gB (51), LM142 (Millipore) and L230 (49) (American Type Culture Collection) to integrin αv, and LS-C44264 to placental AP (LifeSpan BioScience). CL59, E1D1, CL40, F-2-1, CL55, and L230 were purified by affinity chromatography on protein A columns coupled to agarose (RepliGen). Rabbit antibody to a peptide corresponding to residues 518 to 528 of gH (33) was affinity purified both on protein A and on a peptide coupled to Affigel-15 (Bio-Rad). The binding of this antibody to gHtgL was determined by SPR to have a KD (dissociation constant) of 1.2 × 10−6 M. Sheep anti-mouse coupled to phycoerythrin (Jackson ImmunoResearch) was also used.
Integrin and gHtgL purification.
Integrins αvβ3, αvβ6, and αvβ8 were harvested from the spent culture medium of 293 cells that had been switched to serum-free medium after reaching confluence. Integrin αvβ5 was harvested from spent culture medium of Sf9 cells that had been infected for 72 h at a multiplicity of 2 with equal ratios of baculoviruses expressing αv and β5. Culture media were concentrated by ultrafiltration through a YM-30 membrane (Millipore), and integrins were isolated by affinity chromatography on Affigel-10 (Bio-Rad) conjugated to MAb L230. Integrins were allowed to bind to affinity resin for 12 to 16 h at 4°C on a rocking platform, and beads were then placed in a chromatography column and washed with 20 column volumes of 20 mM Tris-HCl, pH 7.2, 1 mM CaCl2, 1 mM MgCl2, and 1 mM MnCl2 containing 300 mM NaCl and with 20 column volumes of the same buffer containing 600 mM NaCl. Protein was eluted with 1% acetic acid, and the pH was immediately adjusted to 7.0 with 1.5 M Tris-HCl buffer, pH 9.0. Eluted integrins were dialyzed against phosphate-buffered saline (PBS) and then against 20 mM HEPES-NaOH buffer (pH 7.4) containing 150 mM NaCl. Protein was concentrated, aliquoted, and stored at −80°C. Soluble gHtgL was purified by affinity chromatography on lentil lectin Sepharose (Sigma) and eluted with buffer containing 10 mM methyl-α-mannopyranoside (Sigma) as previously described (8).
Assay of gHtgL binding to cells.
Binding of gHtgL to AGS cells was done as described previously (5). Briefly, AGS cells were trypsinized, allowed to recover for 1 h at 37°C in growth medium, and incubated for 1 h with protein on ice. Cells were then incubated sequentially with MAb CL59 and sheep anti-mouse antibody coupled to phycoerythrin, with washes between incubations and before flow cytometric analysis.
Assay of fusion.
Epithelial cell fusion was measured as described previously (8, 51). Briefly, CHO-K1 cells were transfected with plasmids pCAGGS-gB, pCAGGS-gH, and pCAGSS-gL and overlaid with integrins at 24 h posttransfection, and 20 h later, they were fixed and stained with MAb CL55 to gB and examined visually to count the number of transfected cells and the percentage of transfected cells containing 4 or more nuclei.
Neutralization assays.
Neutralization was measured using Akata-GFP virus. Virus was preincubated for 1 h at 4°C with MAb at a final ratio of 1.5 × 109 genome copies to 100 μg of antibody, whole molecule, or Fab′ and then added to 5 × 105 EBV-negative Akata cells or SVKCR2 cells grown to approximately 30% confluence in a 4-chamber BD Falcon slide (Fisher). Virus and cells were incubated for 2 h at 37°C in a minimal volume of medium, and then growth medium was added. Two days later, the percentage of Akata cells expressing GFP was measured by flow cytometry, and the percentage of SVKCR2 cells expressing GFP was counted under a fluorescence microscope. Neutralization was expressed as the percentage of cells expressing GFP in the absence of antibody (% control).
SPR spectroscopy.
Kinetic measurements of the interaction between gHtgL and integrins were made with a Biacore 2000 instrument (Biacore AB). MAb LS-C44264 to AP was used to capture integrins αvβ3AP, αvβ6AP, and αvβ8AP on the surface of a sensor chip, and the nonblocking monoclonal antibody LM142 to αv integrin was used to attach integrin αvβ5 fos/jun to the surface. These two antibodies, as well as monoclonal antibodies E1D1, CL59, and CL40, were immobilized on a research-grade CM-5 sensor chip by amino coupling using an immobilization wizard, with 1,200 relative units (RU) as a target for immobilization. The first flow cell (FC1) was always used as a reference (no antibody immobilized), and the response to FC1 was automatically subtracted by Biacore software from data obtained from the other three flow cells. Measurements were made at 25°C. Integrins were injected at a flow rate of 10 to 20 μl/min; gHtgL was injected immediately after integrin capture by using the low-dispersal injection setting (kinject) at a flow rate of 50 μl/min. The Biacore instrument is designed to inject a certain volume of a protein solution at a certain flow rate. When injection is finished, the instrument monitors the dissociation phase. After every run, the sensor chip surface was regenerated by injection of 6 M guanidine chloride in 25 mM HEPES-NaOH, pH 7.2 (50 μl at a flow rate of 100 μl/min), followed by additional washing. The baseline remained stable during 30 to 40 runs. Integrins and gHtgL were centrifuged before use and diluted in running buffer if necessary (10 mM HEPES-NaOH, pH 7.4, 150 mM NaCl, 0.005% surfactant P20 [Biacore, GE Healthcare]). Each sample was degassed before injection. The cognate peptide used to block integrin binding was CYRVTEKGDEHVLSL, and the scrambled peptide was CKVTRSLYEGVHELD (8).
Acrylodan and IANBD labeling.
Labeling with acrylodan (6-acryloyl-2-dimethylaminonaphthalene; AnaSPec) was done as described previously (17). At all steps, the acrylodan and the labeled protein were handled with minimal exposure to light. Prior to mixing with acrylodan, the gHtgL solution was saturated with air for 5 to 10 min to oxidize cysteines forming S-S bridges. Labeling was conducted at 4°C for 3 h on a rocking platform. Unbound acrylodan was separated from protein by size-exclusion chromatography, followed by dialysis against PBS. The average stoichiometry of labeling (gHtgL to acrylodan) was 1:1.3 (n = 4). Labeling with N,N′-dimethyl-N-(iodoacetyl)-N′-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)ethylenediamine (IANBD amide; Invitrogen) was done according to the manufacturer's instructions. The stoichiometry of labeling (gHtgL to IANBD) was 1:0.85. Before labeling, the protein solution was oxidized with air. Unbound probe was removed by size-exclusion chromatography followed by dialysis against PBS.
Fluorescence spectroscopy.
Steady-state emission spectra and “time-based” emission spectra were measured at room temperature or at 25°C, using a Photon Technology Inc. Strobe Master Lifetime spectrometer with an SE-900 steady fluorescence option. The excitation bandwidth was set at 3 nm, and the emission bandwidth was set at 5 nm. The excitation wavelength for acrylodan was set at 359 nm, with emission at 380 to 600 nm; the excitation wavelength for IANBD was set at 505 nm, with emission at 515 to 700 nm. Proteins in buffer were maintained in a 1-cm quartz cuvette (Starna Cells) with constant stirring. The buffer spectrum was subtracted from all obtained protein spectra; an integrin or MAb spectrum was subtracted from spectra of integrin-gHtgL or MAb-gHgL reactions, and all data shown represent gHtgL fluorescence alone. Subtracted emission spectra and obtained “time-based” spectra were corrected by PTI software for energy differences and for spectral intensity distribution to render them free of excitation artifacts. No difference in resulting emission spectra was observed if a binding reaction was conducted in PBS or HEPES-buffered saline.
Curve fitting.
BIAevaluation 4.1 software was used for the Biacore trace alignments and to zero the baseline. All traces were in full accordance with a 1:1 Langmuir model. To decide whether or not integrin-gHtgL binding is a one-step or two-step reaction, the formation part of each trace was fitted to single- and double-exponential functions by using the curve-fitting program Kaleida Graph 3.1 (Synergy Software, PA). The single-exponential fit was equally good compared to the double-exponential one and was accepted as a model. The “on” and “off” rate constants were derived with BIAevaluation 4.1 software. Eight to 16 sensorgrams were analyzed simultaneously. Residuals of the single- or double-exponential fitting, as well as least-squares fitting of data to the linear equation, were obtained with Kaleida Graph 3.1.
Fab′ fragment cleavage and purification.
Fab′ fragments of CL59 (IgG2a) and CL40 (IgG1) were generated according to the manufacturer's instructions by digestion with papain or ficin, using a Pierce Fab′ preparation kit or mouse IgG1 Fab′ preparation kit (Pierce Chemical Company), respectively, and were purified by chromatography on protein A Sepharose. Purified proteins were dialyzed against PBS and analyzed by polyacrylamide gel electrophoresis under nonreducing conditions and without boiling.
RESULTS
In addition to integrins αvβ6 and αvβ8, a third integrin, αvβ5, can also bind to gHgL and trigger fusion.
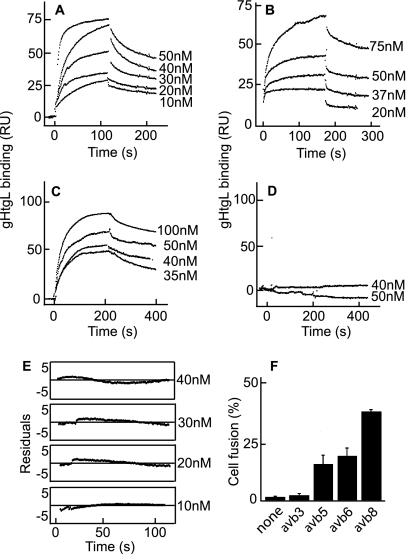
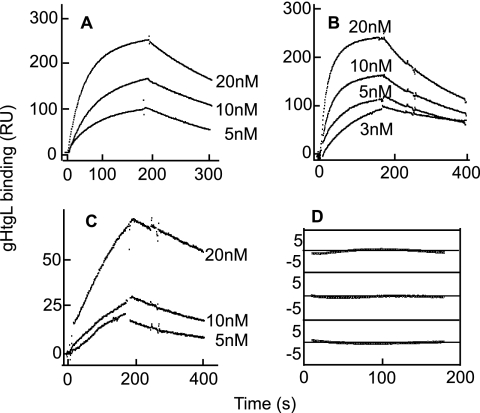
We previously reported that soluble forms of integrins αvβ6 and αvβ8, but not αvβ3, can bind to residues of gH which include a KGD motif and are part of an exposed loop in domain II. Application of function-blocking antibodies suggested but could not confirm that the one other KGD/RGD-binding integrin that might bind gHgL was αvβ5. To test this possibility further, we obtained baculoviruses that expressed truncated αv and truncated β5. Purified αvβ5 was captured by a non-function-blocking antibody to αv coupled to a Biacore sensor chip, and purified αvβ3, αvβ6, and αvβ8 were captured by an antibody to AP. A kinetic analysis of binding to increasing concentrations of soluble gHtgL indicated that gHtgL interacted with integrin αvβ5 as well as αvβ6 and αvβ8 (Fig. 2A to C). No interaction was detected with αvβ3 (Fig. 2D). Integrin αvβ5 was also able to trigger cell-to-cell fusion of epithelial cells, mediated by gB and gHgL (Fig. 2F). To determine if integrin binding occurred in one or more than one step, the formation sections of the sensorgrams were fitted to single- or double-exponential functions by using the curve-fitting program Kaleida Graph 3.1. Comparison of residuals, representing the difference between the theoretical curve and the experimental data, showed that the data fit to a single-exponential function (Fig. 2E).
Fig. 2.
Integrin αvβ5 in addition to integrins αvβ6 and αvβ8 binds to gHtgL with single-exponential kinetics and triggers cell-to-cell fusion mediated by gHgL and gB. SPR analysis was performed to evaluate binding of different concentrations of gHtgL to integrin αvβ5 (A), integrin αvβ6 (B), integrin αvβ8 (C), and integrin αvβ3 (D) immobilized to sensor chips by antibody. (E) Differences between the theoretical fit to a single-exponential function and experimental data (residuals) for the formation portion of each SPR trace at different concentrations of gHtgL added to αvβ5. (F) Fusion of CHO-K1 cells transfected with plasmids expressing gB and gHgL and overlaid for 20 h with integrins. Cells were fixed and stained with MAb to gB, and the percentage of cells expressing gB that contained 4 or more nuclei (% fusion) was calculated.
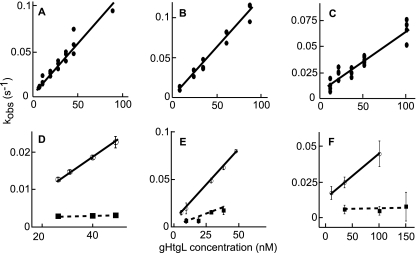
To obtain rate constants of gHtgL binding and to calculate the KD, kobson values were plotted against gHtgL concentrations (Fig. 3). Data were compiled from a minimum of 10 independent experiments for each integrin. The dependence of kobson on the concentration of gHtgL was described by a linear function, which confirmed that it represents a single-step reaction. The intersection with the y axis corresponds to the dissociation rate constant (koff), and the slope corresponds to the associate rate constant (kon) (Fig. 3A to C). The rate constants and thermodynamic KD values were similar for αvβ5, αvβ6, and αvβ8 (Table 1) and affinities were much higher than those of vitronectin, a natural ligand of these integrins which binds with a KD of 10−7 M (46). No correlation between affinity and induction of fusion could be found. To confirm that the KGD motif of gH can compete for binding to αvβ5, as shown previously for αvβ6 and αvβ8 by inhibition of cell binding and infection, gHtgL binding to integrins was also analyzed by SPR with a 40 μM concentration of a peptide corresponding to residues 184 to 196 of gH or a scrambled form of the same peptide. In the presence of the scrambled peptide, the kinetic characteristics of gHtgL binding were similar to those in the absence of peptide (Fig. 3D to F). In contrast, the KGD peptide inhibited the interaction between gHtgL and αvβ5, in addition to that with αvβ6 and αvβ8.
Fig. 3.
Linear plot of kobson versus gHtgL concentration for the gHtgL interaction with αvβ5, αvβ6, or αvβ8. Cognate but not scrambled peptides inhibited binding. Sensorgrams show gHtgL binding to αvβ5 (A and D), αvβ6 (B and E), and αvβ8 (C and F) submitted to global fit analysis (12 to 24 traces at a time), with each point representing a result from one analysis by the global fit function of BIAevaluation. Closed circles in upper panels represent binding of gHtgL alone to integrins. Open circles in lower panels represent binding in the presence of scrambled peptide, and squares represent binding in the presence of cognate peptide. Correlation coefficients for the least-squares fits are 0.97 (A), 0.99 (B), and 0.96 (C). In panels D to F, the correlation coefficient for the scrambled peptide is 0.99, and those for the cognate peptide are 0.99, 0.93, and 0.99, respectively.
Table 1.
Kinetic constants for EBV gHtgL binding to human soluble truncated integrinsa
| Integrin | kon (M−1 s−1) | koff (s−1) | KD (M) |
|---|---|---|---|
| αvβ5 | 930,000 ± 56,000 | 0.0040 ± 0.0002 | (4.3 ± 0.47) × 10−9 |
| αvβ6 | 1,109,600 ± 30,000 | 0.0027 ± 0.0015 | (2.4 ± 1.30) × 10−9 |
| αvβ8 | 640,000 ± 50,000 | 0.0040 ± 0.0002 | (6.0 ± 0.72) × 10−9 |
| αvβ3 | No binding |
Data are means ± standard deviations (SD).
A slow step of gHtgL interaction with integrins can be detected by fluorescence spectrometry.
The model for fusion mediated by HSV gHgL and gB proposes that it is initiated with a conformational change in gD induced by receptor binding. Similarly, B cell fusion mediated by EBV gHgL and gB is thought to be initiated with a conformational change in gp42 induced by binding to HLA class II molecules. It therefore seemed plausible that the integrin-mediated initiation of epithelial fusion by EBV glycoproteins might result in a conformational change in gHgL. If so, the gHgL-integrin interaction should occur in a least two steps: binding and a subsequent conformational change. The analysis of binding by SPR detected only a single step. However, this method has a limited ability to measure rate constants of very fast and very slow reactions. To determine whether the interaction between gHtgL and epithelial integrins αvβ5, αvβ6, and αvβ8 produced any conformational changes in gHtgL, gHtgL was labeled with acrylodan, an environmentally sensitive fluorescent probe which can be coupled to the reduced -SH group of a cysteine (17, 36). The average stoichiometry of gHtgL to acrylodan was 1:1.3 (n = 4). This is consistent with the crystal structure of EBV gHgL (27), in which only one cysteine, at position 153 of gH, is reduced under oxidative conditions.
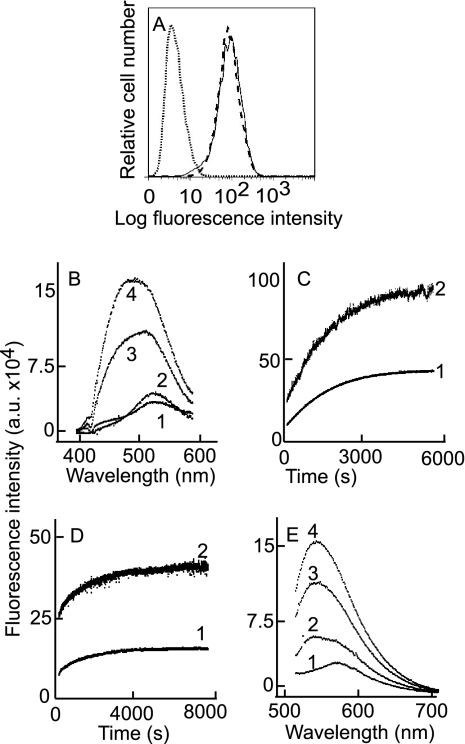
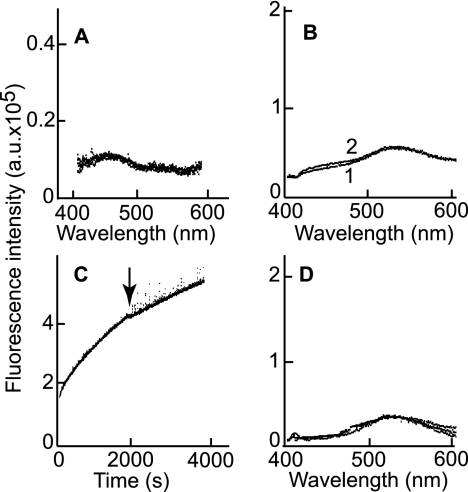
Labeling of gHtgL with acrylodan did not affect its binding to AGS epithelial cells as judged by flow cytometry with CL59, which recognizes a conformational epitope (51) (Fig. 4A), suggesting that it still had the native conformation. The association and dissociation rate constants of binding of integrin αvβ6 to unlabeled gHtgL and acrylodan-labeled gHtgL, as measured by SPR, were also similar. KD values for binding were 2.4 × 10−9 ± 1.3 × 10−9 M and 1.53 × 10−9 ± 0.49 × 10−9 M (n = 9), respectively. Addition of integrin αvβ5, αvβ6, or αvβ8 at 85 nM to 25 nM acrylodan-labeled gHtgL (a molar ratio which would almost eliminate bias introduced by dissociation) produced both a significant increase in fluorescence, which is indicative of an increase in the hydrophobicity of the environment, and a shift in the emission peak from 540 nm, which is typical for acrylodan fluorescence in aqueous solutions, to 504 nm, which is typical of acrylodan fluorescence in hydrophobic solvents (data for integrin αvβ8 are shown in Fig. 4B). The increase in fluorescence over time (the kinetic curve) was monitored at both 504 nm and 540 nm and is described by a single-exponential function for αvβ8 (Fig. 4C) and for αvβ5 (Fig. 4D). One possible explanation for a conformational change would be protein denaturation and aggregation. Four observations made this possibility unlikely. First, the reaction between unlabeled gHtgL and integrin αvβ8 did not show the increase in fluorescence that would have been expected if light scattering had increased fluorescence (Fig. 5A). Second, holding the acrylodan-labeled gHtgL in the quartz cuvette for an hour caused no increase in fluorescence (Fig. 5B). Third, centrifugation of the sample in the middle of the analysis to remove any possible products of aggregation had no effect on fluorescence intensity (Fig. 5C). Finally, incubation of acrylodan-labeled gHtgL with αvβ3 did not lead to an increase in fluorescence (Fig. 5D). The kobs value for the fluorescence change was much lower than that for the reaction measured by SPR (Table 2), which indicated that the conformational change was significantly slower than formation of the intermediate complex.
Fig. 4.
Binding of integrin to acrylodan-labeled gHtgL or IANBD-labeled gHtgL induces an increase in fluorescence and a shift in the fluorescence emission peak. (A) Flow cytometric analysis of binding to AGS cells of acrylodan-labeled gHtgL (dashed line) and unlabeled gHtgL (solid line). The dotted line represents the isotype control. (B) Corrected fluorescence emission spectra at an excitation wavelength of 359 nm and an emission wavelength of 380 to 600 nm for acrylodan-labeled gHtgL alone (1) and 5 min (2), 60 min (3), and 120 min (4) after addition of αvβ8. (C) Corrected time-based emission of acrylodan-gHtgL after addition of αvβ6, measured at an excitation wavelength of 359 nm and an emission wavelength of 504 nm (1) or 540 nm (2). (D) Corrected time-based emission of acrylodan-gHtgL after addition of αvβ5, measured at an excitation wavelength of 359 nm and an emission wavelength of 504 nm (1) or 540 nm (2). (E) Fluorescence emission at an excitation wavelength of 505 nm and an emission wavelength of 515 to 707 nm for IANBD-labeled gHtgL alone (1) and 5 min (2), 30 min (3), and 60 min (4) after addition of αvβ8.
Fig. 5.
Increase in fluorescence of acrylodan-labeled gHtgL after addition of integrin is not a result of protein aggregation. (A) Overlay of emission spectra of unlabeled gHtgL alone and 5 min and 60 min after addition of αvβ8. (B) Overlay of emission spectra of acrylodan-labeled gHtgL measured immediately (1) or after 1 h (2). (C) Time-based emission fluorescence of acrylodan-labeled gHtgL after addition of αvβ8. At the time point indicated by the arrow, the reading was stopped, the sample was centrifuged at 16,000 × g for 15 min, and the sample was returned to the cuvette for resumption of the reading. (D) Overlay of emission spectra of acrylodan-labeled gHtgL alone and 5 min, 60 min, and 120 min after addition of αvβ3. Fluorescence was recorded at an excitation wavelength of 359 nm and an emission wavelength of 380 to 600 nm (A, B, and D) or an excitation wavelength of 359 nm and an emission wavelength of 540 nm (C).
Table 2.
Calculated kobson values for reaction between gHtgL (25 nM) and human soluble integrins (85 nM)
| Integrin |
kobson (×10−3) (mean ± SD) |
|
|---|---|---|
| Step 1: formation of intermediate complex | Step 2: conformational transition | |
| αvβ5 | 28.9 ± 6.1 | 0.66 ± 0.07 (n = 3) |
| αvβ6 | 27.9 ± 6.8 | 0.95 ± 0.04 (n = 3) |
| αvβ8 | 20.1 ± 2.6 | 0.5 ± 0.22 (n = 5) |
To confirm that the conformational change was not unique to acrylodan-labeled gHgL, the complex was also labeled with IANBD, another thiol-reactive fluorescent probe which, because of the sensitivity of its fluorescence properties to solvent accessibility, has been used to detect protein conformational changes (40). The stoichiometry of gHtgL to IANBD was 1:0.85. Addition of each of the three integrins (αvβ5, αvβ6, and αvβ8) again produced both a significant increase in fluorescence and a shift in the emission peak, this time from 573 nm to 542 nm (data for integrin αvβ8 are shown in Fig. 4E).
Neutralizing MAbs also effect a conformational change in gHtgL.
Three MAbs to gH or gHgL are currently available. CL59 maps to the “flap” in domain IV, E1D1 maps to the domain I-domain II interface, and the binding site of CL40 is unknown. We previously reported that all three MAbs block epithelial but not B cell infection (29). Repeating this analysis with a more quantitative assay now revealed that saturating amounts of each antibody can at least reduce B cell infection, though none can completely eliminate infection as is possible for epithelial cells (Table 3). E1D1, whose neutralizing ability may be attributed at least partially to its previously reported ability to reduce integrin binding, had the least effect on B cell infection. Antibody CL40 had the greatest effect. For comparative purposes, the effects of MAb F-2-1 to gp42, which blocks the interaction with HLA class II molecules, and MAb CL55 to gB, which is a nonneutralizing antibody, were also measured. To ensure that the effects of CL59 and CL40 did not simply represent cross-linking of gHgL in the virion envelope, Fab′ fragments were generated and shown to have effects similar to those of the whole molecules.
Table 3.
Neutralization of EBV infection of Akata B cells and SVKCR2 epithelial cells
| Antibody/ligand | % of control infectiona |
|||||
|---|---|---|---|---|---|---|
| Akata B cells |
SVKCR2 epithelial cells |
|||||
| Expt 1 | Expt 2 | Expt 3 | Expt 1 | Expt 2 | Expt 3 | |
| CL55/gB | 94 | 98 | 95 | 95 | 98 | 100 |
| F-2-1/gp42 | 3 | 1 | 1 | 134 | 102 | 98 |
| E1D1/gHgL | 64 | 53 | 68 | 0 | 0 | 0 |
| CL40/gHgL | 19 | 18 | 15 | 0 | 0 | 0 |
| CL59/gH | 36 | 21 | 44 | 0 | 0 | 0 |
| CL40/Fab′ | 12 | ND | ND | 0 | ND | ND |
| CL59/Fab′ | 30 | ND | ND | 0 | ND | ND |
The control was infection in the absence of antibody. Each number represents the average for duplicates. ND, not done.
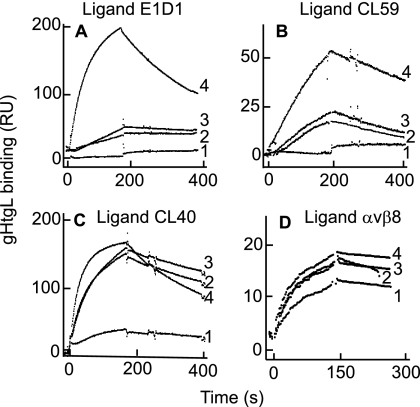
To obtain the rate constants of MAb binding to gHtgL, CL59, E1D1, and CL40 were coupled to a sensor chip, and interaction of different concentrations of gHtgL was monitored by SPR (Fig. 6A to C). Judging by the residuals, the binding curves could be described by a single-exponential function (as an example, the residuals for gHtgL binding to E1D1 are shown in Fig. 6D). All three monoclonal antibodies bound gHtgL with a high affinity, and the kinetic characteristics of this binding are summarized in Table 4. The association rate constant for binding of CL59 was significantly lower than those for both E1D1 and CL40. The interaction of CL40 with gHtgL had a very high value for kon (at the limits of Biacore performance), and its dissociation was also significantly faster than that of the other two MAbs.
Fig. 6.
Binding of MAb against gH or gHgL to gHtgL fits a single-exponential function. SPR analysis was performed to determine the interaction between gHgL and immobilized E1D1 (A), CL40 (B), or CL59 (C). (D) The differences between the theoretical and experimental data (residuals) for the formation portion of each SPR trace of binding to E1D1 were fitted to a single-exponential equation.
Table 4.
Kinetic constants for binding of MAbs E1D1, CL59, and CL40 to EBV gHtgLa
| MAb | kon (M−1 s−1) | koff (s−1) | KD (M) |
|---|---|---|---|
| E1D1 | 857,140 ± 14,800 | 0.0010 ± 0.0001 | (1.20 ± 0.08) × 10−9 |
| CL59 | 154,290 ± 29,600 | 0.0016 ± 0.0004 | (1.04 ± 0.46) × 10−8 |
| CL40 | 1,219,700 ± 249,760 | 0.0120 ± 0.0030 | (9.80 ± 2.20) × 10−9 |
Data are means ± SD.
To determine whether the binding of any one of the MAbs to gHtgL affected the ability of the protein to bind to any of the other antibodies, each was attached in turn to a sensor chip. Following this, gHtgL alone or gHtgL preincubated individually for 1 h on ice with each MAb was injected onto the sensor chip. Prebinding of gHtgL with CL59 or CL40 almost completely ablated the ability to bind to E1D1 (Fig. 7A), and prebinding of gHtgL to E1D1 or CL40 significantly decreased its binding to CL59 (Fig. 7B). In contrast, however, although the association and dissociation rates of gHtgL binding were reduced when the protein was complexed with either E1D1 or CL59, it could still bind to CL40, which together with information already available about domains bound by E1D1 and CL59, indicated that none of the antibodies bound to the same epitope (Fig. 7C). Nonneutralizing rabbit antipeptide antibody to gH was used as a negative control and had no effect on binding to any of the monoclonal antibodies (data not shown). Since all of the MAbs were neutralizing, we also addressed the question of whether any besides E1D1 affected the ability of gHtgL to bind to an integrin. Antibody to AP was coupled to a sensor chip, integrin αvβ8 was captured by the MAb, and gHtgL alone or gHtgL complexed with MAb was injected over the surface of the chip. For complexes with CL40 or CL59, the ability of gHtgL to bind to the integrin was not affected significantly (Fig. 7D, sensorgrams 2 and 3). For complexes with E1D1, consistent with previous findings (29), binding was reduced but not eliminated (Fig. 7D, sensorgram 1).
Fig. 7.
Binding of gHtgL to immobilized E1D1 or CL59, but not to CL40, is affected by prebinding of gHtgL with MAbs to gHtgL, and only E1D1 has an effect on binding to an integrin. (A) Binding to immobilized E1D1 of gHtgL alone (4) or gHtgL complexed with E1D1 (1), CL40 (2), or CL59 (3). (B) Binding to immobilized CL59 of gHtgL alone (4) or gHtgL complexed with CL59 (1), E1D1 (2), or CL40 (3). (C) Binding to immobilized CL40 of gHtgL alone (4) or gHtgL complexed with CL40 (1), E1D1 (2), or CL59 (3). (D) Binding to immobilized αvβ8 of gHtgL alone (4) or gHtgL complexed with E1D1 (1), CL40 (2), or CL59 (3).
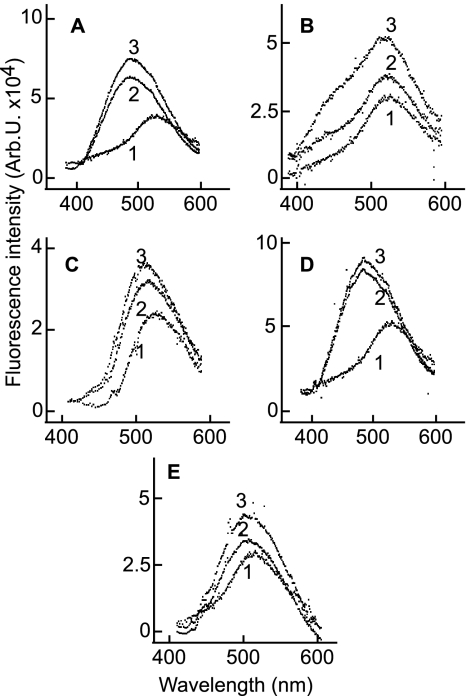
Because prebinding of gHtgL to CL59 and CL40 had a marked effect on the subsequent ability of the protein to bind to E1D1, without significantly affecting its ability to bind to integrins, we considered the possibility that antibody binding itself had elicited a conformational change. Monitoring the fluorescence of acrylodan-labeled gHtgL following addition of whole molecules or Fab′ fragments of MAb CL40 or CL59 revealed an increase in fluorescence, though to a lesser magnitude than that induced by integrin binding. Binding of CL40, however, which increased fluorescence to a greater extent than did binding of CL59, also induced a shift in peak emission spectra similar to that produced by integrin binding (Fig. 8). The rate of change in response to subsequent addition of integrins 100 min after addition of whole antibody or Fab′ fragments of CL40 or CL59 was reduced relative to that seen following addition of integrin in the absence of antibody, and again, CL40 had the greatest effect (Fig. 9). A nonneutralizing antipeptide antibody which binds to residues 518 to 528 of gH with a KD of 1.2 × 10−6 M had no effect, even when added at a saturating concentration of 7 μM.
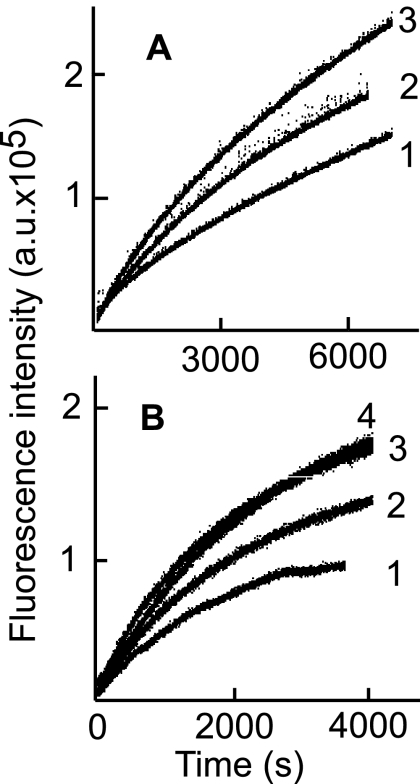
Fig. 8.
Binding of E1D1, CL40, CL59, or Fab′ fragments of antibodies induces an increase in fluorescence of acrylodan-labeled gHtgL. Emission spectra are shown for an excitation wavelength of 359 nm and an emission wavelength of 380 to 600 nm for acrylodan-labeled gHtgL alone (1) and 3 min (2) and 30 min (3) after addition of CL40 (A), CL59 (B), E1D1 (C), or Fab′ fragments of CL40 (D) or CL59 (E).
Fig. 9.
Preincubation of acrylodan-labeled gHtgL with whole antibody or Fab′ fragments of CL40 or CL59 alters the subsequent ability of an integrin to increase fluorescence. Time-based emission is shown for acrylodan-labeled gHtgL preincubated for 100 min at 25°C with CL40 (1), CL59 (2), or no antibody (3) (A) or with Fab′ fragments of CL40 (1) or CL59 (2), no antibody (3), or antipeptide antibody to gH (4) before addition of αvβ8 (B).
DISCUSSION
Previous work has suggested but not formally demonstrated a role for αvβ5 in EBV entry. A function-blocking antibody specific for αvβ5 reduced EBV infection of SVKCR2 epithelial cells by approximately 50% (8). However, at the time that this work was done, we did not have a source of soluble αvβ5. The precise contours of the integrin bound by gHgL are not known, and thus we could not rule out the possibility that the antibody was interfering with recognition of the αv chains of αvβ6 and αvβ8, which are also expressed by SVKCR2 cells. It is now clear that integrin αvβ5 in addition to integrins αvβ6 and αvβ8 can bind and trigger fusion mediated by EBV glycoproteins. None of the other RGD/KGD-binding integrins expressed on human epithelial cells showed any evidence of being able to interact with gHtgL (8), so while there may exist completely different structures that can serve to trigger epithelial cell fusion by EBV gHgL and gB, it appears likely that the integrin triggers are limited to αvβ5, αvβ6, and αvβ8.
Both αvβ6 and αvβ8 are of interest in that they bind to transforming growth factor β1 (TGF-β1) latency-associated peptide and cause local activation of endogenous TGF-β1 (30, 32), an inducer of EBV lytic reactivation in B cells (10, 11, 25). This could potentially increase the extent of transfer of virus from the latent B cell reservoir to epithelial cells, where amplification of virus is thought to occur (14, 19). However, αvβ8 is expressed primarily in basal cells in airway epithelium (41), and αvβ6 is expressed only at low levels on normal epithelial cells, being upregulated during tissue repair and on explantation of cells in tissue culture (6, 7, 47). Thus, although αvβ6 and αvβ8 may be particularly relevant to increased susceptibility of tissues during a variety of disease states and, in the case of αvβ6, inevitably relevant to in vitro analyses in cell lines, they seem less likely to be of importance to infection of normal tonsil epithelium, the probable site of EBV replication in healthy human carriers (35). In this case, αvβ5 may be paramount.
The affinity constants calculated from the SPR analyses of integrin binding to gHtgL were in general agreement with those previously calculated by Scatchard analyses of binding to whole cells, where more than one integrin could contribute to the event (8). The half-life of binding was short, though longer for αvβ8 than for the other two integrins. Labeling of the single available thiol group in gH with acrylodan or IANBD allowed detection of a second, much slower conformational change with a half-life of approximately 12 to 23 min, estimated from the kobs (t1/2 = ln2/kobs), depending on the integrin. To maintain parity with the SPR analyses, the fluorescence measurements were done at 25°C, but by extrapolation, the half-life of this change at 37°C might be expected to convert to as little as 6 min. A pH-induced conformational change in influenza virus hemagglutinin has been measured to have a half-life of approximately 50 min at 25°C and 4 min at 37°C (50). The observation of a ratio of acrylodan or IANBD to gHtgL of close to 1 provided confidence that only one residue had been labeled and that the residue in question was cysteine 153. The fluorescence quantum yields and fluorescence spectra of acrylodan (36) and IANBD (43) are sensitive to their polar environment, and both molecules have been used widely as reporters for conformational change in a protein that involves a shift of the labeled residue into a more hydrophobic environment (40). The increase in quantum yield and the blue shift in fluorescence seen after addition of any of the integrins able to bind to gHtgL were thus consistent with the movement of residue 153 into a more hydrophobic environment and with a potential conformational shift in gHtgL at the domain I-domain II interface. The more limited change in fluorescence intensity seen after addition of αvβ5 may reflect that a smaller proportion of this integrin fused in frame with fos/jun dimerization domains was active than the case for αvβ6 and αvβ8 integrins fused in frame with alkaline phosphatase, but the timing of the increase and the shift in emission wavelength were consistent with those of the other integrins.
Mutations made in domain I, the domain I-domain II interface, and domain IV have been shown to have differential effects on the ability of EBV glycoproteins to mediate fusion with epithelial cells and B cells (34, 51, 52). Some mutations increase fusion, whereas some reduce or abrogate fusion, and for single-point mutants, some may behave differently with each cell type. If it is assumed that the final execution of fusion is not fundamentally different for B cells and epithelial cells, this implies that the triggering interaction transmitted by integrin binding and that triggered by gp42 bound to HLA class II molecules have unique structural requirements. This is mirrored to some extent in the behavior of the three neutralizing MAbs to gH or gHgL. All three completely abrogated infection of an epithelial cell but only partially, to various degrees, neutralized B cell infection. The ability of neither CL59 nor E1D1 to block binding to immobilized CL40 indicated that none of the antibodies bound to the same epitope, and none, not even E1D1, completely blocked gHgL binding to integrins. However, the ability of even CL59, which is known to bind to domain IV, to affect the ability of gHgL to bind to E1D1, which probably binds to surfaces at the domain I-domain II interface (27), strongly suggested that the antibodies induced a conformational change. The ability of each antibody to increase fluorescence of acrylodan-labeled gHtgL and, in the case of E1D1 and CL40, to cause a blue shift, was consistent with this. This presumably means that neutralization results either from a premature conformational change or from a nonfunctional change that prevents a functional change subsequent to integrin binding. The latter possibility is perhaps the most likely, since MAb CL59 can neutralize infection even if it is added after virus has been bound to a cell (S. M. Valencia and L. M. Hutt-Fletcher, unpublished data) and since antibody binding reduced the extent of the change triggered by integrins. Although a change in the environment of cysteine 153 follows either antibody or integrin binding, implicating the same region of gHgL in both, it is not possible to define the changes with any greater precision.
It remains unclear for any herpesvirus how gHgL functions in fusion or even whether it functions as what has been described as “a blue-collar worker for membrane fusion or just a white collar signal mediator” (53). However, it appears that at least in the case of EBV, activation of fusion with an epithelial cell, mediated by interaction with an integrin, involves a conformational shift at the domain I-domain II interface of the complex.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant DE016669 (to L.M.H.-F.) from the National Institute of Dental and Craniofacial Research.
We thank Glen Nemerow for baculoviruses expressing human αv and β5, Stephen Nishimura for 293 cells expressing αvβ3, αvβ6, and αvβ8, Andrew Morgan for baculovirus expressing gHtgL, and Robert Rhoads for use of a Strobe Master Lifetime spectrometer.
Footnotes
Published ahead of print on 28 September 2011.
REFERENCES
- 1. Atanasiu D., et al. 2010. Bimolecular complementation defines functional regions of herpes simplex virus gB that are involved with H/gL as a necessary step leading to fusion. J. Virol. 84:3825–3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Backovic M., et al. 2010. Structure of a core fragment of glycoprotein H from pseudorabies virus in complex with antibody. Proc. Natl. Acad. Sci. U. S. A. 107:22635–22640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Backovic M., Longnecker R., Jardetzky T. S. 2009. Structure of a trimeric variant of the Epstein-Barr virus glycoprotein B. Proc. Natl. Acad. Sci. U. S. A. 106:2880–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Borza C. M., Hutt-Fletcher L. M. 2002. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat. Med. 8:594–599 [DOI] [PubMed] [Google Scholar]
- 5. Borza C. M., Morgan A. J., Turk S. M., Hutt-Fletcher L. M. 2004. Use of gHgL for attachment of Epstein-Barr virus to epithelial cells compromises infection. J. Virol. 78:5007–5014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Breuss J. M., et al. 1995. Expression of the β6 integrin subunit in development, neoplasia and tissue repair suggests a role in epithelial remodeling. J. Cell Sci. 108:2241–2251 [DOI] [PubMed] [Google Scholar]
- 7. Breuss J. M., Gillerr N., Lu L., Sheppard D., Pytela R. 1993. Restricted distribution of integrin β6 mRNA in primate epithelial tissues. J. Histochem. Cytochem. 41:1521–1527 [DOI] [PubMed] [Google Scholar]
- 8. Chesnokova L. S., Nishimura S., Hutt-Fletcher L. 2009. Fusion of epithelial cells by Epstein-Barr virus proteins is triggered by binding of viral proteins gHgL to integrins αvβ6 or αvβ8. Proc. Natl. Acad. Sci. U. S. A. 106:20464–20469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chowdary T. K., et al. 2010. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat. Struct. Mol. Biol. 17:882–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. di Renzo L., Altiok A., Klein G., Klein E. 1994. Endogenous TGF-beta contributes to the induction of the EBV lytic cycle in two Burkitt lymphoma cell lines. Int. J. Cancer 57:914–919 [DOI] [PubMed] [Google Scholar]
- 11. Fahmi H., Cochet C., Hmama Z., Opolon P., Joab I. 2000. Transforming growth factor beta 1 stimulates expression of the Epstein-Barr virus BZLF1 immediate-early gene product ZEBRA by an indirect mechanism which requires the MAPK kinase pathway. J. Virol. 74:5810–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Galdiero M., et al. 1997. Site-directed and linker insertion mutagenesis of herpes simplex virus type 1 glycoprotein H. J. Virol. 71:2163–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gline S. E., Cambier S., Govaerts C., Nishimura S. L. 2004. A 50-Α separation of the integrin αvβ3 extracellular domain C termini reveals an intermediate activation state. J. Biol. Chem. 279:54567–54572 [DOI] [PubMed] [Google Scholar]
- 14. Hadinoto V., Shapiro M., Sun C. C., Thorley-Lawson D. A. 2009. The dynamics of EBV shedding implicate a central role for epithelial cells in amplifying viral output. PLoS Pathog. 7:e10000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hannah B. P., et al. 2009. Herpes simplex virus glycoprotein B associates with target membranes via its fusion loops. J. Virol. 83:6825–6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heldwein E. E., et al. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217–220 [DOI] [PubMed] [Google Scholar]
- 17. Hibbs R. E., Talley T. T., Taylor P. 2004. Acrylodan-conjugated cysteine side chains reveal conformational state and ligand site interactions of the acetylcholine-binding protein. J. Biol. Chem. 279:28483–28491 [DOI] [PubMed] [Google Scholar]
- 18. Hutt-Fletcher L. M. 2007. Epstein-Barr virus entry. J. Virol. 81:7825–7832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jiang R., Scott R. S., Hutt-Fletcher L. M. 2006. Epstein-Barr virus shed in saliva is high in B cell tropic gp42. J. Virol. 80:7281–7283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kirschner A. N., Sorem J., Longnecker R., Jardetzky T. S. 2009. Structure of Epstein-Barr virus glycoprotein gp42 suggests a mechanism for triggering receptor-activated virus entry. Structure 17:223–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krummenacher C., et al. 2005. Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. EMBO J. 24:4144–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lake C. M., Molesworth S. J., Hutt-Fletcher L. M. 1998. The Epstein-Barr virus (EBV) gN homolog BLRF1 encodes a 15 kilodalton glycoprotein that cannot be authentically processed unless it is co-expressed with the EBV gM homolog BBRF3. J. Virol. 72:5559–5564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li Q. X., Turk S. M., Hutt-Fletcher L. M. 1995. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J. Virol. 69:3987–3994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li Q. X., et al. 1992. Epstein-Barr virus infection and replication in a human epithelial system. Nature 356:347–350 [DOI] [PubMed] [Google Scholar]
- 25. Liang C.-L., Chen J.-L., Hsu Y.-P. P., Ou J. T., Chang Y.-S. 2002. Epstein-Barr virus BZLF1 gene is activated by transforming growth factor-β through cooperativity of Smads and c-Jun/c-Fos proteins. J. Biol. Chem. 277:23345–23357 [DOI] [PubMed] [Google Scholar]
- 26. Mathias P., Glalleno M., Nemerow G. R. 1998. Interactions of soluble recombinant integrin alphav beta5 with human adenovirus. J. Virol. 72:8669–8675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matsuura H., Kirschner A. N., Longnecker R., Jardetzky T. S. 2010. Crystal structure of the Epstein-Barr virus (EBV) glycoprotein H/glycoprotein L (gH/gL) complex. Proc. Natl. Acad. Sci. U. S. A. 107:22641–22646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miller N., Hutt-Fletcher L. M. 1992. Epstein-Barr virus enters B cells and epithelial cells by different routes. J. Virol. 66:3409–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Molesworth S. J., Lake C. M., Borza C. M., Turk S. M., Hutt-Fletcher L. M. 2000. Epstein-Barr virus gH is essential for penetration of B cell but also plays a role in attachment of virus to epithelial cells. J. Virol. 74:6324–6332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mu D., et al. 2002. The integrin αvβ8 mediates epithelial homeostasis through MT-1-MMP-dependent activation of TGF-β1. J. Cell Biol. 157:493–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mullen M. M., Haan K. M., Longnecker R., Jardetzky T. S. 2002. Structure of the Epstein-Barr virus gp42 protein bound to the MHC class II receptor HLA-DR1. Mol. Cell 9:375–385 [DOI] [PubMed] [Google Scholar]
- 32. Munger J. S., et al. 1999. The integrin αvβ6 binds and activates latent TGFβ1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell 96:319–328 [DOI] [PubMed] [Google Scholar]
- 33. Oba D. E., Hutt-Fletcher L. M. 1988. Induction of antibodies to the Epstein-Barr virus glycoprotein gp85 with a synthetic peptide corresponding to a sequence in the BXLF2 open reading frame. J. Virol. 62:1108–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Omerovic J., Lev L., Longnecker R. 2005. The amino terminus of Epstein-Barr virus glycoprotein gH is important for fusion with B cells and epithelial cells. J. Virol. 79:12408–12415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pegtel D. M., Middeldorp J., Thorley-Lawson D. A. 2004. Epstein-Barr virus infection in ex-vivo tonsil epithelial cell cultures of asymptomatic carriers. J. Virol. 78:12613–12624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Prendergast F. G., Meyer M., Carlson G. L., Iida S., Potter J. D. 1983. Synthesis, spectral properties and use of 6-acryloyl-2-dimethylaminonaphthalene (Acrylodan). J. Biol. Chem. 258:7541–7544 [PubMed] [Google Scholar]
- 37. Pulford D. J., Lowrey P., Morgan A. J. 1995. Co-expression of the Epstein-Barr virus BXLF2 and BKRF2 genes with a recombinant baculovirus produces gp85 on the cell surface with antigenic similarity to the native protein. J. Gen. Virol. 76:3145–3152 [DOI] [PubMed] [Google Scholar]
- 38. Rickinson A. B., Kieff E. 2007. Epstein-Barr virus, p. 2655–2700 In Knipe D. M., et al.(ed.), Fields virology, 5th ed., vol. 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 39. Roche S., Bressanelli S., Rey F. A., Gaudin Y. 2006. Crystal structure of the low-pH form of the vesicular stomatitis glycoprotein G. Science 313:187–191 [DOI] [PubMed] [Google Scholar]
- 40. Scheibel T., Bloom J., Lindquist S. L. 2004. The elongation of yeast prion fibers involves separable steps of association and conversion. Proc. Natl. Acad. Sci. U. S. A. 101:2287–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sheppard D. 2003. Functions of pulmonary epithelial integrins: from development to disease. Physiol. Rev. 83:673–686 [DOI] [PubMed] [Google Scholar]
- 42. Shimizu N. A., Tanabe-Tochikura A., Kuroiwa Y., Takada K. 1994. Isolation of Epstein-Barr virus (EBV)-negative cell clones from the EBV-positive Burkitt's lymphoma (BL) line Akata: malignant phenotypes of BL cells are dependent on EBV. J. Virol. 68:6069–6073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shore J. D., et al. 1995. A fluorescent probe study of plasminogen activator inhibitor-1: evidence for reactive center loop insertion and its role in the inhibitory mechanism. J. Biol. Chem. 270:5395–5398 [DOI] [PubMed] [Google Scholar]
- 44. Spear P. G. 2004. Herpes simplex virus receptors and ligands for cell entry. Cell. Microbiol. 6:401–410 [DOI] [PubMed] [Google Scholar]
- 45. Spear P. G., Longnecker R. 2003. Herpesvirus entry: an update. J. Virol. 77:10179–10185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Takagi J., Petre B. M., Walz T., Springer T. A. 2002. Global conformational rearrangements in integrin extracellular domains in outside-in and inside out signaling. Cell 110:599–611 [DOI] [PubMed] [Google Scholar]
- 47. Thomas G. J., Nystrom M. L., Marshall J. F. 2006. αvβ6 integrin in wound healing and cancer of the oral cavity. J. Oral Pathol. Med. 35:1–10 [DOI] [PubMed] [Google Scholar]
- 48. Wang X., Kenyon W. J., Li Q. X., Mullberg J., Hutt-Fletcher L. M. 1998. Epstein-Barr virus uses different complexes of glycoproteins gH and gL to infect B lymphocytes and epithelial cells. J. Virol. 72:5552–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weinacker A., et al. 1994. Role of the integrin αvβ6 in cell attachment to fibronectin. J. Biol. Chem. 269:6940–6948 [PubMed] [Google Scholar]
- 50. White J. M., Wilson I. A. 1987. Anti-peptide antibodies detect steps in a protein conformational change: low-pH activation of the influenza virus hemagglutinin. J. Cell Biol. 106:2887–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wu L., Borza C. M., Hutt-Fletcher L. M. 2005. Mutations of Epstein-Barr virus gH that are differentially able to support fusion with B cells or epithelial cells. J. Virol. 79:10923–10930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wu L., Hutt-Fletcher L. M. 2007. Point mutations in EBV gH that abrogate or differentially affect B cell and epithelial cell fusion. Virology 363:148–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yoon T.-Y., Shin Y.-K. 2009. Progress in understanding the neuronal SNARE function and its regulation. Cell. Mol. Life Sci. 66:460–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Young D. F., Carlos T. S., Hagmaier K., Fan L., Randall R. E. 2007. AGS and other tissue culture cells can unknowingly be persistently infected with PIV5: a virus that blocks interferon signalling by degrading STAT1. Virology 365:238–240 [DOI] [PubMed] [Google Scholar]