Abstract
Replication of viral RNA genomes in fruit flies and mosquitoes induces the production of virus-derived small interfering RNAs (siRNAs) to specifically reduce virus accumulation by RNA interference (RNAi). However, it is unknown whether the RNA-based antiviral immunity (RVI) is sufficiently potent to terminate infection in adult insects as occurs in cell culture. We show here that, in contrast to robust infection by Flock house virus (FHV), infection with an FHV mutant (FHVΔB2) unable to express its RNAi suppressor protein B2 was rapidly terminated in adult flies. FHVΔB2 replicated to high levels and induced high mortality rates in dicer-2 and argonaute-2 mutant flies that are RNAi defective, demonstrating that successful infection of adult Drosophila requires a virus-encoded activity to suppress RVI. Drosophila RVI may depend on the RNAi activity of viral siRNAs since efficient FHVΔB2 infection occurred in argonaute-2 and r2d2 mutant flies despite massive production of viral siRNAs. However, RVI appears to be insensitive to the relative abundance of viral siRNAs since FHVΔB2 infection was terminated in flies carrying a partial loss-of-function mutation in loquacious required for viral siRNA biogenesis. Deep sequencing revealed a low-abundance population of Dicer-2-dependent viral siRNAs accompanying FHVΔB2 infection arrest in RVI-competent flies that included an approximately equal ratio of positive and negative strands. Surprisingly, viral small RNAs became strongly biased for positive strands at later stages of infection in RVI-compromised flies due to genetic or viral suppression of RNAi. We propose that degradation of the asymmetrically produced viral positive-strand RNAs associated with abundant virus accumulation contributes to the positive-strand bias of viral small RNAs.
INTRODUCTION
Innate immunity includes distinct mechanisms that provide immediate and broad-spectrum protection against microbial infection and often are conserved across different kingdoms (56). The fruit fly Drosophila melanogaster is a powerful model for studying innate immunity (21). Drosophila innate immunity against bacteria and fungi involves recognition of microbial molecular patterns by germ line-encoded pattern recognition receptors and the production of potent antimicrobial peptides via closely related Toll and IMD signaling pathways (21). In contrast, protection of Drosophila against viruses is mainly mediated by the RNA interference (RNAi) pathway (24, 40, 67, 72, 74) as found in fungi and plants (13, 18, 32).
RNAi and related RNA silencing pathways operate in diverse eukaryotes, including fungi, plants, invertebrates, and mammals, and recruit small RNAs such as small interfering RNAs (siRNAs) and microRNAs (miRNAs) to guide specific silencing of genes by an Argonaute protein in an RNA-induced silencing complex (RISC) or related effector complexes (2, 10, 34). Available data indicate that in Drosophila, double-stranded RNA (dsRNA) replicative intermediates of viruses with an RNA genome are recognized by Dicer-2 (Dcr-2) and further processed into virus-derived siRNAs to guide silencing of the cognate viral RNAs by Argonaute-2 (Ago2) (18, 32). This natural antiviral defense mechanism exhibits features of both innate and adaptive immunity and is referred as RNA-based antiviral immunity (RVI) because virus-specific RNA molecules serve as the inducer, target, and specificity determinant of the immunity mechanism (18). Recent studies indicate that insect viruses and arthropod-borne human viruses (arboviruses) such as Sindbis and dengue viruses are also targeted in mosquitoes by an RVI pathway homologous to the Dcr-2/Ago2 pathway identified in Drosophila (4, 31, 33, 41, 49, 61).
It is known that insect RVI naturally restricts infection of diverse RNA viruses, which induce production of viral siRNAs, accumulate to higher levels, and/or are more virulent in fruit flies and mosquitoes compromised for RNAi (24, 31, 33, 41, 48, 49, 60, 61, 67, 72, 74). An only known exception is Nora virus, to which both wild-type (WT) and RNAi-defective mutant flies exhibited similar susceptibility, although Nora virus-derived siRNAs were detected in Drosophila cells (25, 73). However, since WT flies and mosquitoes are successfully infected by many RNA viruses, it is not known whether RVI alone is sufficiently potent to ensure virus clearance and ultimately terminate infection. Second, several insect positive-strand RNA [(+)RNA] viruses encode viral suppressors of RNAi (VSRs) as found for plant viruses (9, 39). The identified insect VSRs include the B2 protein of Flock house virus (FHV) and Nodamura virus in the Nodaviridae and the 1A protein of cricket paralysis virus (CrPV) and Drosophila C virus in the Dicistroviridae, which target distinct steps of the RNAi pathways (1, 40, 45, 50, 65, 67, 72). Notably, expression of FHV B2 and CrPV 1A from Sindbis virus dramatically enhances the virulence of the arbovirus in adult mosquitoes and fruit flies, respectively (49, 50). VSR-deficient mutants of (+)RNA viruses are defective in the infection of WT plants, but these mutant viruses establish virulent systemic infection in mutant plants defective in antiviral silencing, demonstrating an essential role for the specific activity of VSR during infection of plants with (+)RNA viruses (16, 17, 70, 71). For insect RNA viruses, abolishing VSR expression by using the infectious cDNA clones available only for the nodaviruses, indicates that the VSR activity is indispensable for viral infection in cultured cells of Drosophila and for detectable self-replication of the nodaviral RNA1 in cultured Drosophila/mosquito cells and in transgenic flies (1, 24, 40, 41). However, it is unknown whether a specific, virus-encoded activity to suppress RVI is required for viral infection of adult fruit flies or mosquitoes. In particular, although Sindbis and West Nile viruses do not appear to encode a VSR, they successfully infect both WT fruit flies and mosquitoes (4, 6, 12, 32, 50). Thus, it has been proposed that insect RVI may function to modulate the accumulation level and virulence of RNA viruses including arboviruses but may not be able to terminate infection independently so that VSR may not be essential for infection of adult insects (4, 44).
Recent deep sequencing studies have determined the populations of small RNAs derived from diverse insect viruses with positive-, negative and dsRNA genomes (1, 4, 6, 18, 22, 48, 49, 55, 64, 73). These studies show that insect RNA viruses induce production of virus-derived small RNAs of predominantly 21 nucleotides (nt) in length that are divided approximately equally into positive and negative strands. These findings indicate that insect viral small RNAs are siRNAs processed by Dcr-2 from a dsRNA precursor, which is consistent with the genetic characterization of the RVI pathway in fruit flies (1, 24, 40, 67, 72, 74). However, examining the small RNA populations in Drosophila ovary somatic sheet cells that express both Ago2 and PIWI proteins has revealed production of virus-derived PIWI-interacting RNAs (piRNAs) of 24 to 30 nt in addition to viral siRNAs (73). Similar viral piRNAs were also recently detected in the C6/36 mosquito cells that are Dcr-2 deficient and mosquitoes (7, 28, 62). Major variations between the sequenced populations of insect viral small RNAs in their relative abundance and the distribution of hot spots along the viral RNA genome were also observed (18). It should be pointed out that with one exception the sequenced populations of the insect viral small RNAs were all isolated from WT virus-infected host cells, which support high-level virus accumulation. Therefore, it is not clear what features define the population of viral siRNAs in an insect host in which potent antiviral silencing occurs since some of the observed properties may be detectable only in the viral small RNA populations following efficient suppression of RVI to allow abundant virus accumulation. Cultured Drosophila cells challenged by virions of the FHV mutant (FHVΔB2) that does not replicate to detectable levels due to lack of B2 expression produce a much higher density of positive- and negative-strand viral siRNAs targeting the 5′-terminal region of the viral genomic RNA1 (1). However, it is not clear whether virus infection of adult flies also induces production of viral siRNA hot spots targeting the 5′-terminal region of the viral genome.
In the present study, we characterized the infectivity and siRNA populations of FHVΔB2 in WT and five mutant adult fruit flies. We found that the VSR-deficient mutant virus established highly virulent infection in RNAi-defective adult fruit flies, but its infection was rapidly terminated in WT fruit flies. We conclude therefore that Drosophila RVI is sufficiently potent to terminate viral infection and that the VSR activity is essential for successful infection of adult fruit flies. Since the B2 protein inhibits the biogenesis and antiviral activities of viral siRNAs shown in previous in cell culture and embryo studies (1, 11, 45, 65), our deep sequencing of total small RNAs in FHVΔB2-challenged adult flies revealed key features of virus-derived small RNA populations in RVI-competent and RVI-compromised fruit flies without the interference by the VSR. Notably, we found that abundant virus accumulation at late stages of infection in RVI-compromised fruit flies was associated with a strong bias for the positive-strand viral small RNAs, which did not occur either in RVI-competent fruit flies or in early stages of infection in RVI-compromised fruit flies when virus accumulation was low. A possible contribution of the degradation of the more abundantly produced viral (+)RNAs during (+)RNA virus infection to the positive-strand bias is discussed.
MATERIALS AND METHODS
Fly strains and virus inoculation.
Flies were reared on standard cornmeal-agar medium at room temperature. Canton S (csw) flies were used as the WT genotype in all experiments. ago2 (ago2414) and dcr-2 (dcr-2L811fsX) mutants containing null alleles have been described (38, 53). Use of loqsf00791 and R2D2S165fsX mutants in our initial studies yielded unreproducible results so that two different alleles were combined for both R2D2 (R2D21 and R2D2S165fsX) and loqs (loqsKO and loqsf00791) (23, 43, 57, 72). The loqs-R2D2 double mutant was generated by crossing loqsKO-R2D2S165fsX and loqsf00791-R2D21 mutants as previously described (23, 43, 46, 57, 72). FHV and FHVΔB2 virion suspensions were titrated by using a standard plaque assay, stained with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide. For injection, virion suspensions were diluted in 1× phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.76 mM KH2PO4 [pH 7.4]) to a final concentration of 7 × 106 PFU/ml for FHVΔB2 and 1.3 × 106 PFU/ml for FHV. One day before injection, 10 male and 10 female flies of 3 to 5 days old were transferred into each of the four vials containing fresh medium. Seven days after injection of virions into the thorax of the adult flies with a FemtoJET microinjector (Eppendorf, Germany), flies were collected for the detection of viral RNAs and proteins. Due to their susceptibility to CO2, ago2414 flies were anesthetized on ice instead of CO2. Infectivity experiments were repeated at least three times. For the survival assay, three groups of 20 adult flies from WT flies and ago2 and dcr-2 mutant flies were inoculated with FHVΔB2, and survival was monitored on a daily basis as described previously (72).
Quantitative real-time PCR.
Total RNA was extracted with TRIzol (Invitrogen) according to the manufacturer's protocol. cDNAs were synthesized by using Superscript III reverse transcriptase (Invitrogen) and gene-specific primers complementary to the positive (5′-GGTCCTTGTGTCCCATACCG-3′) or negative (5′-AACTGCTGGTTTCCATCGG-3′) strands of FHV RNA1. Real-time quantitative PCRs were carried out in the presence of iQ SYBR green Supermix (Bio-Rad). The relative abundance of selected RNAs was normalized to an internal control (rp49). Relative abundance was estimated by the ΔCT method. A two-tailed Student t test was used to statistically analyze the differences between the accumulation levels of the positive- or negative-strand RNA1 in WT and mutant flies inoculated with FHVΔB2 or FHV.
Northern and Western blot analyses.
Detection of the viral genomic RNAs and small RNAs by Northern blot hybridization was carried out as described previously (1). To detect small RNAs, high-molecular-weight RNAs in total RNA was precipitated by LiCl and low-molecular-weight RNAs in the supernatant was enriched by isopropanol precipitation. Then, 10-μg portions of low-molecular-weight RNAs were fractionated on a denaturing 15% polyacrylamide gel containing 8 M urea and 0.5× Tris-borate-EDTA, electroblotted onto Hybond-NX membranes, chemically cross-linked (54), and hybridized with [γ-32P]ATP-radiolabeled DNA oligonucleotide mixture specific to either the positive or the negative strands as described previously (1). Each mixture contained a set of 11 40-nt oligonucleotides targeting the B2 region of RNA1, a set of 15 40-nt oligonucleotides targeting the 5′-terminal 460 nt of RNA1, and 4 oligonucleotides targeting RNA2. Mapping of the viral small RNAs were carried out as described previously (1), except that in vitro transcripts of 500 nt corresponding to the positive and negative strands of FHV RNA1were used.
Total proteins extracted from flies were resuspended in 1× sodium dodecyl sulfate (SDS) loading buffer (80 mM Tris-HCl [pH 6.8], 2% SDS, 20% glycerol, 100 mM dithiothreitol, 0.01% bromophenol blue), denatured by boiling for 5 min before fractionation by SDS–12.5% polyacrylamide gel electrophoresis, and transferred to nitrocellulose filters (Bio-Rad). Filters were blocked overnight at 4°C in TBS-T buffer (10 mmol of Tris-HCl [pH 8.0], 150 mmol NaCl, 0.05% Tween 20) containing 5% nonfat milk. Proteins were detected by Western blot analysis with rabbit polyclonal antisera raised against the coat protein and B2 proteins of FHV or monoclonal α-tubulin (Sigma catalog no. T6074) and a secondary goat anti-rabbit/mouse immunoglobulin G conjugated to horseradish peroxidase (Thermo Scientific).
Reference data, small RNA libraries, and data analysis.
FHV RNA1 and RNA2 sequences (GenBank accession numbers NC_004146 and NC_004144) were retrieved from the National Center for Biotechnology Information. Sequences of fruit fly rRNA, endogenous siRNAs, and miRNAs were downloaded from FlyBase (http://flybase.org/). Genome sequences of fruit fly and other annotation data sets were downloaded from UCSC genome browser (http://genome.ucsc.edu/) as described previously (73). Cloning and sequencing of small RNAs by Illumina 2G Analyzer in the core facilities of the UC-Riverside Institute for Integrative Genome Biology and mapping onto target reference sequences by Bowtie was carried out as described previously (37, 73). Prior to analyses, small RNAs that mapped to Drosophila rRNA, tRNA, snRNA, and snoRNA were removed. Subsequently, only reads that perfectly matched the reference sequences (FHV or Drosophila genomes) were used for comparison and downstream analyses, and their sum was considered as the total reads in each library. Virus-derived small RNA reads were normalized per million; to do so, the total number of viral small RNAs ranging between 18 and 28 nt was divided by the millions of total reads in each library.
Accession numbers.
Sequence data from this article can be found at the Gene Expression Omnibus (GEO) data repository under the accession number GSE32011.
RESULTS
Infection of a VSR-deficient mutant virus is terminated in WT flies but is highly virulent in RNAi-defective flies.
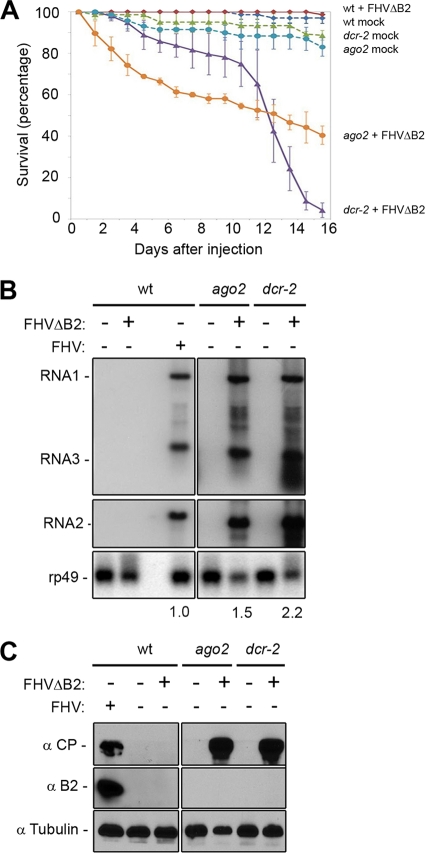
To investigate the true potency of Drosophila RVI, we characterized the infectivity in adult Drosophila of a mutant of FHV not expressing its VSR protein B2. The mutant was designated FHVΔB2 and propagated in cultured Drosophila Schneider 2 (S2) cells following depletion of Ago2 (1), which were shown as free of known viruses by deep sequencing and assembly of total small RNAs (73). In contrast to robust infection by FHV, we found that FHVΔB2 virion inoculation of WT fruit flies, shown to be free of known viruses by deep sequencing (73), had no impact on the survival compared to those mock inoculated with buffer over a time course of 16 days (Fig. 1A). Strikingly, FHVΔB2 was highly pathogenic to mutant flies carrying a loss-of-function allele in Dicer-2 (Dcr-2), required for the dicing of dsRNA into siRNAs (3, 10, 26, 34, 38). Less than 5% of dcr-2 mutant flies survived at 16 days postinocultion (dpi) with FHVΔB2 (Fig. 1A). Since the B2 protein was essential for infection in WT flies but became dispensable in the RNAi-defective mutant flies, our results established a specific role for viral suppression of RNAi in the infection of adult flies.
Fig. 1.
Infection and pathogenicity of FHVΔB2 in WT and RNAi-defective fruit flies. (A) Survival of WT, dcr-2L811fsX (dcr-2), and ago2414 (ago2) fruit flies after inoculation with buffer (mock) or FHVΔB2. Each data point represents the mean value of triplicates, and the error bars indicate the corresponding standard deviation. (B) Detection of the viral positive-strand RNAs (RNAs 1 to 3) by Northern blotting hybridizations in WT and mutant flies 7 dpi with virions of FHVΔB2 or FHV. Drosophila rp49 RNA was used as loading control. (C) Detection of the viral coat protein (CP, upper panel) and B2 protein (lower panel) in the inoculated WT and mutant flies using fly α-tubulin as loading control.
FHV infection produces two genomic (+)RNAs and a subgenomic RNA, RNA3, which acts as the mRNA for B2. FHVΔB2 carries two nucleotide substitutions in the genomic RNA1 to prevent B2 expression, which converted the first and 58th codons of the B2 open reading frame into serine and stop codons, respectively (40). Northern blotting detection of the viral RNAs 1 to 3 (Fig. 1B) and Western blotting detection of the viral coat protein (CP) (Fig. 1C, top panel) revealed that FHVΔB2 replicated to high levels in the dcr-2 mutant flies. Expression of B2 was demonstrated in WT flies infected with FHV (Fig. 1C). However, B2 was not detected in the dcr-2 mutant flies infected with FHVΔB2 (Fig. 1C) 7 dpi when FHVΔB2 replicated to high levels in the mutant flies (Fig. 1A), thereby confirming the genotype of FHVΔB2 used in this work. Since the accumulation levels of FHVΔB2 in dcr-2 flies were comparable to that of FHV in WT flies (Fig. 1B), FHVΔB2 exhibited no obvious defect in the systemic infection of adult flies. In contrast to abundant accumulation of FHVΔB2 in dcr-2 flies, the viral RNAs 1 to 3 and CP were all undetectable in WT flies inoculated with FHVΔB2 (Fig. 1B and C), a finding consistent with the observed resistance of WT flies to FHVΔB2 (Fig. 1A). We conclude therefore that the Dcr2-dependent RVI in adult Drosophila is fully capable of virus clearance and elimination of virus infection in the absence of viral suppression of RNAi.
Properties of the virus-derived siRNAs population capable of directing virus clearance.
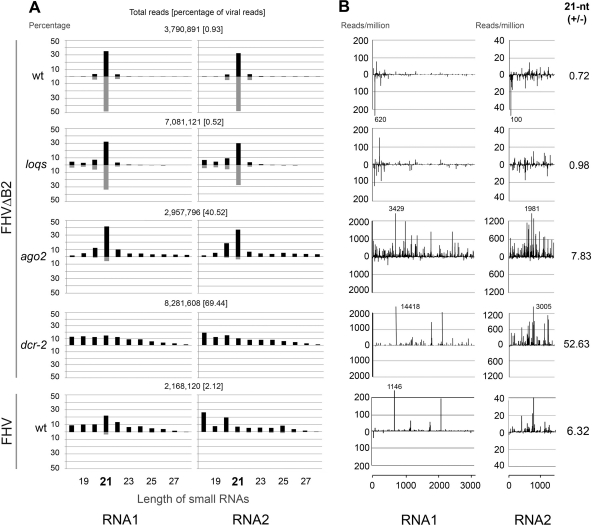
To define the properties of virus-derived siRNAs that could potentially direct virus clearance in adult Drosophila, we sequenced two independent small RNA libraries from FHVΔB2-inoculated WT and dcr-2 flies at 7 dpi using the Illumina platform. In spite of undetectable accumulation of FHVΔB2 in WT flies, we detected accumulation of 18- to 28-nt small RNAs that were 100% identical or complementary to the genomic RNAs 1 and 2 of FHVΔB2 (Fig. 2A). The population of these viral small RNAs exhibited three main properties. These virus-derived small RNAs were divided approximately equally into positive (43%) and negative (57%) strands, unlike the asymmetrical accumulation of the genomic (+)RNAs (35). The most dominant species was 21 nucleotides in length for both positive- and negative-strand small RNAs derived from either RNA1 or RNA2 (Fig. 2A) and a higher density of the 21-nt small RNAs was found to target the 5′-terminal region than those mapped to rest of the viral RNA1 (Fig. 2B). These viral small RNAs produced in adult Drosophila thus share similar properties with the population of viral siRNAs sequenced previously from FHVΔB2-chanllenged S2 cells initially by the 454 platform (1) and more recently by Illumina (see Fig. S1 in the supplemental material). These findings suggest that the viral small RNAs detected from adult Drosophila are siRNAs processed by Dcr-2 from viral dsRNA replicative intermediates produced during the synthesis of the viral progeny (+)RNA.
Fig. 2.
Profiles of virus-derived small RNA pools in WT and mutant fruit flies. (A) The relative abundance of different size classes of positive (top black bars)- and negative (bottom gray bars)-strand small RNAs showing perfect match with the two viral genomic RNAs in WT and loqs, dcr-2, and ago2 mutant flies inoculated with FHVΔB2. The total number of small RNA reads and the percentage of viral reads in each library is shown. (B) Distribution patterns of viral siRNAs along the two viral genomic RNAs in WT and loqs, dcr-2, and ago2 mutant flies inoculated with FHVΔB2. Reads of 21-nt viral siRNAs per million sequenced from WT and mutant flies inoculated with FHVΔB2 were plotted to the positive (top) and negative (bottom) strands of the viral RNA1 (left panels) and RNA2 (right panel). Each bar represents the density of unique small RNAs loci, plotted at the position of the eleventh nucleotide. Note the use of different scales due to the low abundance of viral small RNAs in WT and loqs libraries. The values of those peaks larger than the maximal value in the y axis were given. The ratios of the 21-nt positive- and negative-strand viral siRNAs are given on the right.
Deep sequencing also revealed abundant accumulation of virus-derived small RNAs in dcr-2 mutant flies infected with FHVΔB2 (Fig. 2A). However, FHVΔB2-derived small RNAs cloned from dcr-2 flies exhibited major differences from the viral siRNAs produced in FHVΔB2-challenged WT flies. First, the length distribution of FHVΔB2-derived small RNAs from dcr-2 flies (Fig. 2A) appeared random, and neither positive- nor negative-strand viral small RNAs had a predominant species with a length expected for the products of Dcr-1 (22 nt) and Dcr-2 (21 nt) or for piRNAs (24 to 30 nt) (34, 63). As expected from previous studies (10, 14, 34, 46, 51, 52, 75), the dominant 21-nt siRNA species were detectable from the known endogenous siRNA loci in WT flies either mock or FHVΔB2 inoculated, but not in dcr-2 mutant flies (Fig. 3). Second, the density of the viral small RNAs targeting the 5′-terminal region of RNA1 was not higher than the rest of RNA1 (Fig. 2B). Moreover, we found that 98% of FHVΔB2-derived small RNAs cloned from dcr-2 flies were positive strands (Fig. 2B). Since FHVΔB2 accumulated to high levels and was highly virulent in dcr-2 flies, it is likely that the Dcr2-independent viral small RNAs are nonspecific degradation products of the abundant viral RNAs 1 to 3 and inactive in directing virus clearance by RNAi.
Fig. 3.
Relative abundance of difference size classes of perfectly matched endogenous siRNAs sequenced from mock-inoculated wild-type and mutant Drosophila. Small RNAs in the range of 18 to 28 nt were compared to the following seven representative endogenous siRNAs loci reported in previous studies (30, 51, 52): hp-CG18854, hp-CR32205, hp-pncr009, CG7439, CG14033, esi-1, and esl-2.
Clearance of FHVΔB2 occurs in loqs mutant flies but is inhibited in r2d2 mutant flies.
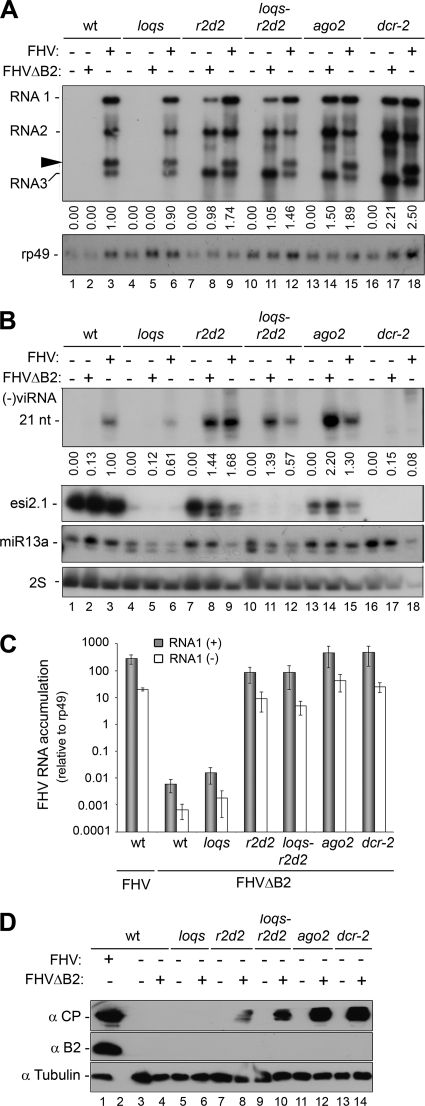
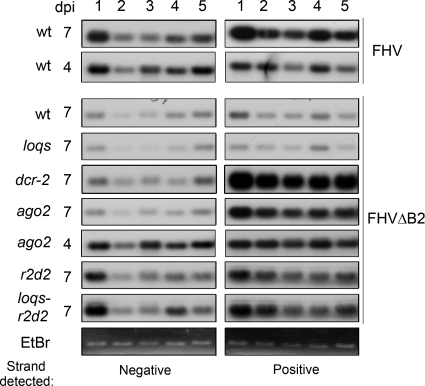
Loquacious (Loqs) and R2D2 proteins contain tandem-repeat dsRNA-binding domains and are the partners of Dicer proteins. Isoforms PB and PD of Loqs are required for the biogenesis of miRNAs and siRNAs by Dcr-1 and Dcr-2, respectively, whereas R2D2 acts together with Dcr-2 to load the exogenous and endogenous siRNAs into Ago2 in Drosophila (23, 43, 46, 57). To determine whether Loqs and R2D2 participated in the silencing of FHVΔB2, we generated loqs (loqsKO/loqsf00791) and r2d2 (R2D2S165fsX/R2D21) single mutants, as well as the loqs-r2d2 double mutant (loqsKO/loqsf00791; R2D2S165fsX/R2D21) as previously described (46). R2D2S165fsX, R2D21, and loqsKO are null alleles, but use of the partial loqsf00791 allele is necessary since it produces sufficient amount of miRNAs essential for development. Northern blotting and Western blotting analyses revealed that FHVΔB2 remained undetectable in the loqs mutant (Fig. 4A and D), indicating that FHVΔB2 was cleared in loqs flies as efficiently as in WT flies. In contrast, FHVΔB2 accumulated to high levels in the r2d2 mutant (Fig. 4A and D), demonstrating that virus resistance in adult flies was defective in r2d2 flies, which is consistent with previous studies (48, 72). Moreover, we found that FHVΔB2 replicated to similar levels in r2d2 and loqs-r2d2 flies (Fig. 4A). These findings together show that presence of the loqs alleles either alone or together with the r2d2 alleles exhibit no detectable negative impact on the antiviral immunity in adult flies.
Fig. 4.
Accumulation of viruses, viral siRNAs, and fly endogenous small RNAs in WT and mutant fruit flies. (A) Accumulation of viral positive-strand genomic and subgenomic RNAs in WT and mutant flies inoculated with either FHV or FHVΔB2 virions at 7 dpi. Fly rp49 RNA was used as loading control. Note that higher accumulation of FHV in RNAi-defective fly mutants than in WT flies is readily detected for coat protein (CP), but the difference is smaller for FHV RNAs at 7 dpi (72). An RNA3 homodimer molecule (20) detected in flies infected with FHV but not with FHVΔB2 is indicated by an arrowhead. (B) Detection of viral negative-strand small RNAs by a mixture of oligonucleotides (top panel) and of the endogenous siRNA2.1 (esi2.1, middle panel) and miRNA13a (miR13a, bottom panel) was performed as previously described (36, 46). Fly ribosomal 2S RNA was used as a loading control. (C) Relative abundance of FHV positive- and negative-strand RNA1 in the FHVΔB2-inoculated flies determined by real-time PCR. WT flies infected with FHV were used as a control. FHVΔB2 RNA1 accumulated to statistically significant lower levels in WT flies compared to all of the fly mutants except the loqs mutant (P < 0.05). (D) Detection of the viral coat protein (CP, upper panel) and B2 protein (lower panel) in the inoculated WT and mutant flies using fly α-tubulin as a loading control.
We further used quantitative real-time PCR to measure the levels of the (+)RNA1 and (−)RNA1 of FHVΔB2 in the inoculated WT and mutant flies (Fig. 4C). FHVΔB2 produced much more abundant (+)RNA1 than (−)RNA1 in WT and mutant flies (Fig. 4C). Similar asymmetrical accumulation of (+)RNA1 was obtained in FHV-infected WT flies (Fig. 4C) as found in S2 cells (35) and as expected for (+)RNA viruses, which produce low levels of (−)RNA as the template for the synthesis of the abundant (+)RNA to be packaged into progeny virions.
Among WT and mutant flies inoculated with FHVΔB2, the highest accumulation of both (+)RNA1 and (−)RNA1 was detected in the dcr-2 mutant flies (Fig. 4A and C). There was an ∼10,000-fold increase of FHVΔB2 in r2d2 and loqs-r2d2 mutant flies compared to WT flies (Fig. 4C). However, the accumulation levels of both the (+)RNA1 and (−)RNA1 of FHVΔB2 remained extremely low in the loqs mutant (Fig. 4C), which was similar to WT flies. These data demonstrate an essential role for R2D2 in the clearance of FHVΔB2 in adult Drosophila. In contrast, the partial genetic defect at the loqs locus had minimal effects on the clearance of FHVΔB2 in adult Drosophila.
Reduced biogenesis of viral siRNAs does not prevent virus clearance in loqs flies.
To understand the contrasting responses of loqs and r2d2 mutant flies to FHVΔB2, we analyzed the accumulation of FHVΔB2-derived small RNAs in WT and mutant flies. The endo-siRNAs were not detectable in dcr-2 flies by Northern blot hybridizations, and their accumulation decreased much more dramatically in loqs and loqs-r2d2 flies than in r2d2 flies compared to WT flies (Fig. 4B). By comparison, presence of the loqs alleles had less effect on the production of the endogenous miRNAs than on the endo-siRNAs (Fig. 4B). These findings were consistent with previous studies (23, 42, 43, 46, 57) and thus verified the genotypes of the fly mutants used in the present study.
Previous attempts to detect viral siRNAs in FHV-infected WT flies by Northern blot hybridization were unsuccessful (72). Use of a chemical cross-linking protocol (54) in this study allowed reproducible detection of the negative-strand viral siRNAs as a discrete 21-nt species in FHV-infected WT flies (Fig. 4B, lane 3). The same protocol failed to detect accumulation of a discrete 21-nt species for either the negative-strand (Fig. 4B, lanes 17 and 18) or positive-strand (data not shown) viral siRNAs following robust infection with either FHV or FHVΔB2 in dcr-2 flies. These findings provide the genetic evidence of the production of the 21-nt viral siRNAs by Dcr-2, which is consistent with the results from deep sequencing. However, the 21-nt viral siRNAs remained undetectable in WT flies inoculated by FHVΔB2 (Fig. 4B, lane 2), indicating that viral siRNAs produced low abundantly in FHVΔB2-challenged WT flies (Fig. 2A) were below the level of detection by the improved small RNA detection protocol. This differed from inoculation of the fly S2 cells with FHVΔB2 virions, which induced abundant accumulation of viral siRNAs readily detectable by Northern blot hybridization (1). The inoculum used in the challenge of S2 cells ensured viral invasion of every cell and the initial rounds of viral RNA replication would yield abundant dsRNA replicative intermediates to be processed into siRNAs, unlike restriction of FHVΔB2 into perhaps a few cells of the inoculated flies.
Production of the 21-nt viral siRNAs was also detected in r2d2, loqs, and loqs-r2d2 mutant flies infected with FHV and in r2d2 and loqs-r2d2 mutant flies infected with FHVΔB2 (Fig. 4B). However, the 21-nt viral siRNAs reproducibly accumulated to higher levels in r2d2 flies than in loqs-r2d2 flies following FHV infection (Fig. 4B, lanes 9 and 12), even though FHV replicated to similar levels in these mutant flies. This indicates a genetic requirement for loqs in the biogenesis of the 21-nt viral siRNAs. Consistently, accumulation of the 21-nt viral siRNAs was higher in WT flies than loqs flies following FHV infection (Fig. 4B, lanes 3 and 6) and a reduced accumulation of the 21-nt viral siRNAs was also detected in loqs-r2d2 flies compared to r2d2 flies following FHVΔB2 infection (Fig. 4B, lanes 8 and 11). By comparison, the genetic defect at the loqs locus had less impact on the biogenesis of viral siRNAs than on the endo-siRNAs, possibly because efficient replication of the viral genomic RNAs produces more siRNA precursors than the transcription of endo-siRNA genes does.
Northern blot hybridization failed to detect accumulation of the 21-nt viral siRNAs in loqs mutant challenged by FHVΔB2 (Fig. 4B, lane 5). Therefore, we sequenced two independent small RNA libraries from loqs flies challenged by FHVΔB2. The results from deep sequencing found accumulation of small RNAs that were 100% identical or complementary to the genomic RNAs 1 and 2 of FHVΔB2 (Fig. 2A). Further analysis showed that the population of the viral small RNAs sequenced from the challenged loqs flies shared similar properties with the viral siRNAs produced in WT flies: they were approximately equally divided into positive and negative strands and contained 21-nt small RNAs as the dominant species and a high density of 21-nt positive- and negative-strand small RNAs mapped to the 5′-terminal region of RNA1.
Our results together illustrate efficient virus clearance and elimination of virus infection in both WT and the loqs mutant flies following FHVΔB2 inoculation in spite of a markedly reduced biogenesis of viral siRNAs in loqs flies. Therefore, an effective RVI in adult Drosophila does not appear to require the full capacity in the production of viral siRNAs, suggesting that RVI is insensitive to the relative abundance of viral siRNAs.
Effective RVI in adult flies requires the Ago2-mediated antiviral activity of the 21-nt viral siRNAs.
FHVΔB2 accumulated to high levels in adult Drosophila homozygous for a loss-of-function allele in Ago2 (ago2414) as indicated by Northern blot (Fig. 1B and 4A) and Western blot (Fig. 1C and 4D) analyses. Real-time PCR further indicated that FHVΔB2 replicated to similar levels in ago2 and dcr-2 mutant flies (Fig. 4C). These findings support an antiviral role of Ago2 reported in previous studies for (40, 48, 67). Unlike in dcr-2 flies, however, the 21-nt viral siRNAs were detected in the FHVΔB2-infected ago2 flies (Fig. 4B) and were in fact the most abundant among the WT and mutant flies inoculated with either FHV or FHVΔB2 (Fig. 4B). A time course analysis further showed that robust infection of ago2 flies with FHVΔB2 was associated with rapid accumulation of the 21-nt viral siRNAs that were readily detectable by Northern blotting (Fig. 5). Thus, efficient dicing of the viral replicative intermediates into siRNAs in the absence of Ago2 failed to suppress FHVΔB2 infection in ago2 flies, indicating a key antiviral role for AGO2 downstream of siRNA biogenesis. The low expression level of Ago2 in adult flies prevents examining siRNA loading by coimmunoprecipitation with Ago2-specific antibodies. However, Ago2 is known to cleave (slice) RNA targets using siRNAs as guides in exogenous RNAi and to load viral siRNAs in S2 cells challenged by FHVΔB2 (1, 26).
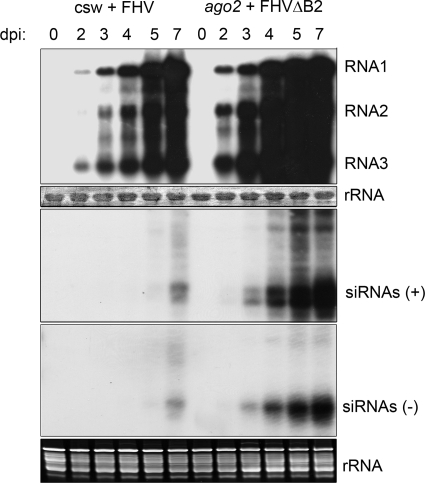
Fig. 5.
Time course analysis of the virus large and small RNAs following fruit fly inoculation with FHV or FHVΔB2. Groups of 20 WT (csw) and ago2 adult flies were inoculated with virions of FHV (1.3 × 106 PFU/ml) and FHVΔB2 (7 × 106 PFU/ml), respectively. Total high- and low-molecular-weight RNAs were extracted from the flies 2, 3, 4, 5, or 7 days postinoculation (dpi) or from mock-inoculated flies (0) and subjected to Northern blot hybridizations to detect either viral positive-strand RNAs 1, 2, and 3 or viral positive (+) and negative (−) siRNAs. Fly rRNAs were used as loading controls.
We found that the average survival rate of ago2 flies 16 dpi with FHVΔB2 was ca. 40% (Fig. 1A), which was higher than the average survival rate of dcr-2 flies (5%). This indicates that presence of Dcr-2 may provide some form of protection against FHVΔB2 in ago2 flies compared to dcr-2 flies even though FHVΔB2 replicated to similar levels in the two fly mutants (Fig. 4C). The Dcr-2-mediated protection may be conferred either by dicing the viral replicative intermediates or, alternatively, by triggering an RNAi-independent defense (15).
Northern blot hybridization detected markedly reduced accumulation of viral siRNAs in ago2 and loqs-r2d2 flies infected with FHV than in those flies infected with FHVΔB2 in spite of comparable virus replication levels (Fig. 4A and B). Similar virus accumulation levels in FHV-infected WT flies and FHVΔB2-infected ago2 flies were also associated with drastically different levels of viral siRNAs (Fig. 5). These results indicate that expression of the B2 protein suppressed production of viral siRNAs in the infected flies, which is consistent with the results from in vitro, cell culture, and embryo studies (1, 11, 45, 65).
Abundant accumulation of the (+)RNA virus is associated with strong bias for the positive-strand virus-derived small RNAs at later stages of infection.
Deep sequencing of the total small RNAs from the FHVΔB2-infected ago2 flies showed that 21-nt species was the most dominant among both the positive and the negative strand small RNAs derived from either RNA1 or RNA2 (Fig. 2A). Thus, these virus-derived small RNAs were siRNAs produced by Dcr-2 in the absence of Ago2. As expected, the most dominant length of the endo-siRNAs was 21 nt (Fig. 3). Strikingly, 92.3% of the FHVΔB2-derived small RNAs sequenced from ago2 mutant flies 7 days after infection were positive strands, unlike FHVΔB2-challenged WT flies in which approximately equal ratios of positive and negative strand viral siRNAs were detected. This strong bias for the positive strands was observed for the 21-nt small RNAs derived from both RNA1 (88%) and RNA2 (91%) of FHVΔB2 in ago2 mutant flies (Fig. 2A).
We used an independent assay (Fig. 6) to determine the relative abundance of the positive- and negative-strand small RNAs derived from FHVΔB2 RNA1 produced in ago2 flies. In this assay, equal amounts of the (+)RNA and (−)RNA corresponding to five consecutive 500-nt regions of FHV RNA1 were transcribed in vitro and blotted onto the same filter for hybridization with the small RNAs of 18 to 28 nt harvested from ago2 mutant flies 7 days after inoculation with FHVΔB2. The harvested small RNAs were dephosphorylated and subsequently labeled at the 5′ ends with [γ-32P]ATP by kinase. The result showed that the hybridization yielded much stronger signal for the panel of the negative-strand segments of RNA1 than the positive-strand segments (Fig. 6), indicating that the FHVΔB2-infected ago2 flies contained more abundant viral positive-strand small RNAs than the viral negative-strand small RNAs. However, labeled small RNAs isolated from either WT or loqs flies 7 days after challenge inoculation with FHVΔB2 detected approximately equal intensities of the hybridization signal for the (+)RNA1 and (−)RNA1 segments (Fig. 6), indicating the absence of strand bias for the viral small RNAs produced in these flies. Therefore, the gel blot hybridization approach independently verified the results from deep sequencing (Fig. 2A), although only the small RNAs with a 5′-monophosphate were cloned for deep sequencing unlike the hybridization approach that examined small RNAs with mono-, di-, and triphosphates at the 5′ end.
Fig. 6.
Relative abundance of the viral positive- and negative-strand small RNAs in WT and mutant adult flies. Positive- and negative-strand RNAs corresponding to the five consecutive 500-nt regions of FHV RNA1 from the 5′ ends were transcribed in vitro by T7 RNA polymerase from PCR-amplified DNA fragments. Note that the 3′-terminal region of FHV RNA1 was not represented in the panel of RNA fragments. Equal amounts of these positive (left panel) and negative (right panel) strand transcripts were fractionated, blotted onto a nylon membrane, and used for hybridization by the same individual probes, which were 5′-end-labeled total small RNAs (18 to 28 nt) gel purified from WT and mutant flies (indicated on the left) 4 or 7 days postinoculation (dpi) with FHVΔB2 or FHV (indicated on the right). The hybridization signal intensities of the right and left panel therefore revealed the relative abundance of the viral positive- and negative-strand small RNAs, respectively, in an infected fly mutant as the harvested small RNAs were used to probe equal amounts of the positive- and negative-strand viral RNAs in the same filter (stained by ethidium bromide in the bottom lane).
The strand ratios of viral small RNAs in r2d2, loqs-r2d2, and dcr-2 mutant flies 7 days after FHVΔB2 inoculation were investigated by the same gel hybridization approach. The results showed that the positive-strand bias occurred in all of those mutant flies (Fig. 6), in which FHVΔB2 replicated to high levels (Fig. 4A). The strongest positive-strand bias was detected in the FHVΔB2-infected dcr-2 flies (Fig. 6), in which the cloned perfect-matched viral small RNAs were mostly positive strands and had a random length distribution expected for nonspecific degradation products (Fig. 2A). Moreover, we found that the positive-strand viral small RNAs were also much more abundant in WT flies 7 days after infection with FHV (Fig. 6). Therefore, the positive strand bias of viral small RNAs occurred in all of the infected flies where there was abundant virus accumulation due to RNAi suppression by either genetic mutation in the RNAi pathway or expression of a VSR. In contrast, the positive- and negative-strand viral small RNAs accumulated to similar levels in FHVΔB2-challenged WT and loqs flies where virus accumulation was undetectable by Northern blot hybridizations (Fig. 4A).
The observed correlation between the accumulation of the (+)RNA virus and of the positive-strand viral small RNAs in the infected flies suggests that the degradation of the asymmetrically accumulated viral genomic (+)RNAs contributes to the viral small RNA positive-strand bias. This idea is consistent with the observation that the positive-strand bias was not associated with a reduced accumulation of the viral negative-strand small RNAs in the infected flies, as shown by Northern blot hybridization (Fig. 4B). To test the idea, we examined the strand ratios of the viral small RNAs in FHV-infected WT flies and FHVΔB2-infected ago2 flies 4 days after inoculation when virus accumulation was much lower than in those flies 7 days after inoculation (Fig. 5). We found no strand bias in the viral small RNAs accumulated in FHV-infected WT flies at 4 dpi in contrast to the strong bias at 7 dpi (Fig. 6). The viral small RNAs from FHVΔB2-infected ago2 flies at 4 dpi exhibited strand bias for the positive strands, which was, however, much weaker than that detected at 7 dpi (Fig. 6). We noted that at 4 dpi FHVΔB2 accumulated to much higher levels in ago2 flies than FHV in WT flies (Fig. 5). Thus, WT and ago2 flies infected, respectively, with FHV and FHVΔB2 accumulated more positive-strand viral small RNAs than the negative strands only at later stages of infection when the (+)RNA virus replicated to high levels. These findings further support a role of the nonspecific degradation of the asymmetrically accumulated viral (+)RNAs in the positive-strand bias of the viral small RNAs detected at later stages of infection.
DISCUSSION
It is known that replication of viral RNA genomes in insect cells or whole organisms induces RVI that reduces the accumulation of the invading virus (24, 33, 40, 41, 49, 50, 60, 61, 67, 72, 74). However, it is unclear whether the induced insect RVI is sufficiently potent to terminate infection so that a specific virus-encoded activity to suppress RVI becomes essential for an RNA virus to establish infection in insects. We show here that the VSR-deficient FHVΔB2 establishes virulent infection in RNAi-defective mutant flies but is rapidly cleared in WT flies by an RVI mechanism that depends on the siRNA pathway defined by Dcr2, R2D2, and Ago2 components. Our work thus demonstrates for the first time that RVI induced by a (+)RNA virus can rapidly terminate infection in adult insects. Since WT flies are efficiently infected by FHV encoding the VSR B2 protein, the expression of which from FHVΔB2 is abolished, our findings also provide the first example for an essential role of viral suppression of RVI in the infection of adult insects. These results are consistent with the genetic studies in plants (16, 17, 70, 71) and extend previous findings carried out in cultured insect cells and adult insects using WT viruses or recombinant viruses expressing a heterologous VSR (1, 24, 40, 41, 49, 50).
Based on these results, we propose that without viral suppression of RVI, virus clearance is an inevitable consequence following replication of viral RNA genomes in insects. This hypothesis is supported by the fact that several insect (+)RNA viruses encode VSR (1, 40, 45, 49, 50, 65, 67, 72). However, this idea is inconsistent with the observations that arboviruses such as Sindbis virus do not exhibit RNAi suppressor activity using available RNAi assays (4, 32, 50). It should be pointed out that identification of some VSRs may require specific assays. For example, potato virus X was not considered to encode VSR activity before the development of the coinfiltration protocol in which a candidate VSR is expressed during the induction of RNA silencing (8, 68, 69). Similarly, the VSR encoded by red clover mottle virus RNA2 was identified by an assay based on complementation of viral movement, but not by the coinfiltration assay (58). However, we cannot rule out an alternative hypothesis that Sindbis virus may sequester their dsRNA intermediates into membrane-enclosed compartments that are inaccessible to Dicer cleavages.
Deep sequencing of small RNAs in WT and loqs mutant flies challenged by FHVΔB2 defined the population of viral siRNAs in an adult insect in which a (+)RNA virus is efficiently being cleared by RVI (Fig. 2A). We found that viral siRNAs sequenced from both WT and loqs flies were divided approximately equally into positive and negative strands unlike the asymmetrical accumulation of the genomic (+)RNAs and that the most dominant species of both the positive- and negative-strand viral siRNAs were 21 nt in length. We also detected a higher density of the 21-nt viral siRNAs targeting the 5′-terminal region of the viral genomic RNA1 compared to the remaining regions of RNA1. None of these features was associated with the perfect match, FHVΔB2-derived, predominantly positive-strand small RNAs sequenced from the dcr-2 mutant flies. These cloned viral small RNAs most likely corresponded to the nonspecific degradation products associated with high-level accumulation of the positive-strand viral genomic and subgenomic RNAs since Northern blot hybridization failed to detect a discrete 21-nt band using probes specific to either the positive or the negative strands. Therefore, our results together show that the population of viral siRNAs detected in WT and loqs flies was produced by Dcr-2 in adult Drosophila. Since dcr-2 flies were highly susceptible to the VSR-defective FHVΔB2, the detected viral small RNAs in dcr-2 flies most likely did not have an antiviral function. However, we do not have direct evidence to rule out this possibility.
Our work also examined the role of Loqs in the silencing of FHVΔB2, which, unlike R2D2 and Ago2, has not been implicated in the RVI against WT viruses in previous studies (24, 40, 48, 67, 72, 74). Consistent with a previous study (46), the presence of the incomplete loss-of-function loqs alleles in fruit flies was associated with a dramatic reduction in the accumulation of endo-siRNAs (Fig. 4B). We also observed only a modestly decreased production of the viral siRNAs in loqs flies compared to WT flies following FHV infection (Fig. 4B), suggesting a role for loqs in the biogenesis of viral siRNAs. However, presence of the loqs alleles either alone in the single mutant or together with the r2d2-null alleles in the double mutant had no detectable inhibitory effect on the clearance of FHVΔB2 in adult flies (Fig. 4A). This indicates that the RVI mechanism may be mediated by the viral siRNAs rather than the endo-siRNAs, which were hardly detectable in loqs flies. This idea is also consistent with the recent demonstration for the gene silencing activity of viral siRNAs produced in infected Drosophila cells (48). It is important to note that the silencing 21-nt viral siRNAs were not abundant in FHVΔB2-challenged WT and loqs flies and detectable by deep sequencing (Fig. 2A) but not by Northern blot hybridization (Fig. 4B). These findings strongly suggest that a large population of viral small RNAs accumulated in an infected cell may not participate in RVI and may instead act as decoy siRNAs to evade the RVI mechanism as proposed recently (5, 22, 64). These decoy siRNAs may include siRNAs processed from the replicating viral defective interfering RNAs (DI RNAs) associated with RNA virus infection, which are known to interfere with the activity of the VSR (27). Viral siRNAs derived from American nodavirus (ANV) failed to guide specific RNAi in S2 cells persistently infected with four other RNA viruses, which may also be caused by decoy siRNAs derived from a 591-nt DI RNA of ANV since 51% of the total viral siRNAs of the S2 cells was mapped to the DI RNA (22, 73).
An unexpected finding of the present study is that although both strands of viral small RNAs accumulated to similar levels in the RVI-competent flies, a strong bias for the positive strands appeared in RVI-compromised flies at later stages of infection when abundant virus accumulation was detected (Fig. 6). These include infection of RNAi-defective fly mutants such as ago2 and r2d2 mutant flies and of WT flies with FHV that expresses a potent VSR. However, no strand bias or only weak strand bias was detected in FHV-infected WT flies or FHVΔB2-infected ago2 flies 4 days after inoculation when the virus accumulation was low. No strand bias was detected for siRNAs derived from vesicular stomatitis virus from infected ago2 flies sequenced at 3 dpi (48), which is consistent with our data. Our findings indicate that the positive strand bias of the viral small RNAs may be caused at least in part by the nonspecific degradation associated with abundant accumulation of (+)RNA viruses, which produce 10-fold or more (+)RNAs than (−)RNAs. Similar positive-strand bias of viral small RNAs has been reported in plants and insects infected with WT (+)RNA viruses (6, 19, 29, 47, 59, 66). Thus, it will be of interest to determine whether the positive-strand bias of viral small RNAs in these examples also occurs at the early stages of infection.
Supplementary Material
ACKNOWLEDGMENT
We thank Richard Carthew for generously providing the mutants used in this study.
This work was supported by NIH grants R01 AI052447 and RCI GM091896 (to S.-W.D.).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 28 September 2011.
REFERENCES
- 1. Aliyari R., et al. 2008. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe 4: 387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baulcombe D. 2004. RNA silencing in plants. Nature 431: 356–363 [DOI] [PubMed] [Google Scholar]
- 3. Bernstein E., Caudy A. A., Hammond S. M., Hannon G. J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- 4. Blair C. D. 2011. Mosquito RNAi is the major innate immune pathway controlling arbovirus infection and transmission. Future Microbiol. 6: 265–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blevins T., et al. 2011. Massive production of small RNAs from a non-coding region of Cauliflower mosaic virus in plant defense and viral counter-defense. Nucleic Acids Res. 39: 5003–5014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brackney D. E., Beane J. E., Ebel G. D. 2009. RNAi targeting of West Nile virus in mosquito midguts promotes virus diversification. PLoS Pathog. 5: e1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brackney D. E., et al. 2010. C6/36 Aedes albopictus cells have a dysfunctional antiviral RNA interference response. PLoS Negl. Trop. Dis. 4: e856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brigneti G., et al. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17: 6739–6746 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9. Burgyan J., Havelda Z. 2011. Viral suppressors of RNA silencing. Trends Plant Sci. 16: 265–272 [DOI] [PubMed] [Google Scholar]
- 10. Carthew R. W., Sontheimer E. J. 2009. Origins and Mechanisms of miRNAs and siRNAs. Cell 136: 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chao J. A., et al. 2005. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat. Struct. Mol. Biol. 12: 952–957 [DOI] [PubMed] [Google Scholar]
- 12. Chotkowski H. L., et al. 2008. West Nile virus infection of Drosophila melanogaster induces a protective RNAi response. Virology 377: 197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Csorba T., Pantaleo V., Burgyan J. 2009. RNA silencing: an antiviral mechanism. Adv. Virus Res. 75: 35–71 [DOI] [PubMed] [Google Scholar]
- 14. Czech B., et al. 2008. An endogenous small interfering RNA pathway in Drosophila. Nature 453: 798–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deddouche S., et al. 2008. The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in drosophila. Nat. Immunol. 9: 1425–1432 [DOI] [PubMed] [Google Scholar]
- 16. Deleris A., et al. 2006. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313: 68–71 [DOI] [PubMed] [Google Scholar]
- 17. Diaz-Pendon J. A., Li F., Li W. X., Ding S. W. 2007. Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell 19: 2053–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ding S. W. 2010. RNA-based antiviral immunity. Nat. Rev. Immunol. 10: 632–644 [DOI] [PubMed] [Google Scholar]
- 19. Donaire L., et al. 2009. Deep-sequencing of plant viral small RNAs reveals effective and widespread targeting of viral genomes. Virology 392: 203–214 [DOI] [PubMed] [Google Scholar]
- 20. Eckerle L. D., Albarino C. G., Ball L. A. 2003. Flock House virus subgenomic RNA3 is replicated and its replication correlates with transactivation of RNA2. Virology 317: 95–108 [DOI] [PubMed] [Google Scholar]
- 21. Ferrandon D., Imler J. L., Hetru C., Hoffmann J. A. 2007. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat. Rev. Immunol. 7: 862–874 [DOI] [PubMed] [Google Scholar]
- 22. Flynt A., Liu N., Martin R., Lai E. C. 2009. Dicing of viral replication intermediates during silencing of latent Drosophila viruses. Proc. Natl. Acad. Sci. U. S. A. 106: 5270–5275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Forstemann K., et al. 2005. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 3: e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Galiana-Arnoux D., Dostert C., Schneemann A., Hoffmann J. A., Imler J. L. 2006. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat. Immunol. 7: 590–597 [DOI] [PubMed] [Google Scholar]
- 25. Habayeb M. S., Ekstrom J. O., Hultmark D. 2009. Nora virus persistent infections are not affected by the RNAi machinery. PLoS One 4: e5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hammond S. M., Boettcher S., Caudy A. A., Kobayashi R., Hannon G. J. 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146–1150 [DOI] [PubMed] [Google Scholar]
- 27. Havelda Z., Hornyik C., Valoczi A., Burgyan J. 2005. Defective interfering RNA hinders the activity of a tombusvirus-encoded posttranscriptional gene silencing suppressor. J. Virol. 79: 450–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hess A. M., et al. 2011. Small RNA profiling of Dengue virus-mosquito interactions implicates the PIWI RNA pathway in antiviral defense. BMC Microbiol. 11: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ho T., Pallett D., Rusholme R., Dalmay T., Wang H. 2006. A simplified method for cloning of short interfering RNAs from Brassica juncea infected with Turnip mosaic potyvirus and Turnip crinkle carmovirus. J. Virol. Methods 136: 217–223 [DOI] [PubMed] [Google Scholar]
- 30. Kawamura Y., et al. 2008. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature 453: 793–797 [DOI] [PubMed] [Google Scholar]
- 31. Keene K. M., et al. 2004. RNA interference acts as a natural antiviral response to O'nyong-nyong virus (Alphavirus; Togaviridae) infection of Anopheles gambiae. Proc. Natl. Acad. Sci. U. S. A. 101: 17240–17245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kemp C., Imler J. L. 2009. Antiviral immunity in drosophila. Curr. Opin. Immunol. 21: 3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Khoo C. C., Piper J., Sanchez-Vargas I., Olson K. E., Franz A. W. 2010. The RNA interference pathway affects midgut infection- and escape barriers for Sindbis virus in Aedes aegypti. BMC Microbiol. 10: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim V. N., Han J., Siomi M. C. 2009. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell. Biol. 10: 126–139 [DOI] [PubMed] [Google Scholar]
- 35. Kopek B. G., Perkins G., Miller D. J., Ellisman M. H., Ahlquist P. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 5: e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853–858 [DOI] [PubMed] [Google Scholar]
- 37. Langmead B., Trapnell C., Pop M., Salzberg S. L. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee Y. S., et al. 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81 [DOI] [PubMed] [Google Scholar]
- 39. Li F., Ding S. W. 2006. Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 60: 503–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li H. W., Li W. X., Ding S. W. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296: 1319–1321 [DOI] [PubMed] [Google Scholar]
- 41. Li W. X., et al. 2004. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl. Acad. Sci. U. S. A. 101: 1350–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu J., Valencia-Sanchez M. A., Hannon G. J., Parker R. 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7: 719–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu Q., et al. 2003. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301: 1921–1925 [DOI] [PubMed] [Google Scholar]
- 44. Lu R., Li H., Li W. X., Ding S. W. 2004. RNA-based immunity in insects, p. 63–74. In Gillespie S. H., Smith G. L., Osbourn A. (ed.), Microbe-vector interactions in vector-borne diseases, vol. 63 Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 45. Lu R., et al. 2005. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature 436: 1040–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marques J. T., et al. 2010. Loqs and R2D2 act sequentially in the siRNA pathway in Drosophila. Nat. Struct. Mol. Biol. 17: 24–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Molnar A., et al. 2005. Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J. Virol. 79: 7812–7818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mueller S., et al. 2010. RNAi-mediated immunity provides strong protection against the negative-strand RNA vesicular stomatitis virus in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 107: 19390–19395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Myles K. M., Wiley M. R., Morazzani E. M., Adelman Z. N. 2008. Alphavirus-derived small RNAs modulate pathogenesis in disease vector mosquitoes. Proc. Natl. Acad. Sci. U. S. A. 105: 19938–19943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nayak A., et al. 2010. Cricket paralysis virus antagonizes Argonaute 2 to modulate antiviral defense in Drosophila. Nat. Struct. Mol. Biol. 17: 547–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Okamura K., Balla S., Martin R., Liu N., Lai E. C. 2008. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nat. Struct. Mol. Biol. 15: 581–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Okamura K., et al. 2008. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 453: 803–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Okamura K., Ishizuka A., Siomi H., Siomi M. C. 2004. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 18: 1655–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pall G. S., Codony-Servat C., Byrne J., Ritchie L., Hamilton A. 2007. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 35: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Parameswaran P., et al. 2010. Six RNA viruses and forty-one hosts: viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog. 6: e1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Parham P. 2003. Innate immunity: the unsung heroes. Nature 423: 20. [DOI] [PubMed] [Google Scholar]
- 57. Park J. K., Liu X., Strauss T. J., McKearin D. M., Liu Q. 2007. The miRNA pathway intrinsically controls self-renewal of Drosophila germline stem cells. Curr. Biol. 17: 533–538 [DOI] [PubMed] [Google Scholar]
- 58. Powers J. G., et al. 2008. The Red clover necrotic mosaic virus RNA-2 encoded movement protein is a second suppressor of RNA silencing. Virology 381: 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Qi X., Bao F. S., Xie Z. 2009. Small RNA deep sequencing reveals role for Arabidopsis thaliana RNA-dependent RNA polymerases in viral siRNA biogenesis. PLoS One 4: e4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sabin L. R., et al. 2009. Ars2 regulates both miRNA- and siRNA-dependent silencing and suppresses RNA virus infection in Drosophila. Cell 138: 340–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sanchez-Vargas I., et al. 2009. Dengue virus type 2 infections of Aedes aegypti are modulated by the mosquito's RNA interference pathway. PLoS Pathog. 5: e1000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Scott J. C., et al. 2010. Comparison of dengue virus type 2-specific small RNAs from RNA interference-competent and -incompetent mosquito cells. PLoS Negl. Trop. Dis. 4: e848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Siomi M. C., Sato K., Pezic D., Aravin A. A. 2011. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell. Biol. 12: 246–258 [DOI] [PubMed] [Google Scholar]
- 64. Siu R. W., et al. 2011. Antiviral RNA interference responses induced by Semliki Forest virus infection of mosquito cells: characterization, origin, and frequency-dependent functions of virus-derived small interfering RNAs. J. Virol. 85: 2907–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sullivan C. S., Ganem D. 2005. A virus-encoded inhibitor that blocks RNA interference in mammalian cells. J. Virol. 79: 7371–7379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Szittya G., et al. 2010. Structural and functional analysis of viral siRNAs. PLoS Pathog. 6: e1000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. van Rij R. P., et al. 2006. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 20: 2985–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Voinnet O., Lederer C., Baulcombe D. C. 2000. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103: 157–167 [DOI] [PubMed] [Google Scholar]
- 69. Voinnet O., Pinto Y. M., Baulcombe D. C. 1999. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. U. S. A. 96: 14147–14152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang X. B., et al. 2011. The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative Argonautes in Arabidopsis thaliana. Plant Cell 23: 1625–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang X. B., et al. 2010. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 107: 484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wang X. H., et al. 2006. RNA interference directs innate immunity against viruses in adult Drosophila. Science 312: 452–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wu Q., et al. 2010. Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc. Natl. Acad. Sci. U. S. A. 107: 1606–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zambon R. A., Vakharia V. N., Wu L. P. 2006. RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cell Microbiol. 8: 880–889 [DOI] [PubMed] [Google Scholar]
- 75. Zhou R., et al. 2009. Processing of Drosophila endo-siRNAs depends on a specific Loquacious isoform. RNA 15: 1886–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.