Abstract
Human cytomegalovirus is a ubiquitous herpesvirus that establishes lifelong latent infection. Changes in immune homeostasis induce the reactivation of lytic infection, which is mostly inapparent in healthy individuals but often causes overt disease in immunocompromised hosts. Based on discrepant tumor necrosis factor receptor 1 surface disposition between human cytomegalovirus AD169 variants differing in the ULb′ region, we identified the latency-associated gene product pUL138, which also is expressed during productive infection, as a selective potentiator of tumor necrosis factor receptor 1, one of the key receptors of innate immunity. Ectopically expressed pUL138 coprecipitated with tumor necrosis factor receptor 1, extended the protein half-life, and enhanced its signaling responses, thus leading to tumor necrosis factor receptor 1 hyperresponsiveness. Conversely, the targeted deletion of UL138 from the human cytomegaloviral genome strongly reduced tumor necrosis factor receptor 1 surface densities of infected cells. Remarkably, the comparison of UL138 deficiency to ULb′ deficiency revealed the presence of further positive modulators of tumor necrosis factor alpha signal transduction encoded within the human cytomegalovirus ULb′ region, identifying this region as a hub for multilayered tumor necrosis factor alpha signaling regulation.
INTRODUCTION
Human cytomegalovirus (HCMV) is a ubiquitous human pathogenic betaherpesvirus persisting as a subclinical, lifelong infection in a large majority of the global human population. Primary infection mostly takes a benign course, resulting in life-long latency from which reactivation occurs under conditions of immune stress or immune deficiency, contingently leading to overt HCMV disease. The lifelong persistence of the HCMV genome exposes the human host to permanent risk for recurrent infection, and HCMV reactivation is a major cause of morbidity and mortality in AIDS patients and immunocompromised individuals, manifested by HCMV pneumonia, hepatitis, colitis, chorioretinitis, and encephalitis (18). Historically, general mechanisms of HCMV biology and pathogenesis have been studied using cell culture-adapted virus strains. Since extensive fibroblast passaging leads to major rearrangements of the viral genome (8, 15, 46), HCMV research focused on gene functions not altered during fibroblast adaptation. The most apparent rearrangements as virus is passaged in cell culture is the loss of 13 to 15 kb at the right end of the long unique (UL) region with the accompanying duplication of sequences from the left end of the viral genome (the replacement of ULb′ by the internal repeat long [IRL] sequence) (8). To unravel the natural determinants of viral pathology, the understanding of the gene functions that are lost during fibroblast adaptation but that are important for in vivo pathogenesis is of growing interest. During productive infection, sophisticated viral immune evasive functions, acquired during millions of years of coevolution with the host, maintain a balanced immune situation allowing lifelong persistence despite a fully functioning host defense system (51). This mutual relationship is constituted by complex interactions between cellular and viral factors altering various host responses. Of the diverse cellular signaling pathways involved in antiviral host response, proinflammatory cytokines are of particular importance. The pleiotropic cytokine tumor necrosis factor alpha (TNF-α) is one of the pivotal factors orchestrating a variety of cellular mechanisms. Through binding to one of its receptors (TNF receptors 1 and 2 [TNRF1 and TNRF2], respectively) (43), TNF-α elicits a wide range of biological effects, such as proliferation, differentiation, apoptosis, inflammation, and immunity (9, 23, 41). TNFR1 engagement leads to both cell death and survival signals (37). TNF-α stimulates complex signaling pathways, resulting in the activation of several transcription factors, among them nuclear factor κB (NF-κB), one of the major players in innate immunity (69). The homo- or heterodimeric NF-κB complex consists of members of the Rel family, with the most common form being composed of p50 and p65/RelA. In unstimulated cells, p50/p65 is sequestered by an inhibitory protein (predominantly IκBα) which masks the nuclear localization site of NF-κB. TNF-α binding to TNFR1 activates NF-κB by phosphorylation, ubiquitination, and the subsequent proteasomal degradation of IκBα to release the transcription factor for the induction of NF-κB-dependent gene expression (33). Since TNF was shown to have cytotoxic and antiviral activities (30, 67), it is not surprising that viruses developed strategies to antagonize TNF and NF-κB signaling (7, 44), but the viral exploitation of cell-intrinsic mechanisms to shape and reroute the profound effects of TNF-α appears to be even more elaborate.
In our study, we were able to assign one of the latency-associated HCMV proteins, pUL138 (21), to a molecular function which is, to our knowledge, unprecedented for any other viral protein: it enhances the surface disposition of TNFR1, one of the key receptors of innate immunity, while keeping it in a ligand-inducible signaling-competent state. The pUL138 function is embedded in a multilayered positive regulation of TNF-α signaling mediated by ULb′ gene products. These viral gene functions seem to be dispensable for fibroblast culture but presumably are important for in vivo replication and pathogenesis.
(Part of this research was presented at the 35th Annual International Herpesvirus Workshop 2010, Salt lake City, UT, 24 to 29 July 2010.)
MATERIALS AND METHODS
Cells.
Human MRC-5 fibroblasts (ATCC CCL-171; passages 8 to 15) as well as HeLa (ATCC CCL-2) and HEK293T (ATCC CRL-11268) cells were grown in Dulbecco's minimal essential medium (DMEM) supplemented with 10% fetal calf serum (FCS), penicillin, streptomycin, and 2 mM glutamine.
Viruses, infection, and virus titration.
The following HCMV strains were used: AD169varATCC, AD169varL (27), the endotheliotropic strain TB40/E (60), which was reconstituted from its respective bacterial artificial chromosome (BAC) clone TB40-BAC4 (59), and Towne (ATCC VR-977). Virus stocks were prepared as described previously by propagating HCMV in MRC-5 fibroblasts (27). HCMV infection was enhanced by centrifugation at 800 × g for 30 min. Virus titers were determined by standard plaque assay on MRC-5 cells. Viral genomic DNA was isolated by phenol-chloroform extraction and subsequent isopropanol precipitation from virions purified and concentrated from supernatants of infected cells.
Cytokines, inhibitors, deglycosylation, and transfection.
The activation of NF-κB signaling was induced by treatment with tumor necrosis factor alpha (TNF-α) or interleukin-1β (IL-1β) (R&D Systems) as indicated. The inhibition of translation was achieved by using cycloheximide (100 μg/ml if not indicated; Sigma). Ganciclovir (50 μM; Sigma) was used to prevent herpesviral DNA replication. Protein glycosylation was analyzed by the overnight treatment of protein lysates with endoglycosidase H (EndoH; Roche) and N-glycosidase F (PNGase F; Roche). HeLa cells were transfected with plasmids expressing ULb′ proteins or TNFR1 using Superfect reagent (Qiagen) by following the manufacturer's instructions. Stable transfectants were generated by the transfection of pcDNA3.1-UL138HA and subsequent selection in the presence of Geneticin (Invitrogen). HeLa-UL138HA cell clones were obtained using hybridoma cell culture dishes.
Cloning and sequencing.
Viral DNA was digested with HindIII and separated on an agarose gel. The additional 1.6-kbp fragment of AD169varL was excised and cloned into pcDNA3.1 for sequencing. The sequence of the whole AD169varL ULb′ region was determined by primer walking. For the cloning of the expression constructs (hemagglutinin [HA]-tagged ULb′ proteins and TNFR1 without epitope tag), primers containing restriction sites and C-terminal HA epitope tag (see Table S2 in the supplemental material) were used to amplify the respective gene product from total RNA by one-step reverse transcription-PCR (RT-PCR) (Qiagen). RT-PCR fragments were cloned into pcDNA3.1 by use of the introduced restriction sites. Subcloning into pIRES-EGFP was performed to generate flow cytometry-suitable expression constructs. All plasmids were confirmed by the sequence determination of the inserted fragment.
Generation of the AD169varL BAC and construction of recombinant HCMV viruses.
For the cloning of the AD169varL HCMV genome as a BAC in Escherichia coli, we followed described procedures (24). Briefly, MRC-5 fibroblasts were transfected with plasmid pEB1097 (5) containing a tk-gpt-bac cassette flanked with HCMV homologous sequences of US1-US2 (AD169 nucleotides [nt] 192,648 to 193,360; GenBank accession no. X17403) on the right side and US6-US7 (AD169 nt 195,705 to 197,398) on the left side of the cassette. Twenty-four h posttransfection, cells were infected with AD169varL (multiplicity of infection [MOI], 5). Recombinant viruses were enriched by three rounds of selection with 25 μM xanthine and 100 μM mycophenoloic acid. Circular viral genomes were extracted (28) and electroporated into E. coli DH10B. Bacteria were selected on agar plates containing 12.5 μg/ml chloramphenicol. BAC DNA was prepared from colonies for restriction analysis. The BAC-cloned AD169varL genome is referred to as pAD169. Recombinant HCMV mutants were generated according to a previously published procedure (68) using BACs pTB40 (59) and pAD169, respectively. For the construction of the HCMV deletion mutants, a PCR fragment was generated (see Table S3 in the supplemental material for primer sequences) using the plasmid pSLFRTKn (1) as the template DNA. The PCR fragments containing a kanamycin resistance gene were inserted into the parental BAC by homologous recombination in E. coli. Correct mutagenesis was confirmed by Southern blotting and PCR analysis (data not shown). Recombinant parental and mutant HCMVs were reconstituted from HCMV BAC DNA by Superfect (Qiagen) transfection.
Northern blotting and semiquantitative RT-PCR analysis of specific transcripts.
Total RNA was extracted from cells using the RNeasy minikit (Qiagen). Total RNA was subjected to morpholinepropanesulfonic acid (MOPS) gel electrophoresis and transferred to nylon membranes using a TurboBlotter (Schleicher and Schuell). Probes were prepared by PCR with gene-specific primers (see Tables S1 and S2 in the supplemental material) and digoxigenin-labeled dUTP (Roche) for the detection of the indicated transcripts. Hybridization and detection were performed as described by Roche manuals. For semiquantitative RT-PCR, total RNA was digested with DNase I and used as the template for one-step RT-PCR (Qiagen) with gene-specific primers (see Tables S1 and S2).
Immunoblot analysis.
For immunoblotting, cells were lysed (66) and equal amounts of protein were subjected to SDS-PAGE and transferred to nitrocellulose membranes. Immunoblot analysis was performed using mouse monoclonal antibody (MAb) anti-β-actin (Sigma), mouse MAb recognizing HCMV pUL83/pp65 (3A12; AbCam), rabbit polyclonal anti-IκBα (sc-371; Santa Cruz), rabbit MAb anti-phospho-IκBα (2859; Cell Signaling), rabbit MAb anti-TNFR1 (3736; Cell Signaling), rabbit polyclonal anti-Fas (sc-1023; Santa Cruz), and rabbit polyclonal anti-HA (Sigma). The proteins were visualized using peroxidase-coupled secondary antibodies and the enhanced chemiluminescence system (GE Healthcare).
Flow cytometry.
For flow cytometry, cells were detached using Alfazyme (PAA Laboratories), washed, and incubated in 50 μl 3% FCS-phosphate-buffered saline (PBS) with 125 ng of labeled antibody (phycoerythrin [PE]-conjugated anti-TNFR1 or PE-conjugated isotype control, fluorescein isothiocyanate [FITC]-conjugated anti-Fas or FITC-conjugated isotype control, or PE-conjugated anti-TRAILR2 or PE-conjugated isotype control; R&D Systems) for 45 min at 4°C in the dark. Cells were washed twice and measured in the PE or FITC channel of a FACS Canto II (Becton Dickinson). All shown histograms are derived from 4′,6′-diamidino-2-phenylindole (DAPI)-negative cells and generated by the use of FlowJo (Tree Star).
Immunofluorescence microscopy.
Subconfluent HeLa cells were transfected with constructs expressing ULb′ proteins. Twenty-four h posttransfection, cells were fixed with 3% paraformaldehyde-PBS for 20 min before being permeabilized with 0.05% saponin-PBS and incubated with blocking solution (2% goat serum in 0.005% saponin-PBS) for 30 min. Cells were incubated with primary rabbit anti-HA (Sigma) for 1 h, washed with 0.005% saponin-PBS, and incubated for 1 h with secondary antibody conjugated with Cy3 (Jackson ImmunoResearch). Cells were visualized using a Nikon TE2000 microscope and LUCIA 4.60.
IP.
HEK293T cells were transfected (Superfect; Qiagen) with expression plasmids. Twenty-four h posttransfection, cells were lysed (0.1 mM EDTA, 150 mM NaCl, 10 mM KCl, 10 mM MgCl2, 10% [vol/vol] glycerol, 20 mM HEPES [pH 7.4], 0.5% [vol/vol] IGEPAL, 0.1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, 0.4 mM pepstatin A, 0.1 mM Na-vanadate, Complete protease inhibitor [Roche]). Lysates were spun for 30 min at 4°C and 16,000 × g. Immunoprecipitation (IP) antibody was added to the supernatant. Immune complexes were precipitated with protein G-Sepharose (GE Healthcare) and washed with 150, 250, and 400 mM NaCl-containing buffer. Samples were separated by SDS-PAGE for immunoblot analysis.
RESULTS
HCMV-AD169 variants differ in their modulation of TNF-α signaling.
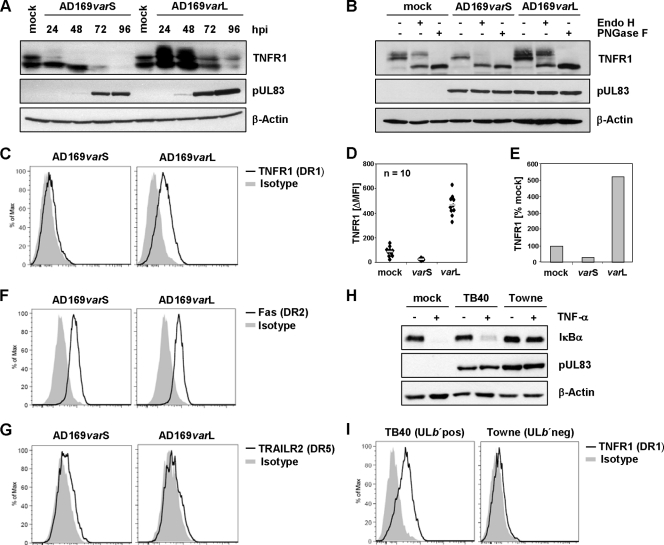
When tracing phenotypic inter- and intrastrain-specific differences in the modulation of innate immune signaling (38), a genomically undefined HCMV variant, AD169varX, was found to exhibit distinct regulation compared to the common AD169varATCC. This discrepancy prompted us to pinpoint the underlying genomic divergence based on DNA restriction fragment polymorphisms. A HindIII fragment found in AD169varX but not in AD169varATCC (Fig. 1A) was identified, cloned, and sequenced, allowing the determination of further adjacent gene sequences. Altogether, 11.8 kbp of AD169varX derived from the ULb′ region, which is present in clinical HCMV isolates but absent from prototypical laboratory strains like AD169varATCC and Towne, was identified, revealing the presence of open reading frames (ORFs) UL148, UL147A, UL147, UL146, UL145, UL139, UL138, UL136, UL135, UL134, UL133, UL148A/B/C/D, UL149, and UL150 from the canonical ULb′ region (Fig. 1B), linking AD169varX to the recently described AD169varUC (6). For convenience, AD169varATCC and AD169varX were denoted AD169varS (short UL region without ULb′) and AD169varL (long UL region containing ULb′), respectively. To identify functions of ULb′-encoded proteins, we tested molecular phenotypes associated with strain- or variant-specific differences. Since the distinct modulation of TNF-α signaling by HCMV AD169 variants has been reported by Montag et al. (45), we examined IκBα degradation upon infection. Strikingly, AD169varL-infected cells exhibited TNF-α-induced IκBα degradation, whereas AD169varS-infected cells did not (Fig. 1C). In contrast, congruent IκBα amounts were found upon IL-1β treatment (Fig. 1D), indicating independent IκBα regulation upon TNF-α and IL-1β signaling, which is consistent with previously published data (31). Despite the substantial discrepancy observed for NF-κB-activating IκBα degradation, infection with both AD169varS and AD169varL suppressed NF-κB target gene ikba transcription with the same efficacy (Fig. 1E). This seeming paradox was most apparent when we determined the kinetics of TNF-α-induced IκBα degradation and its feedback regulation. Although IκBα became phosphorylated and degraded upon TNF-α treatment in AD169varL-infected cells, the negative feedback loop was not initiated to reexpress IκBα, a prototypical NF-κB target gene product (see Fig. S1 in the supplemental material). This finding points to the presence of HCMV-encoded inhibitors of cellular NF-κB-dependent gene expression during lytic infection.
Fig. 1.
HCMV-AD169 variants differ in their modulation of TNF-α signaling. (A) Viral DNA of AD169varATCC and AD169varX (genomically undefined) was digested with HindIII and separated on an agarose gel. (B) Scheme of HCMV genome organization. UL, unique long; US, unique short; TRL, terminal repeat long; IRS, internal repeat short; TRS, terminal repeat short; IRL, internal repeat long; RL, repeat long. (C to E) MRC-5 fibroblasts were mock treated or infected for 72 h (MOI, 5) with AD169varS or AD169varL. (C and D) Cells treated for 20 min with 20 ng/ml TNF-α (C) or 2 ng/ml IL-1β (D) were lysed for the immunoblot analysis of IκBα protein. pUL83 and β-actin served as infection and loading controls, respectively. (E) Cells were treated with 20 ng/ml TNF-α for 3 h, and total RNA was prepared for Northern blot analysis of the indicated transcripts.
ULb′ gene product(s) selectively modulates TNFR1.
Since we observed differential regulation of TNF-α and IL-1β signaling (Fig. 1C and D), we next focused on factors upstream of IκBα degradation. In analyzing cellular TNFR1 protein amount, we found clearly dissimilar levels of TNFR1 protein, i.e., AD169varL-infected cells displayed more TNFR1 protein than AD169varS-infected cells (Fig. 2A). This was especially obvious for fully glycosylated forms (Fig. 2B), inversely correlating with TNF-α-induced IκBα degradation (Fig. 1C). The disparity between AD169varL- and AD169varS-infected cells also included surface-resident TNFR1 molecules (Fig. 2C). To quantify the upregulation of TNFR1 surface densities mediated by AD169varL, we performed 10 independent experiments to measure TNFR1 by flow cytometry and calculated the arithmetic means (Fig. 2D). We found upregulation of ca. 5-fold compared to the level of mock-treated cells (Fig. 2E). The ULb′-mediated modulation of TNFR1 was target selective, since the surface expression of the related death receptors Fas and TRAILR2 did not differ (Fig. 2F and G). The investigation of the HCMV strains TB40 (ULb′pos) and Towne (ULb′neg) (Fig. 2H and I) further confirmed our hypothesis that TNF-α signaling and TNFR1 surface expression must be controlled by genes within the ULb′ region. Consequently, we postulated an HCMV ULb′ gene product to selectively enhance TNFR1 surface density.
Fig. 2.
ULb′ gene product(s) selectively modulate TNFR1. (A and B) MRC-5 fibroblasts were mock treated or infected (MOI, 5) with AD169varS or AD169varL. (A) Cells were lysed at the indicated time points postinfection for the detection of the indicated proteins. (B) Lysates of cells infected for 24 h were treated with endoglycosidase H (EndoH) or N-glycosidase F (PNGase F). The immunoblot analysis of the indicated proteins was performed. (C to G) MRC-5 fibroblasts were infected (MOI, 5) with AD169varS or AD169varL. Cells were stained at 72 hpi with anti-TNFR1 (C), anti-Fas (F), or anti-TRAILR2 (G) and the respective isotype control antibody and were analyzed by flow cytometry. Histograms of DAPI-negative cells are shown. (D) To quantify the upregulation of TNFR1 surface densities by AD169 infection, 10 independent experiments were performed, and the arithmetic means were calculated (gray bar). (E) Changes of TNFR1 surface densities (calculated based on arithmetic mean values). (H) MRC-5 fibroblasts were mock treated or infected (MOI, 5) with TB40 (ULb′pos) or Towne (ULb′neg). Cells were treated at 72 hpi with 20 ng/ml TNF-α for 20 min before being lysed for the immunoblot analysis of the indicated proteins. (I) Cells infected for 72 h with TB40 or Towne were stained with anti-TNFR1 or isotype control antibody and analyzed by flow cytometry. Histograms of DAPI-negative cells are shown.
Identification of pUL138 as an enhancer of TNFR1 surface density.
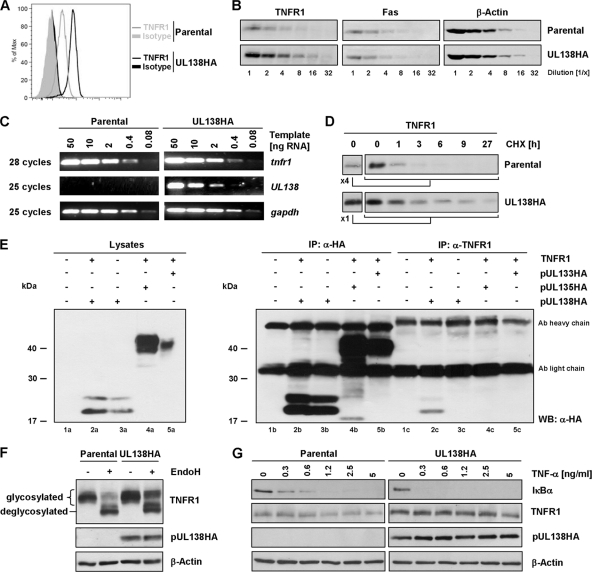
Aiming at the determination of the HCMV-encoded positive regulator of TNFR1 surface disposition, we cloned and expressed HA epitope-tagged versions of all candidate genes (Fig. 3A and B; also see Fig. S2 in the supplemental material) and analyzed TNFR1 surface density upon transfection. This comprehensive approach identified one ULb′-encoded protein, pUL138, to be sufficient to amplify the abundance of TNFR1 on the plasma membrane (Fig. 3C), whereas the ectopic expression of the other tested ULb′ genes was not sufficient (data not shown). Even though pUL138 had deterministic effects on TNFR1, the highest levels of pUL138 expression (gate green fluorescent protein [GFP] high; see Fig. S3A in the supplemental material) resulted in decreased densities of TNFR1 on the plasma membrane. A similar outcome was observed after the transient transfection of increasing amounts of TNFR1 expression plasmid, eventually leading to the hyperexpression of TNFR1. Large amounts of transfected TNFR1 expression plasmid led to an emerging subpopulation of cells totally negative for surface TNFR1 (see Fig. S3B). The coexpression of pUL138 with TNFR1 mimicked the phenotype of hyperexpressed TNFR1 (see Fig. S3C). The lack of surface TNFR1 was not due to diminished overall protein amounts (see Fig. S3D and E), suggesting that cell-intrinsic control precludes excessive levels of TNFR1 molecules at the plasma membrane. When we studied the kinetics of UL138 transcription and upregulation of TNFR1 surface density during productive AD169varL infection, we detected more than one UL138-spanning transcript (Fig. 3D), which is consistent with previous findings (22). UL138 transcripts were expressed as early as 4 h postinfection (hpi) but not under immediate-early conditions (Fig. 3D and E), and sustained transcript levels were detected throughout the replication cycle (Fig. 3D), thus preceding TNFR1 upregulation (Fig. 3G). Only the expression of the longest UL138 transcript was found to be strongly ganciclovir sensitive, classifying this mRNA as a viral late transcript dependent on viral DNA replication (Fig. 3F). Remarkably, UL138 transcription has been detected in natural HCMV latency and suggested to constitute a determinant required for HCMV latency and reactivation (21).
Fig. 3.
Identification of pUL138 as an enhancer of TNFR1 surface density. (A) Scheme of AD169-ULb′ ORFs. Candidate ORFs for TNFR1 upregulation (present in AD169varL and absent from Towne) are depicted in gray. (B) Subcellular localization of ectopically expressed HA-tagged ULb′ candidate proteins. HeLa cells were transfected with the respective expression plasmid. Twenty-four h posttransfection, cells were analyzed by immunofluorescence staining (anti-HA). (C) pUL138 mediates the upregulation of TNFR1 surface expression. HeLa cells were transfected with pIRES-UL138HA-EGFP. Twenty-four h posttransfection, cells were harvested for analysis by flow cytometry. DAPI-negative cells of GFP-positive and GFP-negative gates were analyzed for TNFR1 surface density. TNF-α treatment (20 ng/ml for 30 min) before staining served as a specificity control of TNFR1 detection, since ligand binding competes with MAB225 binding. (D to G) MRC-5 fibroblasts were mock treated or infected with AD169varL (MOI, 5). (D) Cells were harvested for RNA preparation at the indicated times. Equal amounts of total RNA were separated on a MOPS gel, and UL138 transcripts were detected by Northern blotting. (E) Selective transcription of HCMV immediate-early genes was achieved by infection in the presence of cycloheximide (CHX; 50 μg/ml). (F) HCMV DNA replication was prevented by the use of ganciclovir (GCV; 50 μM) to determine GCV-sensitive transcripts. Infected cells were harvested for RNA preparation at the indicated times. Equal amounts of total RNA were separated on a MOPS gel, and UL138-spanning transcripts were detected by Northern blotting. (G) Cells were harvested at the indicated times for the analysis of TNFR1 surface expression by flow cytometry. Histograms of DAPI-negative cells are shown.
pUL138 interacts with TNFR1, leading to stabilization of TNFR1, enhancement of TNFR1 surface disposition, and TNF-α hyperresponsiveness.
To investigate the effect of pUL138 when isolated from further HCMV modulators of NF-κB signaling, we generated cells stably expressing pUL138HA. TNFR1 surface densities were augmented in stably transfected HeLa-UL138HA cells (Fig. 4A). In analyzing HeLa-UL138HA, we observed overall upregulated TNFR1 protein amounts in several independently selected pUL138HA-expressing clones (Fig. 4B and data not shown), while tnfr1 mRNA levels remained unchanged (Fig. 4C). Moreover, TNFR1 half-life was prolonged in HeLa-UL138HA cells compared to that of parental HeLa cells (Fig. 4D). We hypothesized that pUL138 interacts with TNFR1 molecules to enhance TNFR1 protein stability and thus increase its surface disposition. When conducting immunoprecipitation experiments of ectopically expressed TNFR1 in HEK293T cells (Fig. 4E; also see Fig. S4 in the supplemental material) and HeLa cells (data not shown), TNFR1 precipitation recovered coexpressed pUL138 (Fig. 4E, lane 2c) but not pUL133 or pUL135 (Fig. 4E, lanes 4c and 5c), which exhibited a subcellular localization similar to that of pUL138 (Fig. 3B), revealing the complex formation of TNFR1 and pUL138. Based on the fact that pUL138 accumulates at the membrane of the trans-Golgi apparatus (reference 48 and Fig. 3B), we postulated that this interaction occurs in this subcellular compartment. Given that the maturation of oligosaccharide side chains of glycoproteins occurs in the medial- and trans-Golgi apparatuses (35), we compared the ratio of EndoH-sensitive and -resistant forms of TNFR1 in the absence and presence of pUL138. Consistently with our hypothesis, the amount of EndoH-sensitive TNFR1 forms was similar in parental HeLa and HeLa-UL138HA cells, whereas EndoH-resistant forms were significantly enriched in HeLa-UL138HA (Fig. 4F), validating our notion that the stabilization of TNFR1 molecules by pUL138 occurs primarily at post-endoplasmic reticulum (ER)/cis-Golgi compartments. To confirm that the increase in TNFR1 surface densities translates into enhanced TNF-α responsiveness, parental and HeLa-UL138HA cells were treated with grading TNF-α concentrations (Fig. 4G). Indeed, the degradation of IκBα occurred in HeLa-UL138HA transfectants at TNF-α concentrations which did not activate parental cells. Thus, pUL138 was demonstrated to act as a potentiator of TNFR1 surface density, sensitizing cells for TNF-α ligand-induced, TNFR1-dependent signal transduction.
Fig. 4.
pUL138 interaction with TNFR1 leading to the stabilization of TNFR1, enhancement of TNFR1 surface disposition, and TNF-α hyperresponsiveness. (A) DAPI-negative parental HeLa and HeLa-UL138HA cells were gated and analyzed for TNFR1 surface density by flow cytometry. (B) Lysates of parental HeLa and HeLa-UL138HA were log2 diluted to analyze the abundance of the indicated proteins. (C) Increase of TNFR1 protein amount in HeLa-UL138HA cells is not due to the upregulation of tnfr1 transcription. Total RNA from parental and HeLa-UL138HA cells was prepared and treated with DNase I. Graded template amounts were used for the semiquantitative analysis of the indicated transcripts. (D) Cells were treated with 100 μg/ml cycloheximide (CHX), blocking protein neosynthesis for the indicated times. Protein amounts were adjusted to reach the same TNFR1 levels at 0 h to ensure fair comparisons. (E) HEK293T cells were transfected with the indicated expression constructs. Twenty-four h posttransfection, cells were lysed and immunoprecipitation (IP) was performed using TNFR1- and HA-specific antibodies. WB, Western blotting. (F) Lysates of parental HeLa and HeLa-UL138HA cells were treated with endoglycosidase H (EndoH) as indicated, and the immunoblot analysis of indicated proteins was performed. (G) Parental HeLa and HeLa-UL138HA cells were treated with the indicated concentrations of TNF-α for 20 min before being lysed and analyzed by immunoblotting.
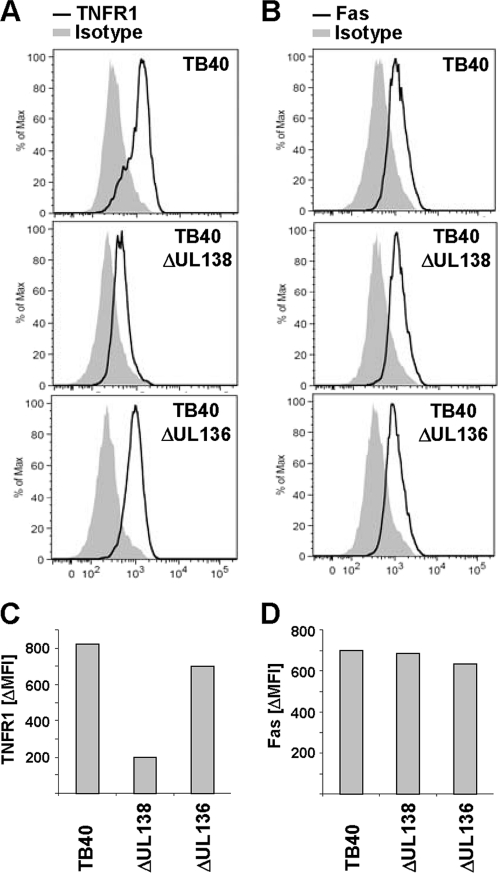
Deletion of UL138 results in decreased TNFR1 surface density on HCMV-infected cells.
To substantiate that pUL138 is required for the upregulation of TNFR1 in the context of HCMV infection, we constructed targeted deletion mutants of the ULb′-positive TB40 strain (59). Since it is known that UL138 is part of a transcription unit comprising UL138, UL136, UL135, and UL133 (48), we also deleted the neighboring gene UL136 to control effects caused by alterations within this transcription unit. The analysis of TB40ΔUL138 revealed that targeted deletion of UL138 resulted in strongly decreased TNFR1 surface amounts, whereas the control mutant TB40ΔUL136 showed unaltered TNFR1 surface densities at high levels (Fig. 5A and C), indicating that pUL138 is both sufficient (Fig. 3C and 4A) and essential (Fig. 5) for the modulation of TNFR1 surface expression. The determination of Fas surface distribution excluded a broad effect on surface receptors (Fig. 5B and D).
Fig. 5.
Deletion of UL138 results in decreased TNFR1 surface density on HCMV-infected cells. (A to D) MRC-5 fibroblasts infected with the indicated virus (MOI, 5) were stained at 72 hpi with anti-TNFR1 (A), anti-Fas (B), or the respective isotype control antibody and analyzed by flow cytometry. Shown are histograms of DAPI-negative cells. The difference between the mean fluorescence intensity (ΔMFI) of TNFR1 (C) and Fas signal (D) and the respective isotype control signal was calculated for better comparison.
Comparison of UL138 and ULb′ deficiencies defines the ULb′ region as a hub for TNF-α signaling regulation.
Interestingly, the TB40-based deletion of UL138 did not entirely mirror the loss of TNFR1 surface expression found for AD169varS- and Towne-infected cells (Fig. 2C and I). Since the comparative analysis of independent virus strains does not allow the unambiguous assessment of involved genes, we generated a bacterial artificial chromosome (BAC) derived from the AD169varL genome (see Fig. S5 in the supplemental material) to allow the AD169-based targeted deletion of UL138. AD169ΔUL138 enabled the direct comparison of UL138 deficiency to ULb′ deficiency, which is otherwise hampered by the fact that ULb′-deficient mutants derived from other HCMV strains exhibit a severely impaired replication efficiency (21, 25). The analysis of AD169-derived mutants concerning TNFR1 and Fas surface densities (Fig. 6A to D) fully validated the conclusion deduced from the TB40 data: the ULb′ region represents the hub for TNFR1 regulation within the HCMV genome, and pUL138 acts as the master regulator enhancing TNFR1 surface disposition. When we analyzed the virus dose dependency of TNFR1 modulation, we found that increasing virus doses (MOI, 2.5 to 10) elevated TNFR1 even further (Fig. 6E and F), indicating a positive UL138 gene-dose effect in the context of productive infection. To dissect the interplay of pUL138-mediated TNFR1 surface expression and TNF-α-induced IκBα degradation in the context of HCMV infection, we monitored IκBα levels in infected cells. Substantially decreased TNFR1 surface molecules on AD169ΔUL138-infected cells (Fig. 6A) were accompanied by reduced TNF-α-induced IκBα degradation (Fig. 6G). Notably, the surface level of TNFR1 on ΔUL138-infected cells (Fig. 5 and 6) seems to be sufficient to allow TNF-α-induced signaling resulting in IκBα degradation (Fig. 6G). Testing graded TNF-α doses revealed that pUL138 enhances IκBα degradation across a wide range of concentrations (see Fig. S6 in the supplemental material). The comparison of TNF-α-induced IκBα degradation in cells infected with AD169ΔUL138 to that of cells infected with AD169varS virus (Fig. 6G; also see Fig. S6) showed that the deletion of UL138 resulted in partial reversion, indicating that the HCMV ULb′ genome region encodes a multilayered and multifactorial positive regulation of TNF-α signaling, with pUL138 as the principle enhancer of TNFR1 surface expression.
Fig. 6.
Comparison of UL138 with ULb′ deficiency defines the ULb′ region as a hub for TNF-α signaling regulation. (A to F) MRC-5 fibroblasts infected with BAC-derived AD169varL, AD169ΔUL138, AD169ΔUL136, or AD169varS HCMV (MOI, 5). Cells were stained at 72 hpi with anti-TNFR1 (A), anti-Fas (B), or the respective isotype control antibody and analyzed by flow cytometry. Histograms of DAPI-negative cells are shown. The difference between the mean fluorescence intensities (ΔMFI) of TNFR1 (C) or Fas signal (D) and the respective isotype control signal are indicated. (E and F) The TNFR1 surface phenotype of ΔUL138 is virus dose independent. MRC-5 fibroblasts infected with the indicated virus (MOI of 2.5, 5, and 10) were stained at 72 hpi with anti-TNFR1, anti-Fas, or the respective isotype control antibody. DAPI-negative cells were analyzed by flow cytometry to calculate the difference between the mean fluorescence intensity (ΔMFI) of TNFR1 (E) or Fas signal (F) and the respective isotype control signal. (G) MRC-5 fibroblasts were mock treated or infected with BAC-derived AD169varL, AD169ΔUL138, AD169ΔUL136, or AD169varS HCMV (MOI, 5). Cells were incubated at 72 hpi with 20 ng/ml TNF-α for 15 min before being lysed for the immunoblot detection of IκBα protein. pUL83 and β-actin served as infection and loading controls, respectively.
DISCUSSION
Here, we document the intriguing finding of TNFR1 upregulation in cells infected with a human pathogenic virus, HCMV. Prompted by distinct phenotypic differences observed between variants of the HCMV laboratory strain AD169, an approach of systematic screening was pursued to identify the HCMV latency-associated gene product pUL138 as a master regulator of TNFR1-p55. Located within the ULb′ region of the unique long segment of the HCMV genome, UL138 became artificially lost in many highly passaged, fibroblast-adapted HCMV laboratory strains, while its sequence is consistently found and strictly conserved among all clinical virus isolates (54). As confirmed by our findings, cells infected with HCMV strains lacking the ULb′ region as represented by AD169varS and Towne were reported to exhibit a pronounced reduction of TNFR1 surface expression due to a relocalization of TNFR1 from the cell surface (2). This phenotype, which results from the secondary loss of UL138 and ULb′ genes, is sharply contrasting from the primary phenotype invariably observed in ULb′-positive viruses. Based on the fact that the UL138 gene is found only in the human and chimpanzee CMV genomes (13), the acquisition of UL138 appears to constitute a relatively recent event in the long phylogeny of the cytomegalovirus genus within the Betaherpesvirinae. Remarkably, the UL138-encoded transmembrane protein interacts selectively with TNFR1, leading to an extended half-life of its target protein, upregulated plasma membrane densities, and increased receptor-induced IκBα degradation upon ligand binding. Based on these findings, we propose for pUL138 the name viral potentiator of TNFR1 signaling.
The function of the pUL138 protein is exceptional, and to our knowledge no other microbial factor potentiating TNFR1 function has been reported to date. Given the cell death-inducing capabilities and the immunostimulatory, antiviral, and proinflammatory bioactivities of the pleiotropic cytokine TNF-α (23, 30, 42, 43, 67), the pUL138-mediated upregulation of TNFR1 leading to the sensitization of infected cells to TNF appears hazardous at first glance. For example, the human adenoviruses have devoted no less than four proteins to protect infected cells from TNF-mediated cytolysis, i.e., E1B/19K (70) and the E3 polypeptides 14.7K, 10.4K, and 14.5K (7, 40). Specifically, the cytoplasmic 14.7K protein of Ad2 was demonstrated to block ligand-induced TNFR1 internalization and recruitment of adaptor proteins to the death domain, although 14.7K itself was not detectable in the TNFR1 fractions, suggesting little or no direct interaction with proteins at the site of the activated receptor (56). Nevertheless, interactions between 14.7K and caspase 8 as well as with NEMO have been documented (10, 34, 39), indicating potential indirect interactions with TNFR1 in TNFR1-associated complexes.
The adenoviral proteins 10.4K and 14.5K form a heterotrimeric complex, composed of two 10.4K molecules and one 14.5K molecule (62), which blocks TNF-induced apoptosis (19, 20). Regarding their effect on different surface-resident transmembrane receptors (e.g., EGFR, FAS, TRAIL-R1, and TRAIL-R2), these molecules are termed the receptor internalization and degradation (RID) complex (4, 57, 64, 65). RID downregulates TNFR1 surface levels and is necessary and sufficient to block TNF-induced NF-κB and AP-1 activation (16). RID interacts with TNFR1 and acts by clathrin-dependent TNFR1 internalization and subsequent endolysosomal degradation (11) without affecting the rate of endocytosis itself (12). In contrast, the transmembrane protein pUL138 is directed to the trans-Golgi compartment (48), where it forms complexes with TNFR1 before TNFR1 molecules are released in a signaling-competent state to the plasma membrane, allowing potentiated ligand-triggered downstream signaling. Thus, pUL138 appears to represent an antipode of the adenoviral RID complex.
The data shown in Fig. 4 give the first mechanistic insights into the pUL138-mediated upregulation of TNFR1 surface densities: pUL138 interacts with TNFR1 to stabilize TNFR1 protein and to increase levels of mature TNFR1 molecules. Since other death domain (DD)-containing receptors are not influenced by pUL138, the rather conserved DD seems to be dispensable for the pUL138 effect. Truncation mutants of both proteins could be useful tools to further analyze the requirements for the pUL138 effect on TNFR1.
Given the proapoptotic and antiviral effects mediated by TNFR1, its upregulation by pUL138 is surprising and deserves comment. First, at the level of cellular target gene expression HCMV exhibits a stringent inhibition (Fig. 1E). Second, pUL138 is the principal HCMV-encoded cause of upregulated TNFR1 surface densities, but further viral modulators of TNF-α signaling must exist (Fig. 6), including the known HCMV proteins regulating NF-κB (49, 63).
These HCMV gene products may be both antagonizing and activating. AD169varS-infected cells reflect the fully antagonized situation, with the complete inhibition of TNF-α signaling at multiple levels of the signaling cascade (e.g., low TNFR1 surface densities and protein amount, lack of ligand-induced IκBα degradation, and blocked cellular target gene expression). The effects of the viral NF-κB inhibitors also are present in AD169varL-infected cells, but the ULb′-encoded positive regulators of TNF-α signaling and NF-κB activation appear to dominate or bypass the inhibitory proteins. pUL138 seems to take part in a complex viral network driving the active modulation and rerouting of TNF-α signaling. In fibroblast culture, the negative modulation of TNF responses might be more beneficial than positive regulation, leading to the counterselection of the ULb′ region. However, the presence of immune cells could reverse the situation and render a more complex regulation of cellular signaling pathways necessary. The precise molecular mechanisms, the functional hierarchy, and potential redundancy or division of labor of the ULb′-encoded gene products participating in the regulation network await extensive investigation. The interplay of NF-κB signals could widely differ depending on the cellular and viral context (cell type, environmental factors like TNF, stage of HCMV gene expression, etc.), leading to various dynamics and strengths of signal.
Considering that UL138 is one of the very few HCMV genes active in experimental as well as natural latency (21, 26), a relevance of upregulated TNFR1 surface densities for latency and reactivation seems obvious. During latency, HCMV gene transcription falls under cellular control, and the immediate-early genes ie1 and ie2 are repressed (55). To initiate transcriptional programs leading to productive infection, genes must be reactivated by cytokines or the viral activation of transcription factors like NF-κB (14, 61, 71). The mouse cytomegalovirus (MCMV) model supports a role for TNF-α in reactivation from latency (29, 58), although cells productively infected with MCMV display downregulated levels of TNFR1 (50). Four NF-κB target sites are found in the core and enhancer region of the HCMV major immediate-early promoter (MIEP) stimulating, for example, genes upon TNF-α exposure (52). This genetic make-up of the MIEP might facilitate or fully open entry into a productive mode of infection when sensing TNFR1 ligands. Assigning one of the very few latency transcripts of HCMV (i.e., UL81-82/LUNA, UL138/pUL138, or UL111.5A/LA-cmvIL-10) (3, 21, 32) to contrive TNFR1 hyperresponsiveness implies a central role of TNFR1 in the immune regulation of HCMV latency and progression to recurrent infection. Our findings could explain the long-standing observation that inflammation and immunological stress responses producing increased levels of TNFR1 signals readily trigger HCMV reactivation and subsequent clinical disease (17, 36, 47, 53).
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Messerle and E. Borst for advice and helpful discussions and S. Determann and A. Voges for excellent technical assistance.
This work was supported by DFG grant He2526/7-2.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 5 October 2011.
REFERENCES
- 1. Atalay R., et al. 2002. Identification and expression of human cytomegalovirus transcription units coding for two distinct Fc gamma receptor homologs. J. Virol. 76:8596–8608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baillie J., Sahlender D. A., Sinclair J. H. 2003. Human cytomegalovirus infection inhibits tumor necrosis factor alpha (TNF-alpha) signaling by targeting the 55-kilodalton TNF-alpha receptor. J. Virol. 77:7007–7016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bego M., Maciejewski J., Khaiboullina S., Pari G., St. Jeor S. 2005. Characterization of an antisense transcript spanning the UL81-82 locus of human cytomegalovirus. J. Virol. 79:11022–11034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benedict C. A., et al. 2001. Three adenovirus E3 proteins cooperate to evade apoptosis by tumor necrosis factor-related apoptosis-inducing ligand receptor-1 and -2. J. Biol. Chem. 276:3270–3278 [DOI] [PubMed] [Google Scholar]
- 5. Borst E. M., Hahn G., Koszinowski U. H., Messerle M. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320–8329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bradley A. J., et al. 2009. High-throughput sequence analysis of variants of human cytomegalovirus strains Towne and AD169. J. Gen. Virol. 90:2375–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burgert H. G., et al. 2002. Subversion of host defense mechanisms by adenoviruses. Viral Proteins Counteract. Host Defenses 269:273–318 [DOI] [PubMed] [Google Scholar]
- 8. Cha T. A., et al. 1996. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 70:78–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen G. Q., Goeddel D. V. 2002. TNF-R1 signaling: a beautiful pathway. Science 296:1634–1635 [DOI] [PubMed] [Google Scholar]
- 10. Chen P., Tian J., Kovesdi I., Bruder J. T. 1998. Interaction of the adenovirus 14.7-kDa protein with FLICE inhibits Fas ligand-induced apoptosis. J. Biol. Chem. 273:5815–5820 [DOI] [PubMed] [Google Scholar]
- 11. Chin Y. R., Horwitz M. S. 2005. Mechanism for removal of tumor necrosis factor receptor 1 from the cell surface by the adenovirus RIDalpha/beta complex. J. Virol. 79:13606–13617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chin Y. R., Horwitz M. S. 2006. Adenovirus RID complex enhances degradation of internalized tumour necrosis factor receptor 1 without affecting its rate of endocytosis. J. Gen. Virol. 87:3161–3167 [DOI] [PubMed] [Google Scholar]
- 13. Davison A. J., et al. 2009. The order Herpesvirales. Arch. Virol. 154:171–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. DeMeritt I. B., Milford L. E., Yurochko A. D. 2004. Activation of the NF-kappa B pathway in human cytomegalovirus-infected cells is necessary for efficient transactivation of the major immediate-early promoter. J. Virol. 78:4498–4507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dolan A., et al. 2004. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 85:1301–1312 [DOI] [PubMed] [Google Scholar]
- 16. Fessler S. P., Chin Y. R., Horwitz M. S. 2004. Inhibition of tumor necrosis factor (TNF) signal transduction by the adenovirus group C RID complex involves downregulation of surface levels of TNF receptor 1. J. Virol. 78:13113–13121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fietze E., et al. 1994. Cytomegalovirus infection in transplant recipients-the role of tumor necrosis factor. Transplantation 58:675–680 [PubMed] [Google Scholar]
- 18. Fishman J. A., Rubin R. H. 1998. Infection in organ-transplant recipients. N. Engl. J. Med. 338:1741–1751 [DOI] [PubMed] [Google Scholar]
- 19. Gooding L. R., et al. 1991. The 10,400-Dalton and 14,500-Dalton proteins encoded by region E3 of adenovirus function together to protect many but not all mouse-cell lines against lysis by tumor necrosis factor. J. Virol. 65:4114–4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gooding L. R., Sofola I. O., Tollefson A. E., Duerksenhughes P., Wold W. S. M. 1990. The adenovirus E3-14.7K protein is a general inhibitor of tumor necrosis factor-mediated cytolysis. J. Immunol. 145:3080–3086 [PubMed] [Google Scholar]
- 21. Goodrum F., Reeves M., Sinclair J., High K., Shenk T. 2007. Human cytomegalovirus sequences expressed in latently infected individuals promote a latent infection in vitro. Blood 110:937–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grainger L., et al. 2010. Stress-inducible alternative translation initiation of human cytomegalovirus latency protein pUL138. J. Virol. 84:9472–9486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grivennikov S. I., Kuprash D. V., Liu Z. G., Nedospasov S. A. 2006. Intracellular signals and events activated by cytokines of the tumor necrosis factor superfamily: from simple paradigms to complex mechanisms. Int. Rev. Cytol. 252:129–161 [DOI] [PubMed] [Google Scholar]
- 24. Hahn G., et al. 2002. The human cytomegalovirus ribonucleotide reductase homolog UL45 is dispensable for growth in endothelial cells, as determined by a BAC-cloned clinical isolate of human cytomegalovirus with preserved wild-type characteristics. J. Virol. 76:9551–9555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hahn G., et al. 2004. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J. Virol. 78:10023–10033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hargett D., Shenk T. E. 2010. Experimental human cytomegalovirus latency in CD14(+) monocytes. Proc. Natl. Acad. Sci. U. S. A. 107:20039–20044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hengel H., Esslinger C., Pool J., Goulmy E., Koszinowski U. H. 1995. Cytokines restore MHC class-I complex formation and control antigen presentation in human cytomegalovirus-infected cells. J. Gen. Virol. 76:2987–2997 [DOI] [PubMed] [Google Scholar]
- 28. Hirt B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365–369 [DOI] [PubMed] [Google Scholar]
- 29. Hummel M., et al. 2001. Allogeneic transplantation induces expression of cytomegalovirus immediate-early genes in vivo: a model for reactivation from latency. J. Virol. 75:4814–4822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ito M., Omalley J. A. 1987. Antiviral effects of recombinant human tumor necrosis factor. Lymphokine Res. 6:309–318 [PubMed] [Google Scholar]
- 31. Jarvis M. A., et al. 2006. Human cytomegalovirus attenuates interleukin-1 beta and tumor necrosis factor alpha proinflammatory signaling by inhibition of NF-kappa B activation. J. Virol. 80:5588–5598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jenkins C., Abendroth A., Slobedman B. 2004. A novel viral transcript with homology to human interleukin-10 is expressed during latent human cytomegalovirus infection. J. Virol. 78:1440–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Karin M., Ben-Neriah Y. 2000. Phosphorylation meets ubiquitination: the control of NF-kappa B activity. Annu. Rev. Immunol. 18:621–663 [DOI] [PubMed] [Google Scholar]
- 34. Kim H. J., Foster M. P. 2002. Characterization of Ad5 E3-14.7K, an adenoviral inhibitor of apoptosis: structure, oligomeric state, and metal binding. Protein Sci. 11:1117–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kornfeld R., Kornfeld S. 1985. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54:631–664 [DOI] [PubMed] [Google Scholar]
- 36. Kutza A. S. T., Muhl E., Hackstein H., Kirchner A., Bein G. 1998. High incidence of active cytomegalovirus infection among septic patients. Clin. Infect. Dis. 26:1076–1082 [DOI] [PubMed] [Google Scholar]
- 37. Lavrik I., Golks A., Krammer P. H. 2005. Death receptor signaling. J. Cell Sci. 118:265–267 [DOI] [PubMed] [Google Scholar]
- 38. Le V. T. K., Trilling M., Wilborn M., Hengel H., Zimmermann A. 2008. Human cytomegalovirus interferes with signal transducer and activator of transcription (STAT) 2 protein stability and tyrosine phosphorylation. J. Gen. Virol. 89:2416–2426 [DOI] [PubMed] [Google Scholar]
- 39. Li Y. G., et al. 1999. Identification of a cell protein (FIP-3) as a modulator of NF-kappa B activity and as a target of an adenovirus inhibitor of tumor necrosis factor alpha-induced apoptosis. Proc. Natl. Acad. Sci. U. S. A. 96:1042–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lichtenstein D. L., Toth K., Doronin K., Tollefson A. E., Wold W. S. M. 2004. Functions and mechanisms of action of the adenovirus E3 proteins. Int. Rev. Immunol. 23:75–111 [DOI] [PubMed] [Google Scholar]
- 41. Liou H. C. 2002. Regulation of the immune system by NF-kappa B and I kappa B. J. Biochem. Mol. Biol. 35:537–546 [DOI] [PubMed] [Google Scholar]
- 42. Lucin P., et al. 1994. Late-phase inhibition of murine cytomegalovirus replication by synergistic action of interferon-gamma and tumor necrosis factor. J. Gen. Virol. 75:101–110 [DOI] [PubMed] [Google Scholar]
- 43. MacEwan D. J. 2002. TNF receptor subtype signalling: differences and cellular consequences. Cell. Signal. 14:477–492 [DOI] [PubMed] [Google Scholar]
- 44. Mohamed M. R., McFadden G. 2009. NF kappa B inhibitors strategies from poxviruses. Cell Cycle 8:3125–3132 [DOI] [PubMed] [Google Scholar]
- 45. Montag C., Wagner J., Gruska I., Hagemeier C. 2006. Human cytomegalovirus blocks tumor necrosis factor alpha- and interleukin-1 beta-mediated NF-kappa B signaling. J. Virol. 80:11686–11698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Murphy E., et al. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 100:14976–14981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mutimer D., Mirza D., Shaw J., ODonnell K., Elias E. 1997. Enhanced (cytomegalovirus) viral replication associated with septic bacterial complications in liver transplant recipients. Transplantation 63:1411–1415 [DOI] [PubMed] [Google Scholar]
- 48. Petrucelli A., Rak M., Grainger L., Goodrum F. 2009. Characterization of a novel Golgi apparatus-localized latency determinant encoded by human cytomegalovirus. J. Virol. 83:5615–5629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Poole E., King C. A., Sinclair J. H., Alcami A. 2006. The UL144 gene product of human cytomegalovirus activates NF kappa B via a TRAF6-dependent mechanism. EMBO J. 25:4390–4399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Popkin D. L., Virgin H. W. 2003. Murine cytomegalovirus infection inhibits tumor necrosis factor alpha responses in primary macrophages. J. Virol. 77:10125–10130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Powers C., DeFilippis V., Malouli D., Fruh K. 2008. Cytomegalovirus immune evasion. Curr. Top. Microbiol. Immunol. 325:333–359 [DOI] [PubMed] [Google Scholar]
- 52. Prösch S., et al. 1995. Stimulation of the human cytomegalovirus IE enhancer promoter in HL-60 cells by TNF-alpha is mediated via induction of NF-kappa-B. Virology 208:197–206 [DOI] [PubMed] [Google Scholar]
- 53. Prösch S., et al. 2000. A novel link between stress and human cytomegalovirus (HCMV) infection: sympathetic hyperactivity stimulates HCMV activation. Virology 272:357–365 [DOI] [PubMed] [Google Scholar]
- 54. Qi Y., et al. 2009. Human cytomegalovirus UL138 open reading frame is highly conserved in clinical strains. Chin. Med. Sci. J. 24:107–111 [DOI] [PubMed] [Google Scholar]
- 55. Reeves M. B., Macary P. A., Lehner P. J., Sissons J. G. P., Sinclair J. H. 2005. Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc. Natl. Acad. Sci. U. S. A. 102:4140–4145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schneider-Brachert W., et al. 2006. Inhibition of TNF receptor 1 internalization by adenovirus 14.7K as a novel immune escape mechanism. J. Clin. Investig. 116:2901–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shisler J., Yang C., Walter B., Ware C. F., Gooding L. R. 1997. The adenovirus E3-10.4K/14.5K complex mediates loss of cell surface Fas (CD95) and resistance to fas-induced apoptosis. J. Virol. 71:8299–8306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Simon C. O., Seckert C. K., Dreis D., Reddehase M. J., Grzimek N. K. A. 2005. Role for tumor necrosis factor alpha in murine cytomegalovirus transcriptional reactivation in latently infected lungs. J. Virol. 79:326–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sinzger C., et al. 2008. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J. Gen. Virol. 89:359–368 [DOI] [PubMed] [Google Scholar]
- 60. Sinzger C., et al. 1999. Modification of human cytomegalovirus tropism through propagation in vitro is associated with changes in the viral genome. J. Gen. Virol. 80:2867–2877 [DOI] [PubMed] [Google Scholar]
- 61. Söderberg-Nauclér C., Fish K. N., Nelson J. A. 1997. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell 91:119–126 [DOI] [PubMed] [Google Scholar]
- 62. Stewart A. R., Tollefson A. E., Krajcsi P., Yei S. P., Wold W. S. M. 1995. The adenovirus E3 10.4K and 14.5K proteins, which function to prevent cytolysis by tumor necrosis factor and to down-regulate the epidermal growth factor receptor, are localized in the plasma membrane. J. Virol. 69:172–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Taylor R. T., Bresnahan W. A. 2006. Human cytomegalovirus IE86 attenuates virus- and tumor necrosis factor alpha-induced NFKB-dependent gene expression. J. Virol. 80:10763–10771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tollefson A. E., et al. 1998. Forced degradation of Fas inhibits apoptosis in adenovirus-infected cells. Nature 392:726–730 [DOI] [PubMed] [Google Scholar]
- 65. Tollefson A. E., et al. 2001. Inhibition of TRAIL-induced apoptosis and forced internalization of TRAIL receptor 1 by adenovirus proteins. J. Virol. 75:8875–8887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Trilling M., et al. 2009. Gamma interferon-induced interferon regulatory factor 1-dependent antiviral response inhibits vaccinia virus replication in mouse but not human fibroblasts. J. Virol. 83:3684–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Van Herreweghe F., Festjens N., Declercq W., Vandenabeele P. 2010. Tumor necrosis factor-mediated cell death: to break or to burst, that's the question. Cell. Mol. Life Sci. 67:1567–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wagner M., Gutermann A., Podlech J., Reddehase M. J., Koszinowski U. H. 2002. Major histocompatibility complex class I allele-specific cooperative and competitive interactions between immune evasion proteins of cytomegalovirus. J. Exp. Med. 196:805–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wajant H., Pfizenmaier K., Scheurich P. 2003. Tumor necrosis factor signaling. Cell Death Differ. 10:45–65 [DOI] [PubMed] [Google Scholar]
- 70. White E. 1998. Regulation of apoptosis by adenovirus E1A and E1B oncogenes. Semin. Virol. 8:505–513 [Google Scholar]
- 71. Yurochko A. D., Kowalik T. F., Huong S. M., Huang E. S. 1995. Human cytomegalovirus up-regulates NF-kappa-B activity by transactivating the NF-kappa-B P105/P50 and P65 promoters. J. Virol. 69:5391–5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.