Abstract
Human herpesvirus 6 (HHV-6) is a T cell-tropic betaherpesvirus. HHV-6 can be classified into two variants, HHV-6A and HHV-6B, based on differences in their genetic, antigenic, and growth characteristics and cell tropisms. The function of HHV-6B should be analyzed more in its life cycle, as more than 90% of people have the antibodies for HHV-6B but not HHV-6A. It has been shown that the cellular receptor for HHV-6A is human CD46 and that the viral ligand for CD46 is the envelope glycoprotein complex gH/gL/gQ1/gQ2; however, the receptor-ligand pair used by HHV-6B is still unknown. In this study, to identify the glycoprotein(s) important for HHV-6B entry, we generated monoclonal antibodies (MAbs) that inhibit infection by HHV-6B. Most of these MAbs were found to recognize gQ1, indicating that HHV-6B gQ1 is critical for virus entry. Interestingly, the recognition of gQ1 by the neutralizing MAb was enhanced by coexpression with gQ2. Moreover, gQ1 deletion or point mutants that are not recognized by the MAb could nonetheless associate with gQ2, indicating that although the MAb recognized the conformational epitope of gQ1 exposed by the gQ2 interaction, this epitope was not related to the gQ2 binding domain. Our study shows that HHV-6B gQ1 is likely a ligand for the HHV-6B receptor, and the recognition site for this MAb will be a promising target for antiviral agents.
INTRODUCTION
Human herpesvirus 6 (HHV-6) was first isolated from patients with lymphocytic disorders in1986 (36) and was subsequently shown to be the causative agent of exanthem subitum (ES) (48). Currently, HHV-6 can be classified into two variants, HHV-6A and HHV-6B, based on differences in genetic, antigenic, and growth characteristics and cell tropisms (1, 5, 7, 8). HHV-6B causes infant ES, and more than 90% of people have antibodies (Abs) against HHV-6B (31, 38), while the pathogenesis of HHV-6A is still unknown. Recently, it was shown that a reactivation of HHV-6B causes encephalitis in immunocompromised hosts (13, 45, 46) and possibly enhances the severity of drug-induced sensitivity syndrome (14).
Human CD46, a regulator of the complement activation receptor expressed on all nuclear cells, is a receptor for HHV-6 (37), and its viral ligand is the envelope glycoprotein complex gH/gL/gQ1/gQ2 (3, 28). Although this complex can bind CD46 (28), those of some clinical isolates, including laboratory strains of HHV-6B, do not bind it (24, 26). The gQ gene is unique because it is conserved only among HHV-6A, HHV-6B, and HHV-7 (12, 15, 19). Recently, we successfully reconstituted a virus from the HHV-6 genome (43) and found that HHV-6 gQ1 is essential for virus growth and probably for entry.
As monoclonal antibodies (MAbs) against gH and gB inhibit virus-induced cell fusion and infection, gH and gB are thought to be fusogenic candidates (39). In addition, as it is common to herpesviruses generally, gH homologues expressed on viral envelopes form a complex with gL homologues (18, 20, 21).
In addition to gH/gL/gQ1/gQ2, another gH/gL complex, gH/gL/gO, is present in the viral envelopes of both HHV-6 variants (24, 26, 44), and this complex may also be important for virus entry. Since the amino acid identities of gQ1 and gO between the two variants are 76.55% and 73.48%, respectively, the complexes may be important determinants of different viral tropisms between both variants. Human cytomegalovirus also has two gH/gL complexes: gH/gL/UL128-131 and gH/gL/gO. These complexes were shown previously to be related to viral cell tropism for entry processes (33–35, 47).
Because reactivated HHV-6B, and not HHV-6A, causes several diseases in immunocompromised patients (49), and as primary infection by HHV-6B also causes diseases in infants (16, 48), it is essential to identify the viral and cellular molecules mediating HHV-6B infection.
Several MAbs against the HHV-6B glycoproteins gH and gB that neutralize the virus have been established (40, 41). Although the MAb that recognizes gp82-gp105 (gQ1) was shown previously to have neutralizing activity against HHV-6A (32), it is still unknown whether HHV-6B gQ1 functions in viral entry. As described above, since another gH/gL complex, gH/gL/gO, is also present in the viral envelope (26), these two complexes may work for variant-specific cell tropisms.
To determine which viral molecule(s) functions in HHV-6B entry and cellular receptor binding, we generated MAbs that prevent virus entry. Interestingly, the neutralizing MAbs obtained were almost all against gQ1, indicating that as for HHV-6A gQ1, HHV-6B gQ1 plays an essential role in virus entry and is a promising candidate for antiviral therapy.
MATERIALS AND METHODS
Cells and viruses.
The T cell lines MT4 and HSB-2 were cultured in RPMI 1640 medium supplemented with 8% fetal bovine serum. The human embryonic kidney cell line 293T was cultured in Dulbecco's modified Eagle's medium supplemented with 8% fetal bovine serum. Umbilical cord blood mononuclear cells (CBMC) were prepared as described previously (11).
HHV-6 strains GS (HHV-6A) and HST (HHV-6B) and clinical isolate KYO (HHV-6B) were propagated in CBMCs, and the titers of the GS and HST or KYO viruses were estimated by using HSB-2 and MT4 cells, respectively. Virus purification was performed as described previously (27). This study was approved by the ethics committees of all the institutions involved.
Antibodies.
Hybridoma clones producing MAbs were established from the splenocytes of mice immunized with a purified recombinant protein or UV-inactivated HHV-6 virions as described previously (11). MAbs recognizing gQ1 were established as described previously (3, 25). The MAb designated BgQ1-202 was raised against recombinant protein BgQ1-N. BgQ1-N was created by the following procedure. A primer pair (BgQ1bamF [5′-accggatccGTCTACCGAGATGCGGGAAC-3′] and BgQ1pstR [5′-accctgcagCGTGCAGGTTTCCCAATC]) was used to amplify inserts from HHV-6B cDNA (strain HST). The PCR products were inserted into the prokaryotic expression vector pQE30 (Qiagen) at BamHI and PstI restriction sites (ggatcc and ctgcag). The resulting expression plasmid encoded the gQ1 gene product with an N-terminal tag containing six histidine residues and was designated BgQ1-N. The recombinant proteins were expressed in Escherichia coli cells and purified under denaturing conditions as specified by the manufacturer (Qiagen). UV-inactivated virions (HST strain) were used as an antigen for the immunization of mice to raise MAb KH-1. Purified virions (GS strain) were used as an antigen for the immunization of mice to raise MAbs gH1-1 (for gH) and gQ2-4 (for gQ2). MAbs AgL3 (against HHV-6 gL) and OHV-1 (against HHV-6 gB) were described previously (3, 28). The MAb against the hemagglutinin (HA) tag (clone HA-7; Sigma) was purchased. Polyclonal antibodies (Abs) for c-myc (MBL), TGN46 (AbD Serotec), and calnexin (Cell Signaling Technology) were purchased. Secondary Abs were Alexa Fluor 488- or 594-conjugated F(ab′) fragments of donkey anti-mouse, -sheep, or -rabbit immunoglobulin G (IgG) (Invitrogen). The isotype and subtype of MAb KH-1, OHV-1, or BgQ1-202 were determined with the IsoQuick kit for mouse monoclonal isotyping (Sigma).
Immunofluorescence assay.
Indirect immunofluorescence assays (IFAs) were performed as described previously (3). Specific immunofluorescence was observed with a confocal laser scanning microscope (Nikon TE300 Bio-Rad Radiance 2100).
Preparation of metabolically labeled proteins and immunoprecipitation.
MT4 cells were infected with HHV-6B strain HST or were mock infected. At 96 h postinfection, the cells were incubated in methionine-free RPMI medium for 1 h and then radiolabeled with 20 μCi of [35S]methionine (Perkin-Elmer) per ml for 16 h (25). The cells were recovered and lysed in radioimmunoprecipitation assay (RIPA) buffer (0.01 M Tris-HCl [pH 7.4], 0.15 M NaCl, 1 mM EDTA, 1% sodium deoxycholate, 1% Nonidet P-40, 0.05% sodium dodecyl sulfate [SDS]) for 30 min on ice. After centrifugation at 200,000 × g for 1 h, the supernatants were incubated with a MAb (KH-1 or BgQ1-202)-protein G-Sepharose (GE Healthcare Biosciences) complex at 4°C for 4 h. Immunocomplexes were washed with RIPA buffer to remove unbound proteins. Precipitated proteins were solubilized with sample buffer (32 mM Tris-HCl [pH 6.8], 1.5% SDS, 5% glycerol, 2.5% 2-mercaptoethanol) and separated by electrophoresis in Nu-PAGE gels (Invitrogen).
Western blotting.
Western blotting was performed as described previously (3). The bound Abs were detected by chemiluminescence using Western blotting detection reagents (GE Healthcare Biosciences) and were visualized with the LAS minidetection system (GE Healthcare Biosciences).
Plasmid construction.
The HHV-6B gQ1 wild-type, gQ1 carboxyl-terminal deletion mutant, and gQ1 point mutant expression vectors were constructed by using plasmids pCAGGS and pcDNA3.1(+) (Invitrogen). Plasmid pCAGGS was kindly provided by Jun-ichi Miyazaki, Osaka University, Japan (30). HHV-6B gQ1 expression plasmid BgQ1 was generated by inserting the PCR products into pcDNA3.1(+) at the EcoRI restriction site of BgQ1. Plasmids expressing the carboxyl-terminal deletion mutants of HHV-6B gQ1, named BgQ1Δ505gQ1Δ497, BgQ1Δ495, BgQ1Δ484, and BgQ1Δ467, were also generated. The amplified DNA fragments were digested with EcoRV and XhoI and ligated into the digested pcDNA3.1(+) vector. The N-terminal deletions of HHV-6B gQ1, named BgQ1Δ43, BgQ1Δ58, BgQ1Δ70, BgQ1Δ81, and BgQ1Δ102, were also generated. The amplified DNA fragments were digested with BglII and NheI and ligated into digested pFUSE-hIgG1-Fc2vector (Invivogen). The N-terminal deletion mutants were tagged with 6× histidine at the C terminus of the gQ1 sequence. The predicted signal sequence of BgQ1 (amino acids [aa] 1 to 25) was removed, and the interleukin-2 (IL-2) signal sequence in pFUSE-hIgG1-Fc2vector was used instead of the original sequence. HHV-6B gQ1 point mutants were generated by use of a QuikChange Lightning multisite-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. The expression plasmids for myc-tagged HHV-6B gQ2, termed BgQ2-myc, and HA-tagged gQ2, termed BgQ2-HA, were generated by inserting the PCR products into pCAGGS.
Plasmid transfection.
293T cells were transfected by the calcium phosphate method as described previously (22).
Virus neutralization assay.
Virus titers were determined by measuring the 50% tissue culture infective dose (TCID50) (4). One hundred microliters of serial 10-fold dilutions of purified MAb KH-1 (0.4 mg/ml), OHV-1 (0.4 mg/ml), or BgQ1-202 (0.4 mg/ml) was incubated with 100 μl of HST virus solution at 3 × 104 TCID50 at 37°C for 30 min. MT4 cell pellets (5 × 105 cells) were then added to the virus-antibody solution and incubated at 37°C for 1 h. After incubation, cells were washed with RPMI medium and cultured for 4 days. The presence or absence of viral antigen expression was detected by IFA.
RESULTS
MAb KH-1 recognizes HHV-6B but not HHV-6A.
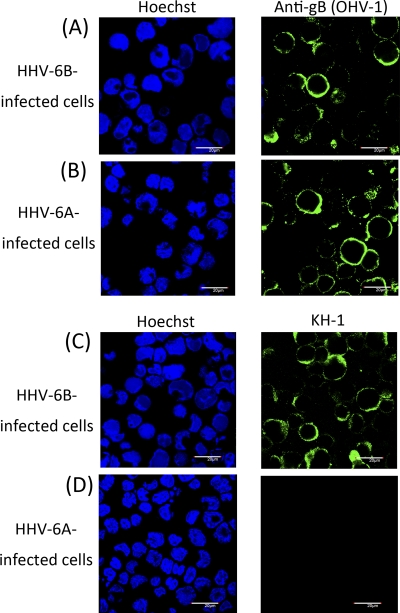
We obtained monoclonal antibody (MAb) KH-1, which blocks HHV-6B entry. The subtype of this MAb was found to be IgG2b. To confirm its specificity, HHV-6A- and HHV-6B-infected cells were treated with MAb KH-1. As shown in Fig. 1, as expected, KH-1 recognized HHV-6B-infected cells (Fig. 1C) but not HHV-6A-infected cells (Fig. 1D), whereas an anti-gB MAb that recognizes both variants reacted with cells infected with both viruses (Fig. 1A and B), confirming that MAb KH-1 recognizes only HHV-6B antigen. In other experiments, KH-1 was found to react with other HHV-6B clinical isolates (data not shown).
Fig. 1.
HHV-6B-specific reactivity to MAb KH-1. MT4 cells infected with HHV-6B strain HST (A and C) or HSB-2 cells infected with HHV-6A strain GS (B and D) were stained with anti-gB MAb OHV-1 (A and B) or MAb KH-1 (C and D) and Hoechst 33258 dye. Scale bar, 20 μm.
Neutralizing monoclonal antibody KH-1 recognizes HHV-6B gQ1.
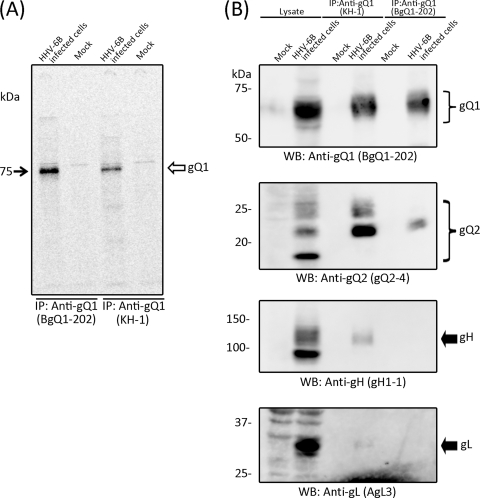
To identify the viral molecule(s) recognized by KH-1, immunoprecipitation was performed with [35S]methionine-labeled HHV-6B-infected cell lysates. As shown in Fig. 2A, most of the precipitate was an approximately 74-kDa protein. Interestingly, a similar band was found in precipitates with MAb BgQ1-202, which recognizes HHV-6B gQ1 (Fig. 2A) and which can be used to detect the protein on Western blots, indicating that the protein precipitated by KH-1 may be gQ1. To confirm this, the KH-1 precipitates were used to generate Western blots and were probed with anti-gQ1 MAb BgQ1-202. As expected, the antibody recognized bands from the precipitate in the range of around 74 kDa, thus indicating that KH-1 recognizes gQ1 (Fig. 2B). Since we previously found that HHV-6A gQ1 (AgQ1) associates with gQ2 in a gH/gL complex (3), we examined whether HHV-6B gQ1 (BgQ1) also associates with a gH/gL/gQ2 complex. We therefore probed Western blots of the KH-1 precipitates with anti-gH, -gL, and -gQ2 MAbs. As expected, gH, gL, and gQ2 were all detected in the precipitates, while the other MAb, BgQ1-202, barely precipitated the gH/gL/gQ1/gQ2 complex under the same conditions (Fig. 2B), indicating that KH-1 recognizes BgQ1 and that this BgQ1 also associates with gH, gL, and gQ2 in infected cells. The specificities of the MAbs used here are shown in Table 1.
Fig. 2.
Characterization of KH-1, a neutralizing MAb against HHV-6B. (A) Mock- or HHV-6B (strain HST)-infected cells were metabolically labeled with [35S]methionine for 16 h, lysed, and immunoprecipitated (IP) with either MAb KH-1 or BgQ1-202, which recognizes HHV-6B gQ1. The immunoprecipitates were resolved by SDS-PAGE (4 to 12% Tris-glycine gel; Invitrogen). (B) Mock- or HHV-6B (strain KYO)-infected cell lysates were immunoprecipitated with MAb KH-1 and subjected to SDS-PAGE, followed by Western blotting (WB), and probed with MAbs BgQ1-202 for gQ1, gH1-1 for gH, AgL3 for gL, and gQ2-4 for gQ2. Numbers beside the panels show molecular masses in kilodaltons. The specificities of the MAbs used here are shown in Table 1.
Table 1.
MAbs that recognize the HHV-6 glycoproteins
| MAb | HHV-6 glycoprotein recognized by MAb | Specificity toward HHV-6A and HHV-6B variants |
|
|---|---|---|---|
| HHV-6A | HHV-6B | ||
| KH-1 | gQ1 | − | + |
| BgQ1-202 | gQ1 | − | + |
| gH1-1 | gH | + | + |
| gQ2-4 | gQ2 | + | + |
| AgL3 | gL | + | + |
| OHV-1 | gB | + | + |
MAb KH-1 neutralized HHV-6B but not HHV-6A. We further examined the ability of KH-1 to neutralize HHV-6B. In this assay, we found that the antibody had a neutralizing activity and that 100 μl of a 1,000-fold dilution of purified MAb KH-1 (0.4 mg/ml) completely blocked infection by HHV-6B strain HST at 3 × 104 TCID50, while the control MAbs (OHV-1 [IgG2b] and BgQ1-202 [IgG2a]) never blocked its infection even at a 10-fold dilution of them (0.4 mg/ml).
Here we generated MAbs by immunizing mice with UV-inactivated HHV-6B virions. As results, we obtained 7 kinds of MAbs against HHV-6 envelope glycoproteins; 6 of the 7 MAbs had neutralizing activity, and 5 of the 6 neutralizing MAbs were against HHV-6B gQ1 (data not shown).
Cloning of HHV-6B gQ1 and gQ2 cDNAs and confirmation of MAb KH-1 specificity.
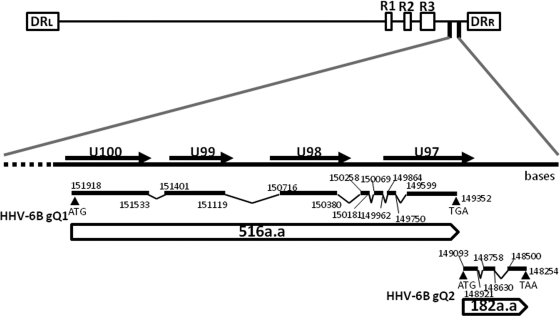
To confirm that the MAb recognizes BgQ1, we isolated gQ1 cDNAs in order to generate protein. BgQ1 cDNAs were amplified by reverse transcription (RT)-PCR and sequenced. As HHV-6B gQ1 transcripts produce multiple splice variants (data not shown), the major transcript homologue of AgQ1 was chosen and cloned into expression vectors (Fig. 3). HHV-6B gQ2 (BgQ2) cDNA was also amplified and sequenced and is shown in Fig. 3. The BgQ1 and BgQ2 cDNAs are predicted to encode proteins of 516 and 182 amino acid residues, respectively.
Fig. 3.
Schematic representation of the HHV-6B genome and the HHV-6 BgQ gene region. All exons are drawn as black lines, and all introns are shown as lines connected to exons. Numbers indicate the nucleotide positions in the HHV-6B strain HST genome. Black arrowheads indicate the start and stop codons of HHV-6B gQ1 and gQ2. White boxes indicate the predicted open reading frames (ORFs) in the genom. The direct repeat termini (DRL and DRR).
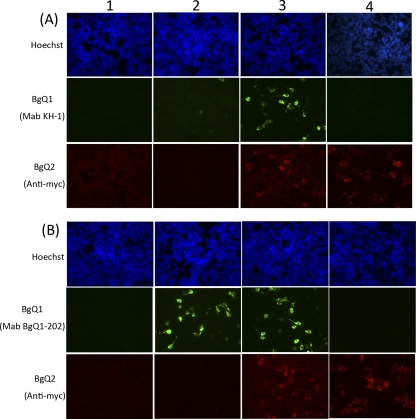
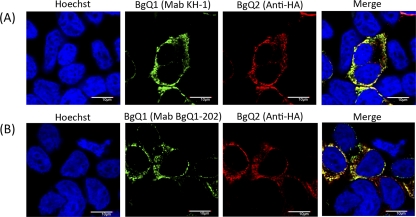
The BgQ1 expression plasmid was transfected into 293T cells, and the cells were harvested, fixed, and stained using either KH-1 (Fig. 4A) or BgQ1-202 (Fig. 4B). As expected, BgQ1 transfectants (Fig. 4A2 and B2) but not the empty vector (Fig. 4A1 and B1) transfectants were recognized by KH-1, although the signal was very weak, while BgQ1-202 reacted with BgQ1 much more clearly. Interestingly, the reaction of KH-1 with BgQ1 antigen was much greater when BgQ1 was coexpressed with BgQ2 (Fig. 4A3). In contrast, the reaction of BgQ1-202 with BgQ1 antigen was not changed when coexpressed with BgQ2. The BgQ1 recognized by KH-1, but not by BgQ1-202, appeared to absolutely increase by the addition of BgQ2. However, BgQ2 colocalized with the versions of BgQ1 recognized by both MAbs (Fig. 5).
Fig. 4.
Analysis of the protein recognized by neutralizing MAb KH-1. 293T cells were either mock transfected (panels 1) or transfected with the pcDNA3.1(+)-BgQ1 vector (panels 2), pCAGGS-BgQ2-myc (panels 4), or pcDNA3.1(+)-BgQ1 with pCAGGS-BgQ2-myc (panels 3). The cells were harvested at 72 h posttransfection and then fixed and stained for IFA with MAb KH-1 (A) or MAb BgQ1-202 (B) and with c-myc antibody for gQ2 and Hoechst 33258 dye.
Fig. 5.
Localization of gQ1 and gQ2 proteins recognized by MAb KH-1 or BgQ1-202 in transfected cells. 293T cells were transfected with the pcDNA3.1(+)-BgQ1 vector and pCAGGS-BgQ2-HA. The cells were harvested at 72 h posttransfection and then fixed and stained for IFA with MAb KH-1 (A) or BgQ1-202 (B) and with HA antibody for gQ2 and Hoechst 33258 dye. Scale bar, 10 μm.
KH-1 did not react with BgQ2 alone (Fig. 4A4), confirming that it recognized BgQ1 in this assay.
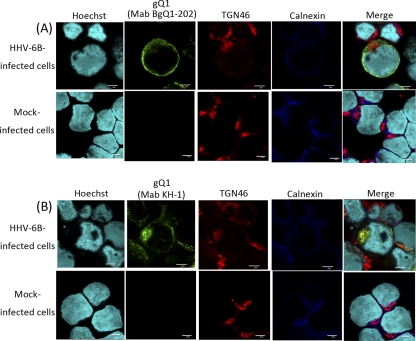
Since our evidence suggests that the KH-1 recognition of BgQ1 is sensitive to the BgQ1 conformation and that this conformation is altered by interactions with BgQ2, we sought to determine whether there were conformational differences within infected cells by comparing stainings with KH-1 and BgQ1-202, which recognizes a linear epitope in BgQ1, in HHV-6B-infected cells at a late infectious phase. Interestingly, BgQ1 stained with KH-1 was colocalized with TGN46, a trans-Golgi network (TGN) marker, in addition to calnexin, an endoplasmic reticulum (ER) marker (Fig. 6B), while BgQ1 stained with BgQ1-202 was colocalized mainly with calnexin (Fig. 6A).
Fig. 6.
Localization of the gQ1 protein by MAb KH-1 or BgQ1-202 in HHV-6B-infected cells. HHV-6B-infected MT4 cells or mock-infected cells were harvested at 96 h postinfection and then fixed and stained for IFA. The cells were costained with antibodies against gQ1 (MAb BgQ1-202), TGN46, and calnexin (A) or gQ1 (MAb KH-1), TGN46, and calnexin (B). Scale bar, 5 μm.
Generation of BgQ1 carboxyl- or amino-terminal truncation mutants and their reactivity with the MAb.
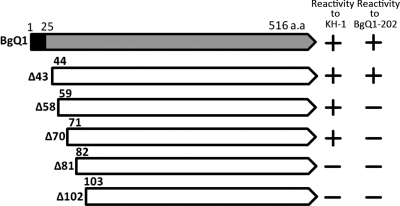
To identify the recognition site(s) for KH-1, BgQ1 carboxyl-terminal (C-terminal) truncation mutants were generated, and their reactivity with the KH-1 and BgQ1 was examined by IFA. As shown in Fig. 7, the mutant with a C-terminal truncation beginning at residue 495 was recognized with MAb KH-1, while the mutants truncated beginning at residue 484 were not recognized, suggesting that the important site(s) for MAb KH-1 recognition exists between residues 484 and 496. In contrast, BgQ1-202 reacted with all truncation mutants shown in Fig. 7. We also constructed amino-terminal (N-terminal) truncation mutants and examined their reactivity with the MAb (Fig. 8). The mutant with an N-terminal truncation at residues 1 to 70 was recognized by MAb KH-1, while the truncation mutant at residues 1 to 81 was not recognized, suggesting that the important site(s) at the N terminus for MAb KH-1 recognition exists between residues 71 and 81. The recognition site(s) for BgQ1-202 was found to be between residues 44 and 58.
Fig. 7.
Schematic diagram of HHV-6B gQ1 mutants with various carboxy-terminal truncations and their reactivities to MAb KH-1. 293T cells were cotransfected with a plasmid expressing gQ1 or individual gQ1 truncation mutants and a plasmid expressing gQ2. The cells were stained for IFA at 72 h posttransfection with MAb KH-1 or BgQ1-202. The reactivities of KH-1 and BgQ1-202 for each gQ1 variant are shown at the right.
Fig. 8.
Schematic diagram of HHV-6B gQ1 mutants with various amino-terminal truncations and their reactivities to MAb KH-1. 293T cells were cotransfected with a plasmid expressing gQ1 or individual gQ1 truncation mutants and a plasmid expressing gQ2. The cells were stained for IFA at 72 h posttransfection with MAb KH-1 or BgQ1-202. The reactivities of KH-1 and BgQ1-202 for each gQ1 variant are shown at the right. The black box indicates the predicted signal sequence (aa 1 to 25) on HHV-6B gQ1. The N-terminal deletion mutants without the signal sequence (aa 1 to 25) containing a 6×His tag at the C terminus were fused to the IL-2 signal sequence instead of that of gQ1.
Generation of BgQ1 point mutants and their reactivity with KH-1 and BgQ1-202.
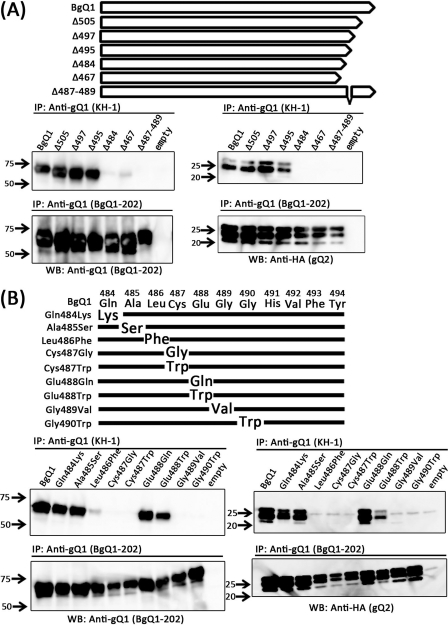
To define in more detail the recognition site(s) for KH-1, point mutants were generated. Before these mutants were generated, the amino acid sequences within the region from residues 484 to 494 in the two variants were compared (Fig. 9). The comparison revealed that Glu488 of HHV-6B was replaced with Gln in HHV-6A (Fig. 10A). Therefore, a mutant BgQ1 in which Glu488 was replaced with Gln was generated (Glu488Gln), and its reactivity with KH-1 was examined by IFA. Unexpectedly, this mutant reacted with KH-1 (Glu488Gln in Fig. 10). Since Gln is a neutral amino acid, the same as Glu, Glu488 was next replaced with Trp, a hydrophobic amino acid; however, this mutant also reacted with KH-1 (Glu488Trp in Fig. 10B). We therefore generated several BgQ1 mutants in which residues surrounding Glu488 were replaced, as shown in Fig. 10. Interestingly, the reactivity with KH-1 significantly decreased with the substitutions Leu486Phe, Gly489Val, and Gly490Trp but not with the substitutions Glu488Gln and Ala485Ser. We therefore next generated and tested a deletion mutant lacking residues 487 to 489. As expected, this mutant did not react with KH-1 at all. In contrast, all mutants reacted with MAb BgQ1-202.
Fig. 9.
Amino acid sequence alignment of HHV-6A and HHV-6B gQ1. Amino acids conserved between the two variants are in shaded boxes. The numbers on the right denote amino acid residues. U1102, HHV-6A variant strain; HST, HHV-6B variant strain.
Fig. 10.
gQ1 amino acid sequence comparison between HHV-6A and HHV-6B and schematic diagram of gQ1 mutants and their reactivities to MAb KH-1. (A) Amino acid sequence comparison of gQ1 (residues 484 to 494) of HHV-6 strains HST and GS. The white arrow-shaped box indicates the direction of the HST gQ1 protein. Amino acid residues 484 to 494 of HST gQ1 were enlarged and are compared with those of GS. Differences in amino acid sequences between the variants are indicated by a box. (B) Substitutions of amino acid residues at the indicated positions of gQ1 were introduced into the gQ1 gene of pcDNA3.1(+)-BgQ1 as described in Materials and Methods. 293T cells were cotransfected with a plasmid expressing gQ1 or each gQ1 point mutant and a plasmid expressing gQ2. The cells were stained for IFA at 72 h posttransfection with MAb KH-1 or BgQ1-202. The reactivity of MAb KH-1 or BgQ1-202 for each gQ1 variant is shown at the right.
On the other hand, a point mutation in the N terminus was also generated. The comparison of the amino acid sequences within the region at residues 71 to 81 in the two variants revealed that Pro72 of HHV-6B was replaced with Ser in HHV-6A (Fig. 9). Therefore, a mutant BgQ1 in which Pro72 was replaced with Ser was generated (Pro72Ser), and its reactivity with KH-1 was examined by IFA. However, this mutant also reacted with KH-1 (data not shown).
The BgQ1 C-terminal mutants that are not recognized by MAb KH-1 interact with BgQ2.
Next, to examine the interaction of BQ1 mutants with BgQ2, we immunoprecipitated BQ1 from extracts of BgQ1 and BgQ2 cotransfectants and examined the immunoprecipitates for the presence of BQ1 and BQ2. Interestingly, the BgQ1 mutants not recognized by KH-1, including both deletion and point mutants, nonetheless interacted with BgQ2 (Fig. 11).
Fig. 11.
Detection of gQ1 mutants by immunoprecipitation with MAb KH-1 and their interactions with gQ2. Shown are diagrams of gQ1 deletion mutants (A) and point mutants (B). 293T cells were cotransfected with a plasmid expressing complete gQ1, an empty vector (without the gQ1 gene), each gQ1 deletion mutant (A), or each gQ1 point mutant (B) with a plasmid expressing gQ2. The cells were lysed and immunoprecipitated with MAb KH-1 or BgQ1-202. The precipitated proteins were resolved by SDS-PAGE and detected after Western blotting with BgQ1-202 or HA antibody for gQ2. Numbers beside the panels show molecular masses in kilodaltons.
DISCUSSION
Envelope glycoproteins of herpesviruses are critical for virus entry (2, 39, 44). Structural analyses have been reported for the herpesvirus glycoproteins gH/gL, gB, gD, and gp42, which all function in this process (6, 9, 10, 17, 23, 29).
In the case of HHV-6A, the envelope glycoprotein complex gH/gL/gQ1/gQ2 has been shown to be critical for virus entry (43) through binding to its cellular receptor, human CD46 (26, 28). In order for receptor binding to occur, complex formation is required, and, moreover, they all are needed for their correct folding and their trafficking (42). Although the gQ1 molecule exists in HHV-6B (26), it was previously unknown whether gQ1 is critical for HHV-6B receptor binding or entry.
By screening for MAbs with neutralizing activity against HHV-6B, we have identified neutralizing MAb KH-1, which we have shown specifically recognizes HHV-6B gQ1, indicating that HHV-6B gQ1 plays a critical role in HHV-6B entry.
By immunoprecipitation using KH-1, we found that the complex gH/gL/gQ1/Q2 is also present in the HHV-6B viral envelope. This complex may also therefore bind to the HHV-6B cellular receptor.
Interestingly, the recognition of BgQ1 by KH-1, but not that by BgQ1-202, which recognizes a linear epitope of BgQ1, was much stronger when BgQ1 was coexpressed with BgQ2. KH-1 likely recognizes a specific conformation of BgQ1, and the conformational change induced by the interaction with BgQ2 is important for its recognition. Thus, KH-1 recognizes a native form of BgQ1 associated with BgQ2, and probably both are required for each other's folding.
Since the BgQ1 that reacted with KH-1 was colocalized with TGN46 in addition to calnexin, in HHV-6B-infected cells, KH-1 appeared to recognize folded BgQ1 associated with gH/gL/gQ2. Since the other MAb, BgQ1-202, barely precipitated the gH/gL/gQ1/gQ2 complex under the same conditions where KH-1 did, KH-1 was confirmed to recognize the folded BgQ1 (Fig. 2B).
From analyses of C-terminal truncation mutants of BgQ1, the determinant(s) for KH-1 was mapped to the interval between residues 484 and 496, inclusive. Unexpectedly, the Glu488Gln point mutation was recognized by KH-1, even though this residue is the only one within the interval from residues 484 to 496 that differs between the two variants. Surprisingly, mutants with substitutions in residues around Glu were little recognized with KH-1. Furthermore, KH-1 did not recognize at all a mutant BgQ1 for which three residues around Glu488 were deleted. It is interesting that although all the mutated residues found to be important for the recognition of BgQ1 from HHV-6B, Leu486, Cys487, Gly489, and Gly490, are conserved between the two variants, KH-1 does not recognize HHV-6A gQ1. The paradox may be explained if the real epitope of KH-1 is hidden by the mutation of a residue(s) around Glu488 through steric hindrance, indicating that there are other residues involved in direct binding to KH-1.
All the BgQ1 mutants that we constructed interacted with BgQ2, indicating that although the KH-1 epitope is present on the intact BgQ1/BgQ2 complex, it is not required for an association with gQ2. Indeed, the results show that BgQ1 has a conformational epitope(s) that is important for virus entry. The determination of the BgQ1 structure will be required to understand the nature of this epitope and how it is influenced by folding and interactions with BgQ2. Since several different neutralizing MAbs against HHV-6B that we generated from different mice were almost found to recognize BgQ1, their epitopes are possibly similar (our unpublished data). Because KH-1 reacted only with HHV-6B, including several clinical HHV-6B strains, the recognition site(s) of KH-1 may be highly conserved among variant B strains, having high immunogenicity. Our study shows that gQ1 of HHV-6B, like that of HHV-6A, is likely a ligand molecule for the virus's cellular receptor, and therefore, gQ1 is one of the key players in determining variant specific cell tropisms. The study also suggests that the epitope for KH-1 is a promising target for antiviral agents, and it will be interesting to determine the gQ1 architecture by structural analysis.
ACKNOWLEDGMENTS
We thank Emi Hayashi (National Institute of Biomedical Innovation, Osaka, Japan) for her technical assistance. We thank Kazusige Adachi (Minoh City Hospital, Minoh, Osaka, Japan) and Hideto Yamada (Department of Obstetrics and Gynecology, Kobe University Graduate School of Medicine, Kobe, Japan) for providing cord blood. We thank Jun-ichi Miyazaki (Division of Stem Cell Regulation Research, Osaka University Graduate School of Medicine) for pCAGGS.
This study was supported in part by a grant-in-aid for scientific research on priority areas from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan and a grant-in-aid for scientific research (B) from the Japan Society for the Promotion of Science (JSPS).
Footnotes
Published ahead of print on 28 September 2011.
REFERENCES
- 1. Ablashi D. V., et al. 1991. Genomic polymorphism, growth properties, and immunologic variations in human herpesvirus-6 isolates. Virology 184:545–552 [DOI] [PubMed] [Google Scholar]
- 2. Akhtar J., Shukla D. 2009. Viral entry mechanisms: cellular and viral mediators of herpes simplex virus entry. FEBS J. 276:7228–7236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Akkapaiboon P., Mori Y., Sadaoka T., Yonemoto S., Yamanishi K. 2004. Intracellular processing of human herpesvirus 6 glycoproteins Q1 and Q2 into tetrameric complexes expressed on the viral envelope. J. Virol. 78:7969–7983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Asada H., Yalcin S., Balachandra K., Higashi K., Yamanishi K. 1989. Establishment of titration system for human herpesvirus 6 and evaluation of neutralizing antibody response to the virus. J. Clin. Microbiol. 27:2204–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aubin J. T., et al. 1991. Several groups among human herpesvirus 6 strains can be distinguished by Southern blotting and polymerase chain reaction. J. Clin. Microbiol. 29:367–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Backovic M., Longnecker R., Jardetzky T. S. 2009. Structure of a trimeric variant of the Epstein-Barr virus glycoprotein B. Proc. Natl. Acad. Sci. U. S. A. 106:2880–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Campadelli-Fiume G., Guerrini S., Liu X., Foa-Tomasi L. 1993. Monoclonal antibodies to glycoprotein B differentiate human herpesvirus 6 into two clusters, variants A and B. J. Gen. Virol. 74(Pt. 10):2257–2262 [DOI] [PubMed] [Google Scholar]
- 8. Chandran B., Tirawatnapong S., Pfeiffer B., Ablashi D. V. 1992. Antigenic relationships among human herpesvirus-6 isolates. J. Med. Virol. 37:247–254 [DOI] [PubMed] [Google Scholar]
- 9. Chowdary T. K., et al. 2010. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat. Struct. Mol. Biol. 17:882–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Connolly S. A., Jackson J. O., Jardetzky T. S., Longnecker R. 2011. Fusing structure and function: a structural view of the herpesvirus entry machinery. Nat. Rev. Microbiol. 9:369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dhepakson P., et al. 2002. Human herpesvirus-6 rep/U94 gene product has single-stranded DNA-binding activity. J. Gen. Virol. 83:847–854 [DOI] [PubMed] [Google Scholar]
- 12. Dominguez G., et al. 1999. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J. Virol. 73:8040–8052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Drobyski W. R., Knox K. K., Majewski D., Carrigan D. R. 1994. Fatal encephalitis due to variant B human herpesvirus-6 infection in a bone marrow-transplant recipient. N. Engl. J. Med. 330:1356–1360 [DOI] [PubMed] [Google Scholar]
- 14. Gentile I., Talamo M., Borgia G. 2010. Is the drug-induced hypersensitivity syndrome (DIHS) due to human herpesvirus 6 infection or to allergy-mediated viral reactivation? Report of a case and literature review. BMC Infect. Dis. 10:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gompels U. A., et al. 1995. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology 209:29–51 [DOI] [PubMed] [Google Scholar]
- 16. Hall C. B., et al. 1994. Human herpesvirus-6 infection in children. A prospective study of complications and reactivation. N. Engl. J. Med. 331:432–438 [DOI] [PubMed] [Google Scholar]
- 17. Heldwein E. E., et al. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217–220 [DOI] [PubMed] [Google Scholar]
- 18. Hutchinson L., et al. 1992. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J. Virol. 66:2240–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Isegawa Y., et al. 1999. Comparison of the complete DNA sequences of human herpesvirus 6 variants A and B. J. Virol. 73:8053–8063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kaye J. F., Gompels U. A., Minson A. C. 1992. Glycoprotein H of human cytomegalovirus (HCMV) forms a stable complex with the HCMV UL115 gene product. J. Gen. Virol. 73(Pt. 10):2693–2698 [DOI] [PubMed] [Google Scholar]
- 21. Khattar S. K., van Drunen Littel-van den Harke S., Attah-Poku S. K., Babiuk L. A., Tikoo S. K. 1996. Identification and characterization of a bovine herpesvirus-1 (BHV-1) glycoprotein gL which is required for proper antigenicity, processing, and transport of BHV-1 glycoprotein gH. Virology 219:66–76 [DOI] [PubMed] [Google Scholar]
- 22. Koshizuka T., Ota M., Yamanishi K., Mori Y. 2010. Characterization of varicella-zoster virus-encoded ORF0 gene—comparison of parental and vaccine strains. Virology 405:280–288 [DOI] [PubMed] [Google Scholar]
- 23. Krummenacher C., et al. 2005. Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. EMBO J. 24:4144–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mori Y. 2009. Recent topics related to human herpesvirus 6 cell tropism. Cell. Microbiol. 11:1001–1006 [DOI] [PubMed] [Google Scholar]
- 25. Mori Y., Akkapaiboon P., Yang X., Yamanishi K. 2003. The human herpesvirus 6 U100 gene product is the third component of the gH-gL glycoprotein complex on the viral envelope. J. Virol. 77:2452–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mori Y., et al. 2004. Discovery of a second form of tripartite complex containing gH-gL of human herpesvirus 6 and observations on CD46. J. Virol. 78:4609–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mori Y., et al. 2008. Human herpesvirus-6 induces MVB formation, and virus egress occurs by an exosomal release pathway. Traffic 9:1728–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mori Y., Yang X., Akkapaiboon P., Okuno T., Yamanishi K. 2003. Human herpesvirus 6 variant A glycoprotein H-glycoprotein L-glycoprotein Q complex associates with human CD46. J. Virol. 77:4992–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mullen M. M., Haan K. M., Longnecker R., Jardetzky T. S. 2002. Structure of the Epstein-Barr virus gp42 protein bound to the MHC class II receptor HLA-DR1. Mol. Cell 9:375–385 [DOI] [PubMed] [Google Scholar]
- 30. Niwa H., Yamamura K., Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199 [DOI] [PubMed] [Google Scholar]
- 31. Okuno T., et al. 1989. Seroepidemiology of human herpesvirus 6 infection in normal children and adults. J. Clin. Microbiol. 27:651–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pfeiffer B., et al. 1993. Identification and mapping of the gene encoding the glycoprotein complex gp82-gp105 of human herpesvirus 6 and mapping of the neutralizing epitope recognized by monoclonal antibodies. J. Virol. 67:4611–4620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ryckman B. J., Chase M. C., Johnson D. C. 2008. HCMV gH/gL/UL128-131 interferes with virus entry into epithelial cells: evidence for cell type-specific receptors. Proc. Natl. Acad. Sci. U. S. A. 105:14118–14123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ryckman B. J., Chase M. C., Johnson D. C. 2010. Human cytomegalovirus TR strain glycoprotein O acts as a chaperone promoting gH/gL incorporation into virions but is not present in virions. J. Virol. 84:2597–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ryckman B. J., et al. 2008. Characterization of the human cytomegalovirus gH/gL/UL128-131 complex that mediates entry into epithelial and endothelial cells. J. Virol. 82:60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Salahuddin S. Z., et al. 1986. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science 234:596–601 [DOI] [PubMed] [Google Scholar]
- 37. Santoro F., et al. 1999. CD46 is a cellular receptor for human herpesvirus 6. Cell 99:817–827 [DOI] [PubMed] [Google Scholar]
- 38. Saxinger C., et al. 1988. Antibody reactivity with HBLV (HHV-6) in U.S. populations. J. Virol. Methods 21:199–208 [DOI] [PubMed] [Google Scholar]
- 39. Spear P. G., Longnecker R. 2003. Herpesvirus entry: an update. J. Virol. 77:10179–10185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Takeda K., et al. 1997. Identification of a variant B-specific neutralizing epitope on glycoprotein H of human herpesvirus-6. J. Gen. Virol. 78(Pt. 9):2171–2178 [DOI] [PubMed] [Google Scholar]
- 41. Takeda K., Okuno T., Isegawa Y., Yamanishi K. 1996. Identification of a variant A-specific neutralizing epitope on glycoprotein B (gB) of human herpesvirus-6 (HHV-6). Virology 222:176–183 [DOI] [PubMed] [Google Scholar]
- 42. Tang H., Hayashi M., Maeki T., Yamanishi K., Mori Y. 2011. Human herpesvirus 6 glycoprotein complex formation is required for folding and trafficking of the gH/gL/gQ1/gQ2 complex and its cellular receptor binding. J. Virol. 85:11121–11130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tang H., et al. 2010. Human herpesvirus 6 encoded glycoprotein Q1 gene is essential for virus growth. Virology 407:360–367 [DOI] [PubMed] [Google Scholar]
- 44. Tang H., Mori Y. 2010. Human herpesvirus-6 entry into host cells. Future Microbiol. 5:1015–1023 [DOI] [PubMed] [Google Scholar]
- 45. Tiacci E., et al. 2000. Fatal herpesvirus-6 encephalitis in a recipient of a T-cell-depleted peripheral blood stem cell transplant from a 3-loci mismatched related donor. Haematologicaae 85:94–97 [PubMed] [Google Scholar]
- 46. Wainwright M. S., et al. 2001. Human herpesvirus 6 limbic encephalitis after stem cell transplantation. Ann. Neurol. 50:612–619 [DOI] [PubMed] [Google Scholar]
- 47. Wille P. T., Knoche A. J., Nelson J. A., Jarvis M. A., Johnson D. C. 2010. A human cytomegalovirus gO-null mutant fails to incorporate gH/gL into the virion envelope and is unable to enter fibroblasts and epithelial and endothelial cells. J. Virol. 84:2585–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yamanishi K., et al. 1988. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet i:1065–1067 [DOI] [PubMed] [Google Scholar]
- 49. Zerr D. M., et al. 2005. Clinical outcomes of human herpesvirus 6 reactivation after hematopoietic stem cell transplantation. Clin. Infect. Dis. 40:932–940 [DOI] [PubMed] [Google Scholar]