Abstract
Recent studies indicate that hepatitis B virus (HBV) may induce autophagy to enhance its replication in cell cultures. To understand whether autophagy can indeed enhance HBV replication in vivo, we generated HBV transgenic mice with liver-specific knockout of the Atg5 gene, a gene critical for the initiation of autophagy. Immunoblot analyses confirmed the inhibition of autophagy in the livers of Atg5 knockout mice. This inhibition of autophagy slightly reduced HBV gene expression and affected nuclear localization of the HBV core protein. It also reduced the HBV DNA level in sera by more than 90% and the level of the HBV DNA replicative intermediate in the mouse liver to an almost undetectable level. Our results thus demonstrate that autophagy is important for HBV replication in vivo and raise the possibility of targeting this pathway to treat HBV patients.
TEXT
Hepatitis B virus (HBV) is a hepatotropic virus that can cause severe liver diseases, including liver cirrhosis and hepatocellular carcinoma. This virus chronically infects approximately 350 million people in the world, causing significant morbidity and mortality. HBV is a small DNA virus with a partially double-stranded and circular DNA genome that has a length of about 3.2 kb. After the infection of hepatocytes, this DNA is repaired to form a covalently closed circular DNA (cccDNA) molecule, which then directs the transcription of viral mRNAs. The mRNA of the viral core protein is larger than the genome length. This core protein mRNA, which is also termed the pregenomic RNA (pgRNA), is packaged by the core protein to form the viral core particle. It is subsequently converted to the partially double-stranded viral genome by the viral RNA polymerase, which is also packaged in the core particle. The core particle subsequently interacts with the viral envelope proteins for the formation of the mature virion, which is then released from infected cells (for a review, see reference 1).
Recently, we demonstrated that HBV can induce autophagy in cell cultures, in the mouse liver, and during natural infection (18). We also demonstrated that autophagy can enhance HBV replication primarily at the step of viral DNA replication in cell cultures (17, 18). However, whether autophagy can indeed enhance HBV replication in vivo remains unclear. Autophagy plays an important role in maintaining cellular homeostasis. In the initial stage of autophagy, membrane crescents, known as isolation membranes or phagophores, appear in the cytoplasm. These membranes will extend and eventually form enclosed double-membrane structures called autophagosomes. The autophagosomal lumen may contain protein aggregates, damaged organelles, such as mitochondria, and microbial pathogens. Autophagosomes mature by fusing with lysosomes to form autolysosomes. The contents of autophagosomes will subsequently be digested by lysosomal enzymes for recycling. Two distinct ubiquitin-like protein conjugation systems are required for autophagy. One involves the conjugation of Atg5 and Atg12 and the subsequent recruitment of Atg16 to form oligomers for the elongation of isolation membranes. The other involves the conjugation of LC3 to phosphatidylethanolamine for the formation of autophagosomes (for a review, see reference 9).
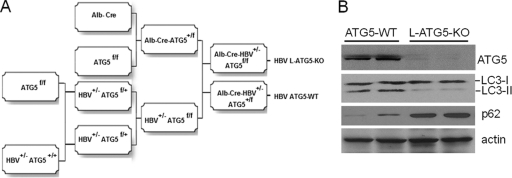
Subsequent to our report, another group also reported that autophagy induced by HBV positively affected HBV replication in cell cultures but primarily at the step of envelopment (11). To resolve this discrepancy and to understand whether autophagy indeed affects HBV replication in vivo, we decided to use HBV transgenic mice that we previously established in our laboratory for studies. The HBV Tg05 mouse line, which contains the 1.3-mer HBV genome and productively replicates HBV in the liver (21), was crossed with the mouse line that contains the floxed Atg5 gene (Atg5f/f). The crossbred offspring mice were subsequently crossed with Alb-Cre mice, which express the Cre recombinase in the liver under the control of the albumin promoter. These breeding procedures resulted in the production of HBV transgenic mice with the liver-specific knockout of Atg5 (Alb-Cre HBV+/− Atg5f/f) and their control littermates without the Atg5 knockout (Alb-Cre HBV+/− Atg5+/f and HBV+/− Atg5f/f). The detailed steps for the production of these mice are illustrated in Fig. 1A. As Atg5 is essential for autophagy, its liver-specific knockout is expected to abolish autophagy in the mouse liver. Indeed, as shown in Fig. 1B, HBV mice with Atg5 knocked out expressed little Atg5 in their livers and an almost undetectable level of lipidated LC3, which is required for the formation of autophagosomes. To further analyze whether autophagy was impaired in the livers of Atg5 knockout mice, we performed Western blot analysis on p62/SQSTM1, a protein that binds to LC3 and is removed by autophagy (9). As shown in Fig. 1B, the p62 levels were significantly increased in the livers of Atg5 knockout mice, indicating a defect in autophagy. Mice with the Atg5 knockout had enlarged hepatocytes and livers (Fig. 2A to C), results which were also consistent with the results of previous reports (7, 16, 20). Taken together, these findings confirmed the successful knockout of Atg5 and the inhibition of autophagy in the HBV transgenic mouse liver.
Fig. 1.
Production of HBV transgenic mice with liver-specific knockout of Atg5. (A) Schematic illustration for the production of HBV transgenic mice with liver-specific knockout of Atg5 and their control littermates. The HBV transgenic mouse line Tg05 has been described previously (21). The floxed Atg5 mouse line was a generous gift of Noboru Mizushima (5), and the Alb-Cre mouse line was obtained from Jackson Laboratory (13). All three mouse lines had the C57BL/6 genetic background. The final breeding step also produced HBV+/− Atg5f/f control mice without Alb-Cre (not shown). The use of either Alb-Cre HBV+/− Atg5+/f or HBV+/− Atg5f/f control mice generated similar results for HBV. HBV ATG5-WT, HBV transgenic mice with the wild-type ATG5 gene; HBV L-ATG5-KO, HBV mice with liver-specific knockout of the ATG5 gene. (B) Western blot analysis for autophagy in the mouse liver. ATG5 represents the conjugated heterodimer of Atg5-Atg12, as ATG5 was not detectable by itself in mouse livers (16). LC3-I and LC3-II are nonlipidated and lipidated LC3, respectively. Lipidated LC3 has a higher electrophoretic mobility than nonlipidated LC3 (18). The p62 protein was analyzed for measuring the autophagic protein degradation, and α-actin served as the loading control. Two 2-month-old male mice were used in each group for confirmation of the results.
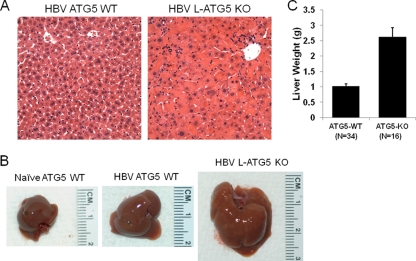
Fig. 2.
Morphological analysis of the HBV transgenic mouse liver. (A) Hematoxylin and eosin staining of the liver tissue sections of HBV mice without (left) and with (right) the Atg5 knockout. (B) Comparison of the livers of naïve, wild-type mice (left panel) and HBV mice without (middle panel) and with (right panel) the Atg5 knockout. (C) Comparison of the average liver weights of HBV transgenic mice with and without the Atg5 knockout. The numbers under the bars indicate the numbers of mice analyzed. The average mouse liver weights without and with the Atg5 knockout were 1.03 ± 0.07 g and 2.62 ± 0.29 g, respectively. HBV by itself did not significantly alter the morphology of hepatocytes or liver size. However, the Atg5 knockout significantly enlarged hepatocytes and the liver. Two-month-old male mice were used in this study and our subsequent studies unless otherwise indicated.
To investigate the possible effect of autophagy on HBV in vivo, we analyzed HBV e antigen (HBeAg), surface antigen (HBsAg), and DNA levels in the mouse sera. As shown in Fig. 3A and B, the suppression of autophagy by liver-specific Atg5 knockout reduced HBeAg and HBsAg levels in the sera by roughly 50% and 60%, respectively. However, as shown in Fig. 3C, this suppression of autophagy reduced the HBV DNA level by more than 90%.
Fig. 3.
Analysis of HBV markers in mouse sera. Mice were bled from the tail vein, and the sera were used for the analysis of HBeAg (A), HBsAg (B), or HBV DNA (C). The analyses of HBeAg and HBsAg were conducted using enzyme-linked immunosorbent assay (ELISA), and the HBV DNA was analyzed by real-time PCR. Details of these experimental procedures have been described previously (21). The levels of HBeAg, HBsAg, and HBV DNA of mice with wild-type Atg5 were arbitrarily defined as 100%. The numbers under the bars indicate the numbers of mice analyzed. Asterisks indicate statistical significance (P < 0.05), which was determined by Student's t test. Only male mice were used for the studies for consistency.
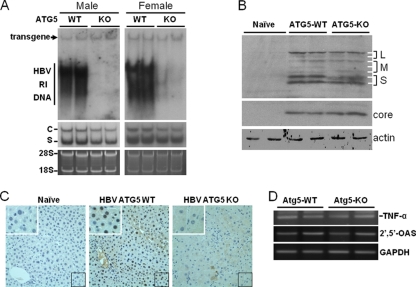
To further understand how autophagy might have reduced the circulating HBV DNA levels in the mouse sera, we isolated mouse livers and performed Southern blot analysis to examine the HBV DNA replicative intermediates (RI). As shown in Fig. 4A, the loss of Atg5 and autophagy reduced the HBV RI DNA to an almost undetectable level in the livers of both male and female mice. We also performed Northern blot analysis to examine the HBV RNA levels. As shown in Fig. 4A, the Atg5 knockout slightly reduced both the HBV C gene and S gene RNA levels, again independently of the genders. Western blot analysis of HBsAg proteins revealed a similar slight reduction of HBsAg proteins by Atg5 knockout (Fig. 4B). However, such reduction was not observed with the HBV core protein (Fig. 4B). We further investigated the subcellular localization of the HBV core protein by performing immunohistochemistry staining using the anti-HBcAg antibody. The HBV core protein was localized primarily to the nuclei of wild-type mouse hepatocytes, with some cytoplasmic localization also visible, particularly in hepatocytes in the perivascular regions. This observation is consistent with the results of a previous report (4). This nuclear localization of the core protein was significantly weakened in the livers of the Atg5 knockout mice, with a concomitant diffuse appearance of the core protein in the cytoplasm. These results indicated that autophagy may affect the subcellular localization of the HBV core protein in vivo.
Fig. 4.
Analysis of HBV replication in mouse livers. (A) Southern (top panel) and Northern (middle panel) blot analyses. For the isolation of HBV RI DNA, mouse livers were homogenized in the radioimmunoprecipitation assay (RIPA) solution (10 mM Tris-HCl [pH 7.0], 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate), treated with proteinase K, and phenol extracted using our previous procedures (21). The DNA was then subjected to Southern blot analysis using 32P-labeled HBV DNA as the probe. The HBV transgene served as the loading control. For the isolation of total RNA, mouse livers were homogenized in TRIzol (Invitrogen), and RNA was isolated as previously described (21). C and S denote the HBV C gene and S gene transcripts, respectively. The 28S and 18S ribosomal RNAs were used as the loading controls for Northern blot analysis (bottom panel). Left panels, male mice; right panels, female mice. (B) Western blot analysis of HBsAg (upper panel) and HBV core protein (lower panel). Liver homogenates as mentioned above were subjected to Western blot analysis. The α-actin protein was also analyzed to serve as the loading control. L, M, and S denote the locations of large, middle, and small HBsAg proteins. The two L protein bands and the two S protein bands represent the glycosylated and the nonglycosylated forms of their respective protein species. The three M protein bands represent doubly glycosylated, monoglycosylated, and nonglycosylated M protein forms, as previously reported (15, 19). (C) Immunohistochemistry analysis of the HBV core protein. Liver tissue sections were stained with the mouse anti-HBcAg antibody (Abcam) and the alkaline phosphatase-conjugated rabbit anti-mouse secondary antibody. A naïve mouse without HBV was used as the control in the staining experiment. The area boxed in the lower right corner of individual panels was enlarged and is shown in the upper left corner of that particular panel. (D) Semiquantitative reverse transcription (RT)-PCR analysis of TNF-α and 2′,5′-OAS RNAs in the mouse liver. The GAPDH (glyceraldehyde-3-phosphate dehydrogenase) RNA was also analyzed to serve as a control. Details of the semiquantitative RT-PCR analysis for the RNAs of 2′,5′-OAS and GAPDH have been described previously (21). The forward and reverse primers used for the TNF-α RT-PCR analysis were 434-CCCACGTCGTAGCAAACCAC-453 and 610-CGTAGTCGGGGCAGCCTTGTC-590, respectively.
Our previous cell culture studies indicated that autophagy had only a slight effect on HBV RNA transcription and pregenomic RNA packaging but that it was required for efficient HBV DNA replication. Our results obtained from transgenic mice are in good agreement with our previous cell culture results and demonstrate that autophagy is also required for efficient HBV DNA replication in vivo. This effect of autophagy on HBV DNA replication in vivo is unlikely to have involved inflammatory cytokines, as the analysis of the RNA levels of tumor necrosis factor alpha (TNF-α), an inflammatory cytokine, and 2′,5′-oligo(A) synthase (2′,5′-OAS), an interferon-stimulated gene product, did not reveal the activation of these genes in the liver (Fig. 4D). The Atg5 knockout resulted in the reduction of the HBeAg and HBsAg levels in sera by approximately half, which may be attributed to the slight reduction of the HBV RNA levels in the mouse liver (Fig. 4A). Indeed, our Western blot analysis also revealed a slight reduction of the HBsAg protein level in the livers of Atg5 knockout mice (Fig. 4B). In contrast, there was no significant reduction of the core protein level in the livers of mice with the Atg5 knockout (Fig. 4B). This may be caused by the lack of release of mature HBV virions due to the inhibition of HBV DNA replication, which then resulted in the accumulation of core protein in hepatocytes and compensated for the reduction of the core protein expression. Interestingly, the core protein was localized primarily to the nucleus in wild-type mouse hepatocytes and diffusely to the cytoplasm in mice with the Atg5 knockout. This result suggests that autophagy may also regulate the subcellular localization of the core protein in vivo. The carboxy terminus of the core protein contains both the nuclear localization signal and the nuclear export signal (2, 6, 10, 22), which overlap several phosphorylation sites (12). Since the phosphorylation of the core protein at these sites has been shown to regulate the nuclear localization of the core protein (6, 10, 12), it is conceivable that autophagy may affect the activities of cellular kinases, which then affect the phosphorylation of the core protein and its nuclear import and export.
Recent studies by Li et al. (11) indicated that autophagy was required primarily for the envelopment of HBV in cell cultures. Their results were not consistent with those of our previous cell culture studies and current mouse studies, as our results clearly demonstrated the requirement of autophagy for HBV DNA replication. The reason for this discrepancy is unclear and may be due to the use of a different HBV strain or a different subline of Huh7 cells in their studies. To resolve this discrepancy, it will be essential to also produce HBV transgenic mice using their specific HBV DNA clone for further analysis in vivo. How autophagy may regulate HBV DNA replication remains unclear. It is possible that autophagic vacuoles may be involved in HBV DNA replication, or alternatively, it is possible that signaling molecules induced by autophagy may regulate the phosphorylation of the HBV core protein, which has been shown to regulate HBV DNA replication (3, 8, 14). In any case, our results, which demonstrate that autophagy is required for the efficient replication of HBV in vivo, indicate that it may be possible to target this particular cellular pathway to treat HBV patients.
Acknowledgments
We thank Michelle McVeigh at the USC Research Center for Liver Diseases for help with the histology work and Noboru Mizushima at the Tokyo Medical and Dental University for providing us with the floxed Atg5 mice.
This research is supported by a research grant from the National Institutes of Health (P01CA123328).
Footnotes
Published ahead of print on 28 September 2011.
REFERENCES
- 1. Beck J., Nassal M. 2007. Hepatitis B virus replication. World J. Gastroenterol. 13:48–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Eckhardt S. G., Milich D. R., McLachlan A. 1991. Hepatitis B virus core antigen has two nuclear localization sequences in the arginine-rich carboxyl terminus. J. Virol. 65:575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gazina E. V., Fielding J. E., Lin B., Anderson D. A. 2000. Core protein phosphorylation modulates pregenomic RNA encapsidation to different extents in human and duck hepatitis B viruses. J. Virol. 74:4721–4728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guidotti L. G., Matzke B., Chisari F. V. 1997. Hepatitis B virus replication is cell cycle independent during liver regeneration in transgenic mice. J. Virol. 71:4804–4808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hara T., et al. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441:885–889 [DOI] [PubMed] [Google Scholar]
- 6. Kann M., Sodeik B., Vlachou A., Gerlich W. H., Helenius A. 1999. Phosphorylation-dependent binding of hepatitis B virus core particles to the nuclear pore complex. J. Cell Biol. 145:45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Komatsu M., et al. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169:425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lan Y. T., Li J., Liao W., Ou J. 1999. Roles of the three major phosphorylation sites of hepatitis B virus core protein in viral replication. Virology 259:342–348 [DOI] [PubMed] [Google Scholar]
- 9. Levine B., Mizushima N., Virgin H. W. 2011. Autophagy in immunity and inflammation. Nature 469:323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li H. C., et al. 2010. Nuclear export and import of human hepatitis B virus capsid protein and particles. PLoS Pathog. 6:e1001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li J., et al. 2011. Subversion of cellular autophagy machinery by hepatitis B virus for viral envelopment. J. Virol. 85:6319–6333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liao W., Ou J. H. 1995. Phosphorylation and nuclear localization of the hepatitis B virus core protein: significance of serine in the three repeated SPRRR motifs. J. Virol. 69:1025–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Machida K., et al. 2010. c-Jun mediates hepatitis C virus hepatocarcinogenesis through signal transducer and activator of transcription 3 and nitric oxide-dependent impairment of oxidative DNA repair. Hepatology 52:480–492 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14. Melegari M., Wolf S. K., Schneider R. J. 2005. Hepatitis B virus DNA replication is coordinated by core protein serine phosphorylation and HBx expression. J. Virol. 79:9810–98120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ou J. H., Bell K. D. 1990. Comparative studies of hepatitis B virus precore and core particles. Virology 174:185–191 [DOI] [PubMed] [Google Scholar]
- 16. Singh R., et al. 2009. Autophagy regulates lipid metabolism. Nature 458:1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sir D., Ann D. K., Ou J. H. 2010. Autophagy by hepatitis B virus and for hepatitis B virus. Autophagy 6:548–549 [DOI] [PubMed] [Google Scholar]
- 18. Sir D., et al. 2010. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc. Natl. Acad. Sci. U. S. A. 107:4383–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Standring D. N., Ou J. H., Rutter W. J. 1986. Assembly of viral particles in Xenopus oocytes: pre-surface-antigens regulate secretion of the hepatitis B viral surface envelope particle. Proc. Natl. Acad. Sci. U. S. A. 83:9338–9342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takamura A., et al. 2011. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 25:795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tian Y., Chen W. L., Ou J. H. J. 2011. Effects of interferon-α/β on HBV replication determined by viral load. PLoS Pathog. 7:e1002159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yeh C. T., Liaw Y. F., Ou J. H. 1990. The arginine-rich domain of hepatitis B virus precore and core proteins contains a signal for nuclear transport. J. Virol. 64:6141–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]