Abstract
The migratory waterfowl of the world are considered to be the natural reservoir of influenza A viruses. Of the 16 hemagglutinin subtypes of avian influenza viruses, the H6 subtype is commonly perpetuated in its natural hosts and is of concern due to its potential to be a precursor of highly pathogenic influenza viruses by reassortment. During routine influenza surveillance, we isolated an unconventional H6N5 subtype of avian influenza virus. Experimental infection of mice revealed that this isolate replicated efficiently in the lungs, subsequently spread systemically, and caused lethality. The isolate also productively infected ferrets, with direct evidence of contact transmission, but no disease or transmission was seen in pigs. Although the isolate possessed the conserved receptor-binding site sequences of avian influenza viruses, it exhibited relatively low replication efficiencies in ducks and chickens. Our genetic and molecular analyses of the isolate revealed that its PB1 sequence showed the highest evolutionary relationship to those of highly pathogenic H5N1 avian influenza viruses and that its PA protein had an isoleucine residue at position 97 (a representative virulence marker). Further studies will be required to examine why our isolate has the virologic characteristics of mammalian influenza viruses but the archetypal receptor binding profiles of avian influenza viruses, as well as to determine whether its potential virulence markers (PB1 analogous to those of H5N1 viruses or isoleucine residue at position 97 within PA) could render it highly pathogenic in mice.
INTRODUCTION
Influenza A viruses are perpetuated in wild aquatic birds, primarily waterfowl, gulls, and shorebirds (17, 23). Genetic analyses have indicated that these viruses exist in a state of evolutionary stasis within the natural reservoir, with most internal genes showing high conservation at the amino acid level. However, two major surface glycoprotein-encoding genes, those for hemagglutinin (HA) and neuraminidase (NA), are much more variable (26). This has allowed the viruses to be classified into multiple antigenic subtypes; to date, 16 HA and 9 NA subtypes have been reported in the natural hosts (29).
Avian influenza viruses are unusual in that they can infect various animal hosts and cause a broad spectrum of disease symptoms, ranging from subclinical infections to highly pathogenic infections with 100% lethality. Avian influenza viruses generally cause asymptomatic infection in the natural hosts, but the transmission and adaptation of these viruses beyond the species barrier to alternate hosts, including other bird species and mammals, may cause anywhere from mild to severe respiratory diseases (1). However, only two HA subtypes of these viruses (H5 and H7) have been shown to occasionally attain high pathogenicity once crossing the mammalian species barrier (27).
During the 2009-to-2010 winter season, we collected a total of 350 fecal samples from migratory waterfowl through influenza surveillance in a wild migratory bird habitat of South Korea. Of these samples, 16 samples (4.6%) were found to be positive for influenza A viruses (H1, H5, H6, and H11 subtypes), as assessed by virus isolation using chicken embryos. Experimental infection of mice with the isolates revealed generally inefficient infection. However, an isolate of the H6N5 subtype productively infected mice with no evidence of adaptation and caused illness and lethality. Here, we report the replicability, pathogenicity, and intraspecies transmissibility of this H6N5 avian influenza isolate in mice and other avian (ducks and chickens) and mammalian (ferrets and pigs) species.
MATERIALS AND METHODS
Viruses.
A total of 350 fecal samples were collected from a stopover site of wild migratory aquatic birds in South Korea, and virus isolation was performed using 10-day-old embryonated chicken eggs. Among the samples, 16 were determined to be positive for influenza A viruses, which were then subtyped. From them, the H6N5 avian influenza isolate (A/Aquatic Bird/Korea/CN5/09 [A/AB/Kor/CN5/09]), which was mouse lethal within 7 days, was selected and subjected to further characterization by genetic and in vivo analyses. A/Aquatic Bird/Korea/CN20/10 (A/AB/Kor/CN20/10; H6N1), A/Aquatic Bird/Korea/CN17/09 (A/AB/Kor/CN17/09; H6N2), A/Aquatic Bird/Korea/W69/05 (A/AB/Kor/W69/05; H6N5), and A/Aquatic Bird/Korea/CN9/09 (A/AB/Kor/CN9/09; H6N8) viruses were used as control H6 viruses in characterizing the A/AB/Kor/CN5/09 (H6N5) virus.
Genetic analyses.
Viral gene amplification and sequencing were carried out as described previously (10), with slight modifications. Briefly, viral RNA was extracted using an RNeasy minikit (Qiagen, Valencia, CA) according to the manufacturer's protocol. One-step reverse transcriptase PCR (RT-PCR) was performed to amplify the viral gene segments, using a OneStep RT-PCR kit (Qiagen, Valencia, CA) with universal primer sets, followed by purification using a QIAquick gel extraction kit (Qiagen, Valencia, CA). The amplified gene segments were commercially sequenced (Cosmogenetech, Seoul, South Korea).
Phylogenetic analyses.
The viral nucleotide sequences were edited using the Lasergene sequence analysis software package (DNASTAR, Madison, WI) and aligned with CLUSTAL V (9). Rooted phylograms were prepared using the neighbor-joining (NJ) algorithm and then plotted with the NJ Plot software (21). The branch lengths were proportional to the estimates of sequence divergence measured using a scale of 0.01 nucleotide changes per site. The entire coding regions of all gene segments were used in the phylogenetic analyses.
Animal studies.
All animal experiments were performed in biosafety level 2-plus (BSL2+) facilities at Korea Research Institute of Bioscience and Biotechnology (Daejeon, South Korea) or BSL3+ facilities at BioLeaders Corporation (Daejeon, South Korea). General animal care was provided as required by the Institutional Animal Care and Use Committee.
Six-week-old C57BL/6 mice (Korea Animal Technology, Pyeongtaek, South Korea) were used. Each group of 26 mice was inoculated via intranasal instillation with 106 50% egg infectious doses (EID50) of each virus, and an equal number of contact mice were caged with the infected mice beginning 1 day postinfection. Eight mice per group were sacrificed at 3 and 5 days postinfection, and lung, brain, heart, kidney, liver, and spleen virus titers were determined. Ten mice per group were monitored daily for 14 days for weight change, morbidity, and mortality. Blood samples were collected prior to infection and at 14 days postinfection, and seroconversion was evaluated (see below).
Three- to 4-month-old ferrets (Triple F Farm, Sayre, PA) were used. Three ferrets were inoculated via intranasal instillation with 106 EID50 of the virus, and two contact ferrets were caged with the infected ferrets beginning 1 day postinfection. Ferrets were monitored daily for 14 days for weight change, temperature, and signs of clinical disease. Nasal washes were collected from each ferret on days 3, 5, 7, and 10 postinfection, and virus titers in the upper respiratory tract were determined.
Four-month-old Yucatan miniature pigs (Medi-Pig Korea, Cheonan, South Korea) were used. Three pigs were inoculated via intranasal instillation with 106 EID50 of the virus, and two contact pigs were caged with the infected pigs beginning 1 day postinfection. Pigs were monitored daily for 14 days for weight change, temperature, and signs of clinical disease. Nasal washes were collected from each pig on days 3, 5, 7, and 10 postinfection, and virus titers in the upper respiratory tract were determined.
Four-week-old white Leghorn chickens (Gallus domesticus) and 4-week-old mallard ducks (Anas platyrhynchos) were purchased from Namdeok Hatchery (Gyeonggi, South Korea) and Central Laboratory Animal, Inc. (Seoul, South Korea), respectively. For each species, six birds were inoculated via intranasal, intraocular, and intratracheal instillation with 106 EID50 of the virus, and an equal number of birds were caged with the infected birds beginning 1 day postinfection. Birds were monitored daily for 14 days for morbidity and mortality. Tracheal and cloacal swabs were collected at 3, 5, 7, and 10 days postinfection, and virus titers were determined.
Immunohistochemistry.
Six-week-old C57BL/6 mice were inoculated via intranasal instillation with 106 EID50 of the virus or phosphate-buffered saline (PBS). Five days later, the mice were sacrificed, and the lungs, brain, heart, kidneys, liver, and spleen were removed from each. The freshly harvested organ samples were washed with PBS, embedded in Tissue-Tek optimal cutting temperature (OCT) compound (Sakura Finetek USA, Inc., Torrance, CA), and sectioned at 8 μm. The sections were stained using a Vectastain ABC kit (Vector Laboratories, Burlingame, CA) according to the manufacturer's protocol, with slight modifications. Briefly, the sections were fixed in 10% neutral buffered formalin for 30 min and rinsed with tap water for 10 min. The endogenous peroxidase activity was quenched with 0.3% H2O2 in methanol for 30 min, and the sections were blocked in blocking solution (1.5% normal goat serum in PBS) for 1 h, followed by incubation with a monoclonal anti-influenza A nucleoprotein antibody (1:500 dilution in PBS; Southern Biotech, Birmingham, AL) for 1 h. The sections were washed with PBS for 15 min, incubated with a biotin-conjugated goat anti-mouse antibody for 30 min, washed again with PBS for 15 min, and then incubated with 1% streptavidin-horseradish peroxidase conjugate in PBS for 30 min. The presence of the viral nucleoprotein was detected with 3,3′-diaminobenzidine substrate (Vector Laboratories, Peterborough, CA), the sections were counterstained with hematoxylin, and the results were examined using an Olympus TH4-200 system microscope attached to a DP72 microscope digital camera (Olympus Corporation, Tokyo, Japan). The capture images were analyzed with the DP2-BSW software (version 2.1; Olympus Corporation, Tokyo, Japan). All staining procedures were performed at room temperature.
Serology.
Serum specimens were collected from mice, ferrets, pigs, chickens, and ducks prior to infection and at 14 days postinfection, and seroreactivity was analyzed using the hemagglutination inhibition (HI) assay, which was performed using chicken erythrocytes according to standard methods (20).
Statistical analyses.
Numerical data are presented as means ± standard errors of the means. Comparisons between groups were performed using a two-tailed Student's t test. The threshold of significance was set at a P value of <0.05.
Nucleotide sequence accession numbers.
Gene sequences of the H6 avian influenza isolates obtained in this study were deposited in GenBank under accession numbers CY088569 to CY088576, CY098219 to CY098242, and CY098529 to CY098536.
RESULTS
Avian influenza virus isolation and infection of mice.
A total of 350 fecal samples were collected from a wild migratory bird habitat in South Korea during the 2009-to-2010 winter migration season. The presence of influenza A viruses was determined by virus isolation using chicken embryos; of the 350 total samples, 16 (4.6%) were positive. Sequence analysis revealed that the isolates represented the H1, H5, H6, and H11 subtypes (data not shown).
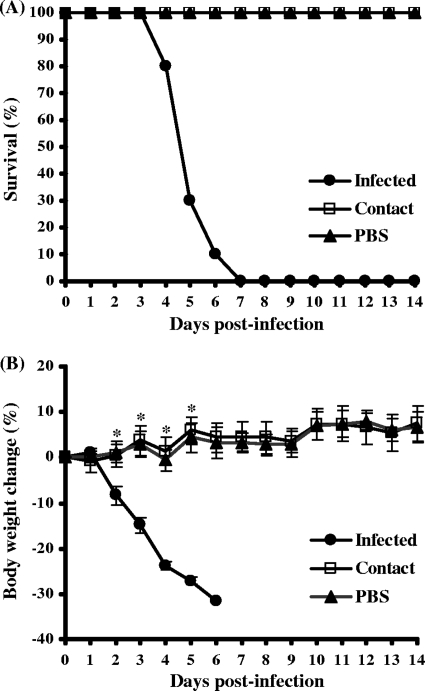
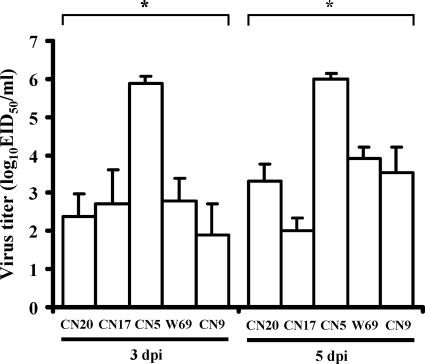
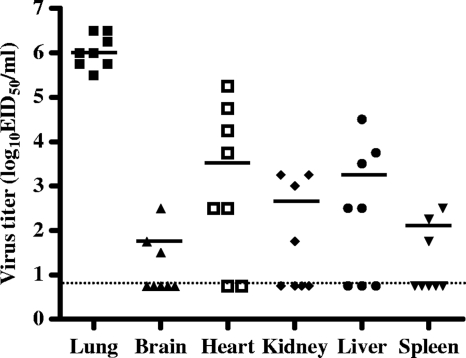
Representative isolates from each subtype were selected and examined for pathogenicity by experimental infection of a mouse model. Most of the isolates failed to infect efficiently (data not shown). However, one of the isolates (A/AB/Kor/CN5/09, H6N5) caused illness and lethality in mice. Mice infected with the H6N5-subtype isolate all showed severe clinical signs, including significant weight loss, ruffled fur, lethargy, and ataxia, and experienced 100% mortality within 7 days (Fig. 1). The isolate was also associated with a high viral load in mouse lungs (mean titers of 6.0 ± 0.3 log10 EID50/ml at 3 and 5 days postinfection) (Fig. 2). In contrast, mice infected with comparison H6 viruses (H6N1, H6N2, H6N5, and H6N8 subtypes) used in this study showed no symptoms of disease (data not shown) and produced significantly lower lung virus titers (Fig. 2). Interestingly, the A/AB/Kor/CN5/09 (H6N5) isolate was also detected in the other organs, including brain, heart, kidney, liver, and/or spleen, proved by virus isolation using chicken embryos (mean titers of 1.5 to 3.7 log10 EID50/ml) (Fig. 3). This extrapulmonary infection was additionally confirmed by immunohistochemical analysis of the lung, brain, heart, and liver (data not shown). None of the comparison H6 viruses caused extrapulmonary infection in mice (data not shown). Although the A/AB/Kor/CN5/09 (H6N5) isolate replicated efficiently in mice, there was no evidence of contact transmission, as the contact mice housed with the infected mice did not show any clinical signs or virus shedding (Fig. 1). These results demonstrated that the H6N5-subtype isolate (A/AB/Kor/CN5/09) from a wild aquatic bird could replicate efficiently in mouse lungs and showed systemic spread and lethality with no evidence of preadaptation, but there was no evidence of contact transmission in mice.
Fig. 1.
Survival rates (A) and body weight changes (B) among mice infected with the A/AB/Kor/CN5/09 (H6N5) virus. Ten mice (C57BL/6) were intranasally inoculated with the virus, and an equal number of contact mice were caged with the infected mice beginning 1 day postinfection. Negative-control mice were inoculated with PBS. All mice were monitored daily for 2 weeks or until death.
Fig. 2.
Lung virus titers of mice infected with the H6 avian influenza viruses. Each group of mice was intranasally inoculated with each virus and sacrificed at 3 or 5 days postinfection, and lungs were removed. Data are presented as log10 50% egg infectious doses per milliliter (log10 EID50/ml). Abbreviations: CN20, A/AB/Kor/CN20/10 (H6N1); CN17, A/AB/Kor/CN17/09 (H6N2); CN5, A/AB/Kor/CN5/09 (H6N5); W69, A/AB/Kor/W69/05 (H6N5); CN9, A/AB/CN9/09 (H6N8).
Fig. 3.
Virus titers in the extrapulmonary organs of mice infected with the A/AB/Kor/CN5/09 (H6N5) virus. The mice were intranasally inoculated with the virus and sacrificed at 5 days postinfection, and organs were removed. Data are presented as log10 50% egg infectious doses per milliliter (log10 EID50/ml).
Infection of ferrets and pigs with the A/AB/Kor/CN5/09 (H6N5) virus.
The replicability, pathogenicity, and contact transmissibility of the mouse-lethal H6N5 avian influenza isolate were examined in two other mammalian species: ferrets and pigs. Experimental infection of ferrets was associated with mild clinical signs, including sluggishness and sneezing, but there were no deaths among the infected animals. Virus isolation from nasal washes obtained at various time points postinfection confirmed replication of the isolate and showed evidence of high viral loads (mean titer of 6.2 ± 0.4 log10 EID50/ml) in the upper respiratory tract of all infected ferrets (Table 1). Furthermore, contact transmission was observed, with one of the two contact ferrets caged with the infected animals showing a high level of virus shedding (peak titer of 6.5 ± 0.0 log10 EID50/ml at 7 days postinfection) in the upper respiratory tract (Table 1). In contrast, a comparison H6 virus (A/AB/Kor/CN9/09) could infect ferrets but showed lower levels of virus shedding (mean titer of 3.6 ± 0.4 log10 EID50/ml) in the upper respiratory tract (data not shown). No contact transmission of the comparison H6 virus was observed. This efficient replication of the H6N5 avian influenza isolate in the upper respiratory tract of infected (100%) and contact (50%; one of two animals) ferrets suggests that the isolate may have zoonotic potential.
Table 1.
Virus titration of nasal washes of ferrets or pigs and oropharyngeal swabs of ducks or chickens infected with A/AB/Kor/CN5/09 (H6N5) virus
| Animal | Group | Mean virus titer (log10 EID50/ml) ± SD (no. of animals shedding virus/no. tested) at indicated day postinfectiona |
|||
|---|---|---|---|---|---|
| 3 | 5 | 7 | 10 | ||
| Ferret | Infected | 6.0 ± 0.3 (3/3) | 6.2 ± 0.4 (3/3) | < (0/3) | < (0/3) |
| Contact | < | < | 6.5 ± 0.0 (1/2) | 5.5 ± 0.0 (1/2) | |
| Pig | Infected | < | < | < | < |
| Contact | < | < | < | < | |
| Duck | Infected | 1.8 ± 1.8 (6/6) | 1.9 ± 1.2 (2/6) | < | < |
| Contact | < | 2.7 ± 1.0 (6/6) | 2.2 ± 1.5 (5/6) | < | |
| Chicken | Infected | 2.5 ± 1.6 (6/6) | 1.5 ± 1.6 (2/6) | < | < |
| Contact | < | < | < | < | |
Virus titers were determined in eggs. Data are titers of positive samples (≥0.75 log10 EID50/ml). <, titer was below the limit of detection (<0.75 log10 EID50/ml).
The ability of the H6N5 avian influenza isolate to infect and cause disease in pigs was also determined. Experimental infection of pigs failed to yield evidence of replication or contact transmission, as evidenced by the lack of detectable virus in the nasal washes and lungs from both infected and contact pigs (Table 1). Seroconversion was detected in the infected pigs but not in the contact animals (data not shown). These results indicate that the H6N5 avian influenza isolate may infect pigs but with very inefficient replicability.
Infection of avian species with the A/AB/Kor/CN5/09 (H6N5) virus.
The wild migratory bird habitat where the samples were collected was largely populated by mallard ducks. Considering them to be the presumable source of the isolates, we assessed the ability of the H6N5 avian influenza isolate to replicate and transmit in mallard ducks. Experimental infection of mallard ducks induced virus shedding predominantly through the respiratory tract in both inoculated and contact birds (mean tracheal titers of 1.8 ± 1.8 to 2.7 ± 1.0 log10 EID50/ml) (Table 1). The isolate was also shed through the fecal route (mean cloacal titers of 1.1 ± 0.5 log10 EID50/ml), but this was seen among only 33% of the infected or contact birds (data not shown). Infection of mallard ducks with the isolate failed to induce clinical signs of illness, and no deaths were observed.
The replication and transmission potentials of the H6N5 avian influenza isolate were also determined in domestic poultry. Experimental infection of chickens induced virus shedding through the respiratory tract but not through the fecal route, and no virus shedding was detected in the contact birds (Table 1). No morbidity or mortality was observed in either infected or contact birds. Taken together, our findings indicate that the H6N5 avian influenza isolate showed low replication efficiencies in wild and domestic birds compared to its replication efficiencies in mice and ferrets.
Genetic and molecular characterization of the A/AB/Kor/CN5/09 (H6N5) virus.
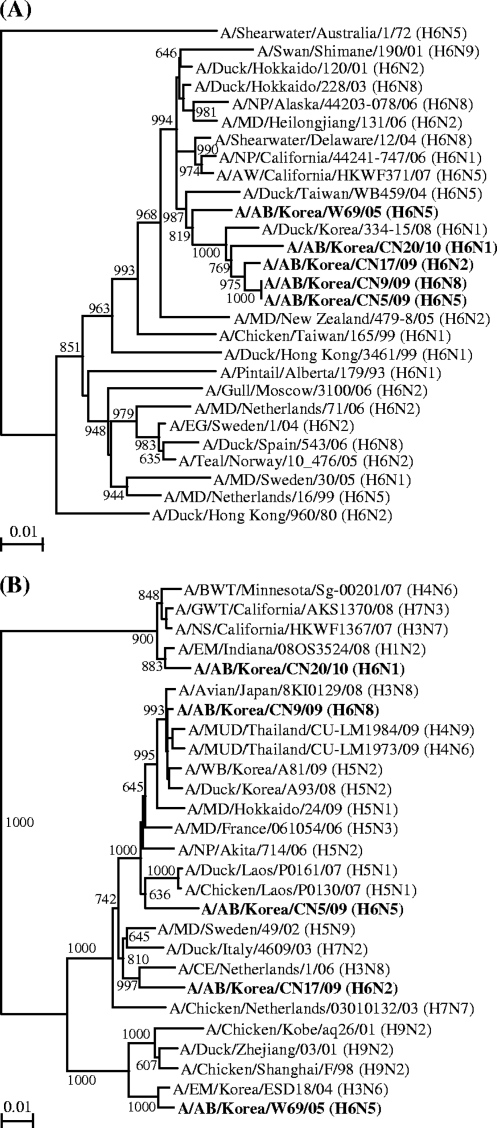
Phylogenetic analysis of the H6N5 avian influenza isolate revealed that the genes encoding surface glycoproteins HA and NA clustered together proximate to those of the avian influenza viruses circulating in South Korea and eastern Asia, respectively (Fig. 4). The phylogenetic topologies of the isolate's internal genes paralleled those of the surface glycoprotein-encoding genes (data not shown). Overall, our phylogenetic analysis indicated that all of the gene segments of the isolate were derived from Eurasian-like avian viruses resident in wild birds migrating along either the East Asian-Australian Flyway or the West Pacific Flyway. The PB1 sequence of the isolate was unique in that it showed its highest evolutionary relationship to the corresponding sequences from H5N1 highly pathogenic avian influenza viruses (e.g., A/Chicken/Laos/P0130/07), while the comparison H6 viruses did not form a clade with any of the H5N1 viruses (Fig. 4). This result implies that the H6N5 avian influenza isolate (A/AB/Kor/CN5/09) would have undergone reassortment or shared a common ancestor for the PB1 gene segment with the H5N1 viruses.
Fig. 4.
Phylogenetic relationships of the HA and PB1 genes of the A/AB/Kor/CN5/09 (H6N5) virus. Phylogenetic trees were constructed using the nucleotide sequences of the HA (A) and PB1 (B) genes from our isolate, along with those of selected influenza A viruses available in GenBank. The phylograms were generated by neighbor-joining (NJ) analysis with 1,000 bootstrapped replicates; the NJ percentage bootstrap values (>600) for each node are shown in each tree. The viruses characterized in this study are shown in boldface. Scale bars show 0.01 nucleotide changes per site. Abbreviations: AB, aquatic bird; AW, American wigeon; BWT, blue-winged teal; CE, common eider; EG, Egyptian goose; EM, environment; GWT, green-winged teal; MD, mallard duck; MUD, muscovy duck; NP, northern pintail; NS, northern shoveler; WB, wild bird.
Comparison of the deduced amino acid sequence of the HA protein of the A/AB/Kor/CN5/09 (H6N5) isolate revealed no insertion of an aspartic acid residue between amino acids 144 and 145 (H3 numbering here and below) and that the receptor-binding site sequences conserved in avian influenza viruses (A138, E190, L194, G225, Q226, and G228) were observed, confirming that the virus originated from aquatic birds (3). The presence of a glutamine residue at position 226 and a glycine residue at position 228 indicated that the isolate would have a binding preference for α-2,3-linked sialic acid receptors (5). The HA protein of the isolate further harbored the archetypal avirulent avian consensus of the HA1-HA2 cleavage site of the HA precursor (324PQRETR/G330) (28), along with the eight potential N-glycosylation sites (Asn-X-Ser/Thr) typical for the H6 subtype of avian influenza viruses (28).
In the NA protein of the A/AB/Kor/CN5/09 (H6N5) isolate, we observed only four deletions of amino acid residues in the stalk region. The lack of a large deletion in the stalk region confirmed that the isolate originated from aquatic birds (28). The NA protein of the isolate retained the conserved catalytic residues (R118, D151, R152, R224, E276, R292, R371, and Y406; N2 numbering here and below) and the framework residues (E119, W178, I222, R156, S179, D198, E227, H274, E277, N294, and E425) without any amino acid substitutions or deletions, suggesting that the isolate may be sensitive to neuraminidase inhibitors. The NA protein of the isolate possessed six potential N-glycosylation sites at positions N46, 52, 69, 86, 146, and 402.
Our molecular analysis of the internal proteins of the A/AB/Kor/CN5/09 (H6N5) isolate failed to reveal any of the known pathogenicity-related mutations in the polymerase complex (e.g., E158G and E627K within the PB2 protein, Y436H within the PB1 protein, and T515A within the PA protein) (7, 13, 31), suggesting that it may not be highly pathogenic in mammalian or avian hosts. However, the PA protein of the isolate possessed an isoleucine residue at position 97, while those of the comparison H6 viruses contained a threonine residue at the same position. The presence of an isoleucine residue at position 97 suggests that the isolate could have enhanced virulence in mice (24). The M2 protein of the isolate contained conserved transmembrane domain residues (25PLVVAASIIGILHLILWIL43), indicating that it may be susceptible to M2 ion channel inhibitors (8, 12).
Erythrocyte-binding preference of the A/AB/Kor/CN5/09 (H6N5) virus.
Erythrocyte agglutination by influenza A viruses may reflect their receptor specificity; typically, avian influenza viruses prefer α-2,3-linked sialic acid receptors, while mammalian (especially human) influenza viruses prefer α-2,6-linked sialic acid receptors (15). Horse and chicken erythrocytes predominantly contain α-2,3-linked sialic acid receptors (there are only α-2,3-linked sialic acid receptors on horse erythrocytes and more α-2,3-linked than α-2,6-linked sialic acid receptors on chicken erythrocytes), whereas turkey and guinea pig erythrocytes and human O cells mainly possess α-2,6-linked sialic acid receptors (15, 25). To predict the receptor specificity of the A/AB/Kor/CN5/09 (H6N5) isolate, we examined the ability of the isolate to agglutinate erythrocytes from five different species: turkey, guinea pig, human (blood type O), chicken, pig, and horse. As shown in Table 2, the isolate agglutinated the erythrocytes of four of the five species (excluding pig) and showed different levels of hemagglutination activity across the species. The highest hemagglutination titer (512 hemagglutination units [HAU]) was obtained with turkey erythrocytes, followed by chicken and guinea pig erythrocytes (256 HAU), human erythrocytes (128 HAU), and finally, horse erythrocytes (64 HAU). In contrast, a comparison H6N5 virus (A/AB/Kor/W69/05) showed reduced agglutination of turkey, guinea pig, and human erythrocytes, implying its lower infection or replication capacity in mammalian hosts. The human and swine viruses agglutinated most of the tested erythrocytes but not horse erythrocytes. Taken together, these results indicate that the A/AB/Kor/CN5/09 (H6N5) isolate may bind to both α-2,3-linked and α-2,6-linked sialic acid receptors but appears to prefer α-2,6-linked sialic acid receptors.
Table 2.
Hemagglutination of erythrocytes from birds, humans, swine, and horses by A/AB/Kor/CN5/09 (H6N5) and comparison viruses
| Virus isolatea | Subtype | Hemagglutination titer (HAU) of erythrocytes from indicated speciesb |
|||||
|---|---|---|---|---|---|---|---|
| Turkeyc | Guinea pigc | Humanc | Chickend | Swinee | Horsef | ||
| Avian isolate | |||||||
| A/AB/Kor/CN5/09 | H6N5 | 512 | 320 | 128 | 256 | <2 | 64 |
| A/AB/Kor/W69/05 | H6N5 | 64 | 64 | 20 | 32 | <2 | 16 |
| Human isolate | |||||||
| A/Puerto Rico/8/34 | H1N1 | 2,048 | 512 | 640 | 1,024 | <2 | <2 |
| A/Philippines/2/82 | H3N2 | 1,024 | 1,024 | 512 | 1,024 | <2 | 32 |
| Swine isolate | |||||||
| A/Swine/Kor/GC0503/05 | H1N1 | 1,024 | 256 | 256 | 512 | <2 | <2 |
| A/Swine/Kor/GC04N7/05 | H3N2 | 256 | 64 | 128 | 256 | 32 | <2 |
AB, aquatic bird; Kor, Korea.
Titers are expressed as the reciprocal of the highest virus dilution that yields complete HA agglutination. HAU, hemagglutination unit.
Neu5Acα2,6Gal > Neu5Acα2,3Gal.
Neu5Acα2,6Gal < Neu5Acα2,3Gal.
Neu5Gcα2,6Gal > Neu5Gcα2,3Gal.
Neu5Gcα2,3Gal.
DISCUSSION
Global surveillance has suggested that avian influenza viruses of the H6 subtype are particularly prevalent in the wild waterfowl of North America and Eurasia, including the Anseriformes and Charadriiformes (16, 19, 30). A recent report found that avian influenza viruses of the H6 subtype were the most abundantly detected in wild birds, comprising 17.8% of all avian influenza isolates obtained through their surveillance program. A previous genetic analysis suggested that the NA gene and the six internal genes of an H6-subtype avian influenza virus (A/Teal/Hong Kong/W312/97 [H6N1]) showed very high (≥97%) nucleotide homologies to the corresponding sequences of a highly pathogenic human influenza virus (A/Hong Kong/156/07 [H5N1]), indicating that the H6N1 virus could donate its gene segments to the H5N1 virus (11). Thus, the prevalence of H6 subtypes in nature and their potential to be precursors of highly pathogenic strains, such as H5N1, mean that the H6 avian influenza viruses are of grave and abiding concern.
In this study, we examined the pathogenicity of the isolates taken as part of avian influenza virus surveillance of wild birds flying into a wild bird habitat in South Korea. The majority of the isolates were nonpathogenic in mice, but an H6N5 subtype (A/AB/Kor/CN5/09) was highly pathogenic, killing all of the infected mice within 7 days. This H6N5 isolate was associated with a high viral load (mean titers of 6.0 ± 0.4 log10 EID50/ml) in mouse lungs up to the point of death, and in some cases extrapulmonary infection was seen in organs, including brain, heart, kidney, liver, and/or spleen. The observed lethality was presumed to be primarily caused by excessive lung inflammation induced by high viral titers, not extrapulmonary spread, because mice showing only lung infection also died. However, extrapulmonary infection, especially brain infection, seemed to contribute to virulence in infected mice, some of which showed neurological symptoms (e.g., ataxia). These findings support a previous observation that the H6 avian viruses could efficiently infect and cause illness in mice without prior adaptation (6). Furthermore, the fact that the H6N5 avian influenza virus could spread systemically in mice should be highlighted in terms of a non-H5N1 case, although the significance of replication in extrapulmonary sites is still unknown.
The unusually high pathogenicity of the H6N5 avian influenza isolate (A/AB/Kor/CN5/09) in mice may be explained by genetic and molecular characteristics. Our phylogenetic analysis showed that the PB1 gene segment of the isolate had the highest evolutionary relationship with that of the A/Chicken/Laos/P0130/07 (H5N1) virus, which was highly pathogenic in chickens and mice (2). Our molecular analysis revealed that the PA protein of the isolate possessed one of the known pathogenic markers examined herein: an isoleucine residue at position 97 that has been associated with enhanced virulence and replication in mice (24). The existence of the isoleucine residue at position 97 in PA of the H6N5 avian influenza isolate may explain its unusual high pathogenicity in mice and its ability to show more efficient replication in mammalian species than in avian hosts. However, further studies with reverse genetics will be required to address whether the possession of a PB1 sequence similar to that of the highly pathogenic H5N1 avian influenza virus or an isoleucine residue at position 97 in PA could confer high virulence to mice.
The ferret is generally regarded as the ideal animal model for studying influenza in humans (18). In order to examine the potential of our H6N5 avian influenza isolate (A/AB/Kor/CN5/09) to replicate in humans, we experimentally infected ferrets. The isolate could productively infect and replicate in ferrets; all three experimentally infected ferrets shed the virus with high titers in the upper respiratory tract. The infected ferrets showed typical symptoms of disease for a few days postinfection (e.g., increased body temperature, decreased body weight, lethargy, and sneezing), but no deaths occurred. A similar pattern, including a high viral load in the upper respiratory tract, was also observed in one of two contact ferrets. The infectivity and replicability of the isolate in ferrets suggest that it may be able to infect and replicate in humans with no preadaptation, and the observed ferret-to-ferret intraspecies transmissibility may indicate the potential for transmission between humans.
Pigs are thought to be mammalian intermediate hosts for the transmission of avian influenza viruses to humans because they are susceptible to infection by both avian and human influenza viruses, enabling these viruses to reassort (29). Pigs may also support the adaptation of avian influenza viruses to mammalian hosts without exchanging gene segments by acquiring human virus-like receptor properties (22). To examine whether pigs are susceptible to infection by the H6N5 avian influenza isolate (A/AB/Kor/CN5/09), we experimentally infected pigs. Although seroconversion was detected in infected pigs at 14 days postinfection, there was no evidence that the isolate was able to replicate in pigs. Thus, pigs appeared to be susceptible to infection but did not support efficient viral replication. This is consistent with previous findings that virological confirmation of infection was not observed in pigs experimentally infected with highly pathogenic avian influenza viruses (14). However, this is somewhat controversial, as nasal excretion of other highly pathogenic avian influenza viruses has been reported in infected (but not contact) pigs (4).
To assess the replicability, transmissibility, and pathogenicity of the H6N5 avian influenza isolate (A/AB/Kor/CN5/09) in representative avian hosts, wild ducks (the presumable sources of the isolates) and chickens (potential avian intermediate hosts) were experimentally infected with the isolate. The ducks and chickens were all susceptible to infection and replication of the isolate; they did not show symptoms of disease, but virus shedding for a relatively short period of time (within 5 days from the beginning of infection) with low viral load (≤2.7 ± 1.0 log10 EID50/ml) was observed in the respiratory tract and, in some cases, feces of the infected birds. Contact transmission was observed among ducks but not chickens. Taken together, the results from our animal studies in mammalian and avian hosts suggest that although our H6N5 avian influenza isolate has an avian origin and a typical avian influenza virus-like receptor-binding preference at the molecular level, the isolate may have a greater ability to infect and replicate in mammalian species (especially mice and ferrets) than in avian species (ducks and chickens).
The unconventional apparent host preference of the H6N5 avian influenza isolate (A/AB/Kor/CN5/09) was confirmed by the results of our erythrocyte-binding specificity test, which showed that the isolate could agglutinate erythrocytes derived from turkey, guinea pig, human (blood type O), chicken, and horse. The overall agglutination results suggested that the isolate can bind to both α-2,3- and α-2,6-linked sialic acid receptors (15, 25). The observation that the highest agglutination occurred with turkey erythrocytes (containing predominantly α-2,6-linked sialic acid receptors) and lower agglutination occurred with horse erythrocytes (containing only α-2,3-linked sialic acid receptors) was consistent with our finding that the isolate is better able to infect and replicate in mammalian than in avian species.
In sum, in the work reported herein, we isolated and characterized an unconventional H6N5 avian influenza virus. The isolate productively infected and caused illness in mice (where it was highly pathogenic) and ferrets (where it showed intraspecies transmission) but not in pigs. In contrast, the isolate showed less efficient replication and transmission and failed to trigger clinical symptoms in birds (ducks and chickens). As such, our H6N5-subtype isolate appears to be a novel mammalian species-infectious and -pathogenic avian influenza virus capable of undergoing systemic spread in mice.
ACKNOWLEDGMENTS
This work was supported by a National Agenda Project grant from the Korea Research Council of Fundamental Science & Technology and by the KRIBB Initiative Program (KGM3111013).
Footnotes
Published ahead of print on 12 October 2011.
REFERENCES
- 1. Alexander D. J. 2007. An overview of the epidemiology of avian influenza. Vaccine 25:5637–5644 [DOI] [PubMed] [Google Scholar]
- 2. Boltz D. A., et al. 2010. Emergence of H5N1 avian influenza viruses with reduced sensitivity to neuraminidase inhibitors and novel reassortants in Lao People's Democratic Republic. J. Gen. Virol. 91:949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chin P. S., et al. 2002. Molecular evolution of H6 influenza viruses from poultry in Southeastern China: prevalence of H6N1 influenza viruses possessing seven A/Hong Kong/156/97 (H5N1)-like genes in poultry. J. Virol. 76:507–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Choi Y. K., et al. 2005. Studies of H5N1 influenza virus infection of pigs by using viruses isolated in Vietnam and Thailand in 2004. J. Virol. 79:10821–10825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Connor R. J., Kawaoka Y., Webster R. G., Paulson J. C. 1994. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology 205:17–23 [DOI] [PubMed] [Google Scholar]
- 6. Gillim-Ross L., et al. 2008. Avian influenza H6 viruses productively infect and cause illness in mice and ferrets. J. Virol. 82:10854–10863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hatta M., Gao P., Halfmann P., Kawaoka Y. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293:1840–1842 [DOI] [PubMed] [Google Scholar]
- 8. Hay A. J., Wolstenholme A. J., Skehel J. J., Smith M. H. 1985. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 4:3021–3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Higgins D. G., Bleasby A. J., Fuchs R. 1992. CLUSTAL V: improved software for multiple sequence alignment. Comput. Appl. Biosci. 8:189–191 [DOI] [PubMed] [Google Scholar]
- 10. Hoffmann E., Stech J., Guan Y., Webster R. G., Perez D. R. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146:2275–2289 [DOI] [PubMed] [Google Scholar]
- 11. Hoffmann E., et al. 2000. Characterization of the influenza A virus gene pool in avian species in southern China: was H6N1 a derivative or a precursor of H5N1? J. Virol. 74:6309–6315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Holsinger L. J., Nichani D., Pinto L. H., Lamb R. A. 1994. Influenza A virus M2 ion channel protein: a structure-function analysis. J. Virol. 68:1551–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hulse-Post D. J., et al. 2007. Molecular changes in the polymerase genes (PA and PB1) associated with high pathogenicity of H5N1 influenza virus in mallard ducks. J. Virol. 81:8515–8524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Isoda N., et al. 2006. Pathogenicity of a highly pathogenic avian influenza virus, A/chicken/Yamaguchi/7/04 (H5N1) in different species of birds and mammals. Arch. Virol. 151:1267–1279 [DOI] [PubMed] [Google Scholar]
- 15. Ito T., et al. 1997. Receptor specificity of influenza A viruses correlates with the agglutination of erythrocytes from different animal species. Virology 227:493–499 [DOI] [PubMed] [Google Scholar]
- 16. Kaleta E. F., Hergarten G., Yilmaz A. 2005. Avian influenza A viruses in birds—an ecological, ornithological and virological view. Dtsch. Tierarztl. Wochenschr. 112:448–456 [PubMed] [Google Scholar]
- 17. Kawaoka Y., Chambers T. M., Sladen W. L., Webster R. G. 1988. Is the gene pool of influenza viruses in shorebirds and gulls different from that in wild ducks? Virology 163:247–250 [DOI] [PubMed] [Google Scholar]
- 18. Maher J. A., DeStefano J. 2004. The ferret: an animal model to study influenza virus. Lab. Anim. (NY) 33:50–53 [DOI] [PubMed] [Google Scholar]
- 19. Munster V. J., et al. 2007. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog. 3:e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Palmer D., Coleman M., Dowdle W., Schild G. 1975. Advanced laboratory techniques for influenza diagnosis. Immunol. Ser. 6:51–52 [Google Scholar]
- 21. Perriere G., Gouy M. 1996. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78:364–369 [DOI] [PubMed] [Google Scholar]
- 22. Rimmelzwaan G. F., et al. 2001. Antigenic and genetic characterization of swine influenza A (H1N1) viruses isolated from pneumonia patients in The Netherlands. Virology 282:301–306 [DOI] [PubMed] [Google Scholar]
- 23. Slemons R. D., Johnson D. C., Osborn J. S., Hayes F. 1974. Type-A influenza viruses isolated from wild free-flying ducks in California. Avian Dis. 18:119–124 [PubMed] [Google Scholar]
- 24. Song M. S., et al. 2009. The polymerase acidic protein gene of influenza A virus contributes to pathogenicity in a mouse model. J. Virol. 83:12325–12335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stephenson I., Wood J. M., Nicholson K. G., Zambon M. C. 2003. Sialic acid receptor specificity on erythrocytes affects detection of antibody to avian influenza haemagglutinin. J. Med. Virol. 70:391–398 [DOI] [PubMed] [Google Scholar]
- 26. Suarez D. L. 2000. Evolution of avian influenza viruses. Vet. Microbiol. 74:15–27 [DOI] [PubMed] [Google Scholar]
- 27. Swayne D. E., Suarez D. L. 2000. Highly pathogenic avian influenza. Rev. Sci. Tech. 19:463–482 [DOI] [PubMed] [Google Scholar]
- 28. Webby R. J., Woolcock P. R., Krauss S. L., Webster R. G. 2002. Reassortment and interspecies transmission of North American H6N2 influenza viruses. Virology 295:44–53 [DOI] [PubMed] [Google Scholar]
- 29. Webster R. G., Bean W. J., Gorman O. T., Chambers T. M., Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Woolcock P. R., Suarez D. L., Kuney D. 2003. Low-pathogenicity avian influenza virus (H6N2) in chickens in California, 2000-02. Avian Dis. 47:872–881 [DOI] [PubMed] [Google Scholar]
- 31. Zhou B., et al. 2011. PB2 residue 158 is a pathogenic determinant of pandemic H1N1 and H5 influenza A viruses in mice. J. Virol. 85:357–365 [DOI] [PMC free article] [PubMed] [Google Scholar]