Abstract
Previous reports have shown that cholesterol depletion of the membrane envelope of the hepatitis B virus (HBV) impairs viral infection of target cells. A potential function of this lipid in later steps of the viral life cycle remained controversial, with secretion of virions and subviral particles (SVP) being either inhibited or not affected, depending on the experimental approach employed to decrease the intracellular cholesterol level. This work addressed the role of host cell cholesterol on HBV replication, assembly, and secretion, using an alternative method to inhibition of the enzymes involved in the biosynthesis pathway. Growing HBV-producing cells with lipoprotein-depleted serum (LPDS) resulted in an important reduction of the amount of cholesterol within 24 h of treatment (about 40%). Cell exposure to chlorpromazine, an inhibitor of the clathrin-mediated pathway used by the low-density lipoprotein receptor for endocytosis, also impacted the cholesterol level; however, this level of inhibition was not achievable when the synthesis inhibitor lovastatin was used. HBV secretion was significantly inhibited in cholesterol-depleted cells (by ∼80%), while SVP release remained unaffected. The viral DNA genome accumulated in LPDS-treated cells in a time-dependent manner. Specific immunoprecipitation of nucleocapsids and mature virions revealed an increased amount of naked nucleocapsids, while synthesis of the envelope proteins occurred as normally. Following analysis of the large envelope protein conformation in purified microsomes, we concluded that cholesterol is important in maintaining the dual topology of this polypeptide, which is critical for viral envelopment.
INTRODUCTION
A large variety of viruses, of which many are important human pathogens, depend on lipid and cholesterol metabolism in host cells during at least one step of their life cycle. Hepatitis C virus, for instance, relies on lipids for entry into target cells (21), RNA replication (22), viral assembly (45), as well as infectivity (1, 37). Cholesterol-rich plasma membrane domains (lipid rafts) are important for HIV entry, assembly, and infectivity (2). In the case of hepatitis B virus (HBV), efficient infection of hepatocytes was shown to be dependent on the cholesterol content of the viral envelope (6, 37); more recently, a role for caveolin-1, a structural protein of lipid rafts, was suggested in HBV entry (30).
HBV is an enveloped member of the Hepadnaviridae family bearing an unusual feature among animal viruses, in that multiple types of virus-related particles are assembled in infected cells. The infectious virions, also called “Dane” particles, are sphere-shaped, 42-nm-diameter particles containing the nucleocapsid surrounded by an envelope composed of cellular lipids and three structural viral proteins. These are designated the large (L), middle (M), and small (S) proteins and derive from the same open reading frame, sharing a common S domain (43). In addition to mature virions, coreless, noninfectious lipoprotein particles occurring in two morphological forms were identified by electron microscopy in human serum (14, 41). These 22-nm-diameter spheres and filament structures, called subviral particles (SVPs), result from the self-assembly of the S protein and are secreted in enormous quantities (up to 106-fold excess over virions). It is estimated that 25% of their mass consists of host cell-derived lipids, of which cholesterol, both free and esterified, is a major component, accounting for approximately 30% of the lipid content (16). Although a role for cholesterol in HBV entry has been clearly shown (6), investigation of virion and SVP secretion from cells treated with cholesterol-lowering agents has led to controversial results. Mammalian cells acquire this lipid through two main pathways: by synthesis from acetyl coenzyme A (acetyl-CoA) via the mevalonate/isoprenoids pathway and by endocytosis of the low-density lipoprotein (LDL)-associated cholesterol from serum, following binding to the LDL receptor (9, 17). These pathways are tightly regulated by sterol regulatory element binding proteins (SREBPs), localized at the endoplasmic reticulum (ER) membrane. Inhibition of cholesterol synthesis in HBV-producing hepatoma cells using lovastatin (Lova), a competitive inhibitor of 3-hydroxy-3-methylglutary-CoA (HMG-CoA) reductase, resulted in impaired secretion of SVPs, while the release of virions was not affected (27). In contrast, an independent study demonstrated that a significant effect on virion but not SVP secretion was obtained following a 6-day treatment of HepAD38 cells with a different inhibitor, NB598 (6). This compound decreases cholesterol synthesis by inhibiting the squalene epoxidase, an enzyme involved in the postisoprenoid synthesis step (19). Isoprenoids are key molecules involved in multiple cellular processes and signaling pathways (18). Thus, the reported discrepancies in HBV and SVP secretion may be accounted for by the isoprenoid synthesis being either perturbed or preserved during treatment with inhibitors of the cholesterol pathway (6).
In this study, we investigated the impact of host cell cholesterol depletion on HBV replication, assembly, and secretion, using a fast and nontoxic approach to decrease the cholesterol levels by preventing its cellular uptake. We found that growing HBV-producing cells with lipoprotein-depleted serum (LPDS) led to a 40% reduction of the intracellular cholesterol level within 24 h of treatment, which was not achievable using the synthesis inhibitor Lova. This effect was dose dependent and similar to that obtained in the presence of chlorpromazine (Cpz), an inhibitor of the clathrin-mediated pathway used by the LDL receptor for internalization (17). Under these conditions, HBV secretion was significantly inhibited (by ∼80%), while SVP release was not affected. Secretion-incompetent viral DNA was found to accumulate in LPDS-treated cells in a time-dependent manner. This was accompanied by an increase of the amount of nonenveloped nucleocapsids, despite synthesis of the envelope proteins occurring normally. Further investigation of the mechanism underlying this effect concluded that cholesterol plays a complex role in maintaining the peculiar, dual topology of the L protein, which is essential for nucleocapsid envelopment and receptor recognition on target cells.
MATERIALS AND METHODS
Cell lines and small interfering RNA (siRNA).
Huh7 and HepG2 cells were grown in RPMI 1640 medium (Euroclone) containing 10% fetal bovine serum (FBS), 50 units/ml penicillin, 50 μg/ml streptomycin, and 2 mM Glutamax medium (Invitrogen). HepG2 2.2.15 cells stably transfected with two copies of the HBV genome were grown as described above, except that the RPMI medium was supplemented with 200 μg/ml of G418 (Gibco). In cholesterol depletion experiments, FBS was replaced by LPDS (Sigma-Aldrich).
The siRNA oligonucleotides designated to silence adipose differentiation-related protein (ADRP; adipophilin) expression were purchased from Sigma-Genosys (sequences 4219141 and 4219142).
Cell transfection.
Cells were seeded in 6-well plates at 60% confluence on the day before transfection. Huh7 cells were transfected with either ADRP-specific or scrambled siRNA oligonucleotides, using Lipofectamine 2000 (Invitrogen). ADRP silencing in HepG2 2.2.15 cells, which are more difficult to transfect, was performed using a reverse transfection protocol and a Lipofectamine siRNA Max kit. At 16 h posttransfection (pt), supernatants were discarded, cells were washed 3 times with phosphate-buffered saline (PBS), and fresh medium was added to the wells. Cell supernatants were harvested at 48 h pt for further analysis.
Chemical inhibition of lipid droplet (LD) biogenesis.
HepG2 2.2.15 cells were treated with 15 μM C75 (Cayman Chemical), a fatty acid synthase (FAS) inhibitor, which was added to RPMI complete medium supplemented with 5% FBS. Following 24 or 48 h of treatment, cell supernatants were harvested and secretion of viral and subviral particles was monitored by real-time PCR and enzyme-linked immunosorbent assay (ELISA).
Chemical inhibition of secretory pathway trafficking.
HepG2 2.2.15 cells were treated with each of the drugs bafilomycin A1 (BAF), brefeldin A (BFA), chloroquine (CQ), and wortmannin (WO) (Calbiochem) for 6 h at concentrations indicated in the figure legends. Viral and SVP secretion was determined by real-time PCR and ELISA.
Quantification of cholesterol levels.
HepG2 2.2.15 cells were seeded in collagen 12-well plates and grown in RPMI medium supplemented with 10% either FBS, LPDS, or a mixture of the two. Where indicated, cells were treated with 10 μM Lova or 10 μg/ml Cpz (Sigma-Aldrich); importantly, this concentration of Cpz is typically used to inhibit endocytosis, while higher concentrations of the drug are necessary to perturb lipid biosynthesis and fluidity (33). Following treatment for 24, 48, or 72 h, cells were washed with PBS and lysed in a buffer containing 10 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2 mM EDTA, 0.5% Triton X-100, and a mixture of protease inhibitors (Sigma-Aldrich). Cell lysates were clarified by centrifugation at 14,000 × g for 10 min, and the cholesterol content was determined using an Amplex Red assay kit (Invitrogen), according to the manufacturer's instructions. Spectrofluorimetric measurements were performed using a Jasco FP-6500 spectrofluorimeter (530-nm excitation/590-nm emission wavelengths), and the values obtained were normalized to the total protein content, as measured by the bicinchoninic acid (BCA) assay (Pierce).
Quantification of HBV secretion by real-time PCR.
HepG2 2.2.15 cells were grown in RPMI medium supplemented with 10% either FBS, 10% LPDS, or a mixture of the two. Where indicated, cells were transfected with ADRP-specific siRNA, as described above. When the effects of cholesterol uptake/synthesis were investigated, cells were treated with 10 μM Lova or 10 μg/ml Cpz for the times indicated. Encapsidated viral DNA was purified from supernatants by phenol-chloroform extraction, as described elsewhere (24). The DNA was quantified using a Corbett Rotor Gene 6000 real-time PCR system and Maxima SYBR green quantitative PCR (qPCR) master mix (Fermentas). Primers were designed to amplify a 279-bp HBV-specific fragment: HBV3575-3854_for (5′-TCCAGGATCCTCAACAACCAGCACG-3′) and HBV3575-3854_rev (5′-TGGCCCCCAATACCACATCATCC-3′). The number of viral genome equivalents was determined using a calibration curve containing known amounts of HBV DNA.
Quantification of HBV replication by Southern blotting.
HepG2 2.2.15 cells were maintained in RPMI complete medium supplemented with 10% either FBS or LPDS for 24, 48, and 72 h. Treatment with 10 μM lamivudine (3TC; Moravek Biochemicals) was also included as a control of inhibition of viral replication. The encapsidated viral DNA was purified from an equal number of cells by phenol-chloroform extraction, as for the real-time PCR. The resulting DNA pellet was resuspended in nuclease-free water, analyzed in a 1.2% agarose gel, and transferred to a Hybond-N+ membrane (GE Healthcare), using a vacuum transfer blotter (Bio-Rad). The blot was further hybridized with a fluorescein-labeled probe obtained by random priming using the HBV DNA genome as template. The HBV-specific DNA bands were detected using antifluorescein alkaline phosphatase (AP)-conjugated monoclonal antibodies (MAbs) and a Gene Images CDP-Star detection kit (GE Healthcare).
Quantification of HBV SVPs by ELISA.
The cells and supernatants used for the quantification of HBV replication and secretion were also analyzed for the amount of HBsAg, using a Monolisa HBsAg ultrakit (Bio-Rad). The results were obtained as ratios of signal to cutoff value and were converted to percentages of HBsAg secretion.
Quantification of intracellular HBV by immunoprecipitation and real-time PCR.
HepG2 2.2.15 cells were maintained in complete RPMI medium supplemented with 10% either FBS or LPDS for 24, 48, and 72 h. Cells were harvested and counted under a microscope, and an equal number was incubated in 5 mM HEPES buffer (pH 7.3) containing 200 mM NaCl and 10 mM CHAPS {3-[(3-chloramidopropyl)-dimethylammonio]-1-propanesulfonate} for 15 min on ice. CHAPS is a weak detergent that preserves the integrity of the virions under the mild treatment conditions used (data not shown). Lysates were cleared by centrifugation at 14,000 × g, divided into equal volumes, and further used for immunoprecipitation. A mixture of anti-core antibodies (Abs), mouse 10E11 and C1 (dilution, 1/1,000; GeneTex) and rabbit anti-HBV core antigen (dilution, 1/1,000; DakoCytomation), was used to pull down nonenveloped HBV nucleocapsids, while enveloped virions were immunoprecipitated with mouse anti-pre-S1 Abs (dilution, 1:500; Santa Cruz Biotechnology). The immune complexes were isolated by incubation of samples with either protein A- or protein G-Sepharose beads (Sigma-Aldrich) overnight at 4°C. In control samples, cell lysates were incubated with beads only. The slurry was washed five times with PBS, and the bound nucleocapsids were eluted by boiling the samples for 10 min in 50 mM Tris-HCl buffer (pH 8) supplemented with 1 mM EDTA and 1% NP-40 with strong shaking. The viral DNA was purified from the isolated nucleocapsids by phenol-chloroform extraction and quantified by real-time PCR. To determine that the assay was quantitative, the supernatants resulting from the first immunoprecipitation were reprecipitated using the same Abs and then analyzed as described above. Only background amounts of HBV DNA were recovered following the second immunoprecipitation, confirming the quantitative pull down.
Preparation of microsomes and trypsin protection assay.
Microsomes were prepared and subjected to the trypsin protection assay to establish the L protein topology, as previously described (39). Briefly, HepG2 2.2.15 cells seeded in 75-cm2 flasks were grown in RPMI medium supplemented with 10% FBS or LPDS. Following 48 h of treatment, cells were harvested and counted. An equal number of cells of each sample was washed with Tris-buffered saline (TBS) containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, followed by incubation with 0.1× TBS for 10 min on ice. Swollen cells were disrupted by Dounce homogenization (20 strokes). Unbroken cells and nuclei were removed by centrifugation for 20 min at 2,500 rpm. The supernatant was layered on 2.7 ml of TBS containing 250 mM sucrose and centrifuged for 30 min at 41,000 rpm in an SW41Ti rotor (Beckman). The microsome pellet was resuspended in TBS and equally divided into three aliquots. Enzymatic proteolysis was performed using 25 μg/ml of trypsin (Gibco), in the presence or absence of 0.5% NP-40, for 2 h on ice. One sample was maintained undigested as a control. Proteolysis was stopped by addition of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, followed by boiling for 10 min at 100°C. The proteins were resolved by SDS-PAGE and identified by Western blotting. The amounts of the L forms protected from proteolysis in intact microsomes were calculated by comparing their intensities with those in samples where no trypsin was added (considered 100%), using Quantity One software from Bio-Rad.
SDS-PAGE and Western blotting.
Cells were lysed as for cholesterol quantification or used for microsome purification as described above. Lysates were clarified by centrifugation for 10 min at 14,000 × g, and the protein content was determined in supernatants using the BCA method (Pierce). Equal amounts of proteins were separated by SDS-PAGE and analyzed by Western blotting following semidry transfer to nitrocellulose membranes. To investigate the efficiency of ADRP silencing, blots were incubated with mouse antiadipophilin Abs (dilution, 1:50; Progen), followed by sheep antimouse HRP Abs (dilution, 1:2,000; GE Healthcare). Trypsin digestion of the HBV envelope proteins was analyzed using either rabbit anti-HBsAg (dilution, 1:1,000; Europa Bioproducts) or mouse anti-pre-S1 Abs (Santa Cruz Biotechnology), followed by goat antirabbit (dilution, 1:2,000; Pierce) or sheep antimouse (dilution, 1:5,000; GE Healthcare) HRP Abs. Expression of the L protein in cell lysates was investigated under reducing conditions, using the mouse anti-pre-S1 Abs, as above. When calnexin or calreticulin was used as a loading control, blots were incubated with either rabbit anticalnexin (dilution, 1:4,000; Santa Cruz Biotechnology) or goat anticalreticulin (dilution, 1:500; Santa Cruz Biotechnology), followed by goat antirabbit (dilution, 1:2,000; Pierce) or donkey antigoat (dilution, 1:2,000; Santa Cruz Biotechnology) HRP Abs. Proteins were detected using an ECL (GE Healthcare) or SuperSignal West Femto maximum sensitivity substrate (Thermo Scientific) detection system according to the manufacturers' instructions.
Fluorescence microscopy.
HepG2 2.2.15 and Huh7 cells were plated on coverslips at 50% confluence and transfected as described above. At 48 h pt, cells were fixed with a mixture of 4% paraformaldehyde and 0.025% glutaraldehyde for 10 min and permeabilized with 0.1% Triton X-100 for 5 min. Cells were sequentially incubated for 30 min with mouse antiadipophilin (dilution, 1:20; Progen), followed by Alexa Fluor 594-labeled goat antimouse Ab (1:500; Molecular Probes). Cells were washed with PBS and mounted with Vectashield mounting medium (Vector Laboratories) containing 4′,6-diamidino-2-phenylindole to visualize the nuclei.
When lipid droplet distribution was investigated, fixed and permeabilized cells were treated with Bodipy 493/503 neutral lipid dye (Invitrogen) for 10 min and then washed with PBS and mounted as described above.
To visualize free cholesterol, cells were fixed with 3% paraformaldehyde for 1 h and incubated with 1.5 mg/ml glycine for 10 min. Following incubation with 0.1 mg/ml filipin (Sigma-Aldrich) for 2 h, cells were washed with PBS and mounted as described above. All samples were analyzed under a Nikon Eclipse E600 microscope, using a ×40 or a ×60 objective.
Cholesterol replenishment of depleted cells.
HepG2 2.2.15 cells were maintained in complete medium supplemented with either 10% FBS or LPDS for 24 h. In replenished samples, cholesterol (10 and 20 μg/ml) was added to LPDS as complexes with methyl β-cyclodextrin (MβCD). These were freshly obtained by mixing ethanol-dissolved cholesterol and MβCD (1:10 molar ratio), followed by 15 min strong vortexing at room temperature (12). Stock solutions of cholesterol-MβCD complexes were passed through a 0.45-μm-pore-size filter before use. The corresponding volume of solvent was added to the LPDS-treated sample as a control. Cells were thoroughly washed with PBS and detached with trypsin, and total cellular cholesterol and virus secretion was quantified by spectrofluorimetry and real time-PCR, respectively, as described above.
RESULTS
LPDS treatment results in rapid decrease of cholesterol levels in HepG2 2.2.15 cells.
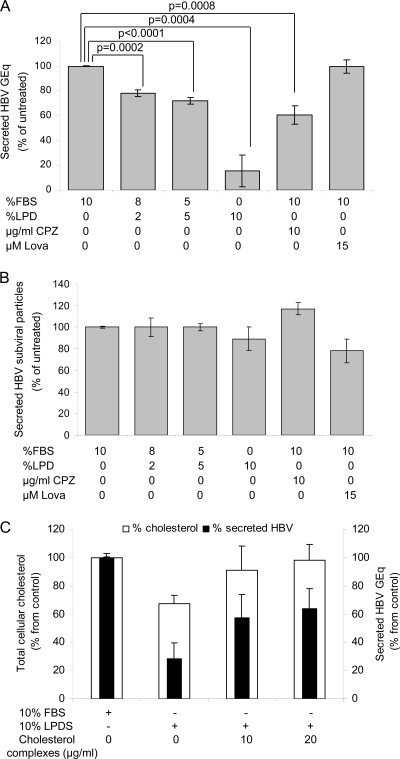
Chemical inhibition of intracellular cholesterol synthesis has led to conflicting results regarding HBV and SVP secretion, depending on the drug and the experimental setting used (6, 27). The duration of treatment required to achieve an effect is usually long, which may result in unwanted effects, such as cell toxicity or upregulation of LDL receptors at the cell surface, as reported for Lova (11). Here, an alternative way to decrease the cholesterol level was used to clarify its role on the late steps of the HBV life cycle, by preventing the uptake of LDL particles into HepG2 2.2.15 cells. To determine the efficiency of this approach, various concentrations of LPDS and FBS were added to the growth medium and cells were incubated for 24 h. As shown in Fig. 1 A, the total cellular cholesterol decreased by up to 40% in the presence of LPDS. The dose-response between the LPDS/FBS ratio and the total cellular cholesterol is not unusual, as the cholesterol level cannot decrease below a certain threshold without significant cellular toxicity. In this experimental setting, cholesterol synthesis is not inhibited and may be upregulated when the intracellular loss becomes significant. Importantly, however, our results show that the lack of exogenous cholesterol cannot be totally compensated for by intracellular synthesis. This inhibition was similar to that observed in another HBV-producing line following a 6-day incubation with a cholesterol synthesis inhibitor (6). Extending the duration of treatment to 48 and 72 h did not alter this level of inhibition (Fig. 1B). Interestingly, the cholesterol level was also reduced following treatment with the clathrin-mediated endocytosis inhibitor Cpz, while Lova had no impact on the cholesterol level within the 24-h time frame. The lack of cholesterol inhibition following Lova treatment is not surprising, as a longer treatment (usually more than 3 days) is required to reduce intracellular cholesterol.
Fig. 1.
LPDS treatment results in rapid decrease of cholesterol levels in HepG2 2.2.15 cells. Cells were grown for 24 h (A and C) or the times indicated (B) in complete medium supplemented with 10% FBS (considered untreated), 10% LPDS, or a mixture of the two. Where indicated, Lova or Cpz was added as a control. The level of total cellular cholesterol was measured by spectrofluorimetry, and results were normalized to total protein levels within the same samples. Data represent the means and standard deviations (SD) of triplicates from two independent experiments and are presented as percentages of the values for untreated samples. Statistical analysis showing P values was performed using Student's unpaired t test (A and B). Free cholesterol was visualized by fluorescence microscopy, following incubation with filipin, using a Nikon E600 microscope (C).
The lower cholesterol content within treated cell membranes was also confirmed by fluorescence microscopy, using filipin, an antibiotic which forms a fluorescent complex with free cholesterol (Fig. 1C). A cell proliferation [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT)] assay revealed no cellular toxicity following either of these treatments (data not shown).
Cholesterol depletion of HBV-producing cells results in reduced levels of secreted HBV virions and nucleocapsids but not SVPs.
The consequences of this fast cholesterol depletion on secretion of both viral and subviral particles were investigated by real-time PCR and ELISA. The quantitative PCR employed detects both enveloped virion and nucleocapsids; as shown in Fig. 2 A, their secretion was inhibited with increasing LPDS/FBS ratios, with the most significant effect measured (∼80% inhibition) when LPDS completely replaced FBS in the cell medium. Under the same treatment conditions, SVP secretion remained unchanged (Fig. 2B). Cpz also inhibited secretion of viral particles, although less efficiently than LPDS, but had no impact on SVP secretion (Fig. 2A and B). The short treatment with Lova resulted in no impact on secretion of either viral or subviral particles, which is consistent with the lack of cholesterol inhibition (Fig. 2A and B).
Fig. 2.
HBV secretion from LPDS-treated cells. HepG2 2.2.15 cells were grown for 24 h in complete medium supplemented with 10% FBS, LPDS, or a mixture of the two. Where indicated, Lova or Cpz was added as a control. (A) The amount of HBV DNA secreted by an equal number of cells was analyzed by real-time PCR. The results were expressed in genome equivalents as percentages of the values for untreated samples. Data represent the means and SDs of duplicates from three independent experiments. Statistical analysis showing P values was performed using Student's unpaired t test. GEq, genome equivalents. (B) Secretion of HBsAg was quantified by ELISA, and values were normalized to the total number of cells. Data represent the means and SDs of triplicates from three independent experiments and are presented as percentages of the values for untreated samples. The cutoff values varied from 0.082 to 0.096. (C) Cholesterol-MβCD complexes were added to 10% LPDS at the concentrations indicated. The levels of total cellular cholesterol and HBV secretion were measured as described in the legends to Fig. 1A and B and for panel A, respectively. Data represent the means and SDs of triplicates from two independent experiments and are presented as percentages of the values for untreated samples.
Growing the HepG2 2.2.15 cells in LPDS supplemented with soluble cholesterol had a minor effect on the total cellular cholesterol level, despite the high concentration used (up to 60 μg/ml; data not shown). However, when cholesterol was added to LPDS as complexes with MβCD, the total cellular amount was almost completely restored in 24 h (Fig. 2C), which is likely a consequence of a more efficient uptake. Under the same conditions, secretion of HBV particles was recovered up to 65% (Fig. 2C).
HBV assembly but not replication is impaired in cholesterol-depleted cells.
The reduced amount of viral particles secreted from cholesterol-depleted HepG2 2.2.15 cells could reflect an inhibition of replication and/or particle assembly or of a trafficking event at the level of the secretory pathway. To discriminate between these possibilities, HBV replication was analyzed by Southern blotting with a specific probe, following 24 h treatment with 10% LPDS. Controls included cells incubated in either complete medium or medium containing the replication inhibitor 3TC. Two major replication forms (RFs) were present in cells grown in either complete or delipidated medium, while their synthesis was significantly inhibited in 3TC-treated cells (Fig. 3 A, top). Interestingly, the intensity of both replication intermediates appeared to increase in LPDS-treated cells; therefore, to further investigate this effect, the treatment was extended to 48 and 72 h. Indeed, an important, time-dependent accumulation of both HBV replication intermediates was observed in treated cells compared to controls (Fig. 3A, top), which was further quantified (Fig. 3A, bottom). The amount of the envelope proteins was quantitatively determined in the same samples by ELISA, revealing a completely different behavior (Fig. 3B).
Fig. 3.
HBV replication and viral envelope protein biosynthesis in LPDS-treated cells. HepG2 2.2.15 cells were grown in complete medium supplemented with 10% FBS or LPDS for 24, 48, and 72 h. When indicated, 3TC treatment was included as a control of replication inhibition. (A) Equal numbers of cells were used for viral DNA purification and further analysis by Southern blotting. The representative autograph of two independent experiments is shown (top). Quantification of the HBV RFs was performed using Quantity One software (bottom). (B) Expression of HBsAg was quantified by ELISA, and values were normalized to the total protein content measured within the same samples. Data represent the means and SDs of triplicates from three independent experiments and are presented as percentages of the values for untreated samples. The cutoff values varied from 0.082 to 0.096. (C) Lysates of HepG2 2.2.15 cells, either grown in LPDS for the times indicated or maintained untreated, were subjected to SDS-PAGE under reducing conditions. Expression of the unglycosylated (p) and glycosylated (gp) L proteins and calnexin (loading control) was analyzed by Western blotting using specific Abs (top) and quantified using Quantity One software (bottom).
Biosynthesis of the L proteins was next analyzed by Western blotting of cell lysates under reducing conditions. The results show a similar expression of the L polypeptides in all samples (Fig. 3C, top), which was further quantified in Fig. 3C, bottom.
In infected cells, HBV DNA replication intermediates can originate from both fully enveloped virions and nucleocapsids. To determine which species accumulate in LPDS-treated cells, samples were split in two and specific immunoprecipitation was performed on each half, using either a mixture of anti-core Abs, to pull down nonenveloped HBV nucleocapsids, or the anti-pre-S1 Abs, to isolate the enveloped virions. The viral DNA was further purified from bound complexes and subjected to real-time PCR.
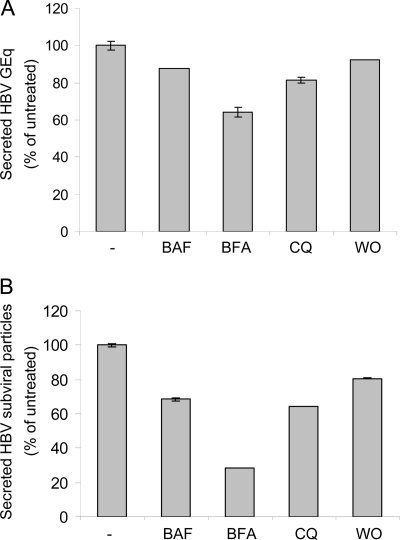
Interestingly, the ratio of nucleocapsids in LPDS-treated to control cells significantly increased, in a time-dependent manner, by 2.5-fold at 72 h of treatment (Fig. 4 A). In contrast, the ratio of virions in LPDS-treated to control cells increased only slightly at 24 h; however, the trend reversed at 48 and 72 h (Fig. 4B). Together, these results point at viral envelopment rather than secretion being perturbed in cholesterol-depleted cells. This conclusion is also supported by the lack of effect of cholesterol depletion on SVP secretion, provided the two types of particles are released from the cell through a common pathway, following assembly at distinct sites. To further investigate this, the effect of several trafficking inhibitors on both HBV and SVP secretion was analyzed. The early secretion steps were inhibited using BFA, a fungal metabolite, which blocks the anterograde transport from the ER to the Golgi apparatus (36). The Golgi apparatus events were interfered with by using the macrolide antibiotic BAF, an inhibitor of vacuolar proton ATPase activity along the Golgi apparatus cisternae (15), and WO, a potent inhibitor of the phosphatidylinositol 3-kinase activity involved in trafficking at the trans-Golgi apparatus level (32). The pH gradient in the Golgi apparatus was also disrupted by treatment with CQ, an acidotropic compound (40). As shown in Fig. 5 A and B, a similar pattern of inhibition of secretion was observed for both virions and SVPs, suggesting that a common export route is used as early as post-ER. The drugs appeared to be more potent in inhibiting SVPs during the short time frame (6 h) of treatment, which could be the consequence of virions and SVPs being trafficked at different rates through the secretory pathway.
Fig. 4.
Quantification of intracellular nucleocapsids and enveloped virions in LPDS-treated cells. HepG2 2.2.15 cells were grown in complete medium supplemented with 10% FBS or LPDS for the times indicated. Cell lysate levels were normalized to the total protein content, and equal volumes were subjected to immunoprecipitation with either anticapsid (A) or anti-pre-S1 (B) Abs. Viral DNA was purified from eluted particles and quantified by real-time PCR. A second immunoprecipitation performed in all samples, using the same Abs, recovered only background amounts of viral DNA. Data represent the means and SDs of duplicates from two independent experiments and are shown as percentages of the values for untreated samples.
Fig. 5.
Secretion of HBV particles in the presence of trafficking inhibitors. HepG2 2.2.15 cells were grown for 6 h in complete medium in the presence of BAF (100 nM), BFA (2.5 μM), CQ (50 μM), or WO (1 μM) or with no treatment. (A) The viral DNA secreted from an equal number of cells was purified and quantified by real-time PCR. The results were expressed in genome equivalents (GEq) as percentages of the values for untreated samples. Data represent the means and SDs of duplicates from two independent experiments. (B) The amount of HBsAg secreted from an equal number of cells was quantified by ELISA. Data represent the means and SDs of triplicates from two independent experiments and are presented as percentages of the values for untreated samples. The cutoff values varied from 0.082 to 0.096.
Impaired HBV assembly is not linked to inhibition of LD morphogenesis in cholesterol-depleted cells.
LDs are ER-derived organelles containing a core of neutral lipids surrounded by a monolayer of phospholipids and several proteins (48). They are found in different cell types, including hepatocytes, under normal conditions. Initially considered simple lipid storage organelles, LDs gained considerable attention when a link between their metabolism and important human diseases, such as obesity, cancer, and viral infections, was clearly established (5, 31, 38).
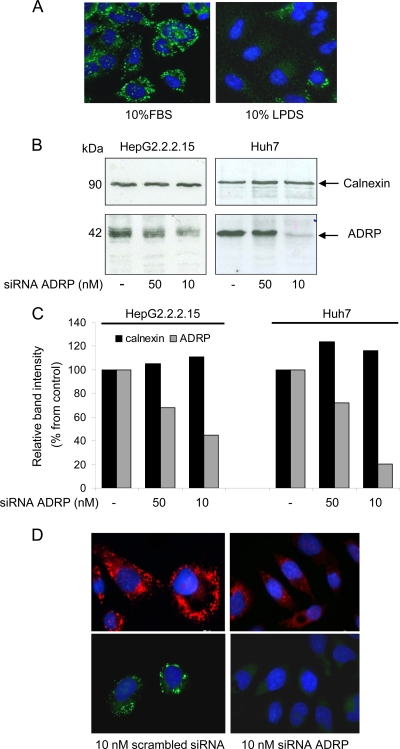
To investigate whether the short treatment of HepG2 2.2.15 cells with LPDS could significantly interfere with biogenesis of these organelles, cells were incubated with the neutral lipid dye Bodipy, and LD abundance and distribution were analyzed by fluorescence microscopy. Indeed, following 24 h of treatment, an important loss of LDs was observed in HepG2 2.2.15 cells compared to controls (Fig. 6 A). Intriguingly, fluorescence microscopy colocalization studies performed in our laboratory often revealed the presence of the S envelope protein in close vicinity of the LD marker ADRP (data not shown). The recent association of LDs with morphogenesis of several enveloped human viruses (34, 42), together with our observation regarding the strong effect of LPDS treatment on both LD accumulation and HBV assembly, prompted a more detailed analysis of a potential link between the two processes.
Fig. 6.
Inhibition of LDs biosynthesis in HBV-producing cells. (A) HepG2 2.2.15 cells were grown for 24 h in complete medium containing either FBS or LPDS. LDs were stained with Bodipy 493/503 and visualized using a Nikon E600 fluorescence microscope. (B) HepG2 2.2.15 and Huh7 cells were transfected with different concentrations of ADRP-specific siRNA or scrambled siRNA (lanes −). Cells were lysed at 48 h pt, and equal amounts of protein were loaded on SDS-polyacrylamide gels. ADRP and calnexin (protein loading control) expression was analyzed by Western blotting using specific Abs. (C) ADRP and calnexin levels were quantified using Quantity One software. (D) HepG2 2.2.15 cells were transfected with 10 nM either ADRP-specific or scrambled siRNA for 24 h. ADRP expression (top) and LD core (bottom) were evidenced by fluorescence microscopy, using ADRP-specific Abs or Bodipy 493/503 and a Nikon E600 fluorescence microscope.
In a first approach, expression of ADRP, which plays a key role in LD structural integrity, was silenced using two concentrations of specific siRNA. The efficiency of ADRP knockdown was monitored by Western blotting, using calnexin expression as a total protein gel-loading control. As shown in Fig. 6B (left), a significant, albeit not complete, inhibition of ADRP was observed in HepG2 2.2.15 cells transfected with 10 nM specific siRNA. Increasing the siRNA concentration to 50 nM resulted in a rather moderate inhibition of ADRP expression. To determine whether this unusual inhibition profile was artifactual, the silencing experiments were also performed in Huh7 cells. Again, transfecting cells with 50 nM siRNA had little effect on ADRP biosynthesis, while the 10 nM concentration resulted in a nearly total loss of expression (Fig. 6B, right). Unlike Huh7 cells, HepG2 2.215 cells are notoriously difficult to transfect, which may explain the lower efficiency of ADRP silencing in these cells, even when using the more potent, 10 nM siRNA concentration.
Consistent with the results obtained by Western blotting (quantified in Fig. 6C), silencing of ADRP expression in HepG2 2.2.15 cells was clearly observed by fluorescence microscopy using anti-ADRP Abs. Moreover, LD lipid core staining with Bodipy revealed a strong reduction of both number and size, suggesting that the level of ADRP inhibition was sufficient to affect LD morphogenesis (Fig. 6D).
In a second approach, HepG2 2.2.15 cells were treated with C75, a fatty acid synthase inhibitor previously reported to reduce the amount of LDs and inhibit preadipocyte differentiation (42). The highest C75 concentration that had no toxic effects in HepG2 2.2.15 cells was established following the MTT assay (data not shown) and used for further experiments.
HBV and SVP secretion was next investigated in C75-treated and ADRP-silenced HepG2 2.2.15 cells. As shown in Fig. 7, neither the drug inhibition of LD accumulation nor the ADRP knockdown had any relevant impact on viral or subviral particle secretion. This demonstrates that the HBV life cycle and LD assembly are not interconnected and the effects of LPDS treatment on HBV rely on a different inhibitory mechanism.
Fig. 7.
Effect of inhibition of LD biogenesis on secretion of HBV particles. HepG2 2.2.15 cells were treated with C75 for the times and at the concentrations indicated (A and B) or transfected with control and ADRP-specific siRNA (C and D), as described in the legend to Fig. 6. The amount of viral DNA secreted from an equal number of cells quantified by real-time PCR is expressed in genome equivalents (GEq) as a percentage of the values for control samples. Data represent the means and SDs of duplicates from two independent experiments (A and C). The amount of HBsAg secreted from an equal number of cells was determined by ELISA. Data represent the means and SDs of triplicates from two independent experiments and are presented as percentages of the values for untreated samples. The cutoff values varied from 0.082 to 0.096 (B and D).
Topology of the L envelope protein is altered in cholesterol-depleted cells.
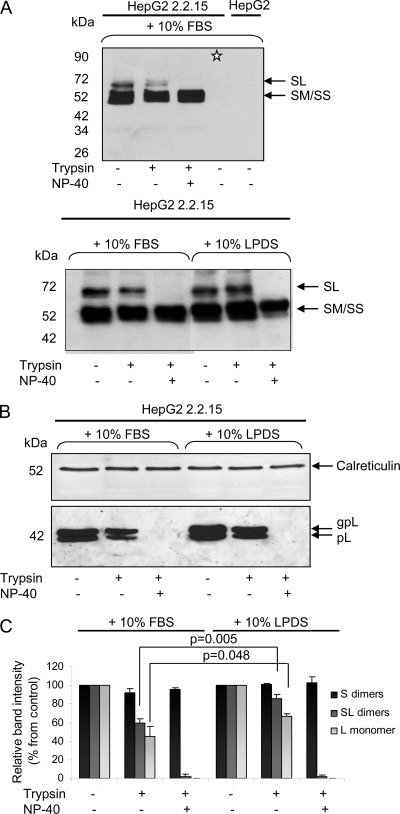
HBV envelopment is believed to occur at post-ER/pre-Golgi apparatus membranes, where the cytosolic nucleocapsids are packaged inside a lipid envelope containing the S, M, and L transmembrane proteins (20). A crucial role in this process is played by the L protein, which performs a matrix-like function at the virion budding site, in addition to mediating receptor binding during viral attachment to the target cell. The exceptional, multifunctional nature of L depends on its dual transmembrane topology at the ER membrane, established after polypeptide synthesis. This is the consequence of the partial translocation of the pre-S domain across membranes, with the mature L protein disposing this domain oriented to both the cytosolic and the luminal sides of the ER (23). Unlike mature virions, SVPs assemble and secrete normally in the absence of the L protein (7). Therefore, a potential modification of L conformation in cholesterol-depleted cells could account for the poor assembly of virions, while SVPs remain unaffected. To test this hypothesis, microsomes were purified from LPDS-treated and control cells, and L protein topology was investigated by determination of the susceptibility of the pre-S domain to trypsin digestion in the absence and the presence of a nondenaturing detergent (NP-40). While the pre-S domain of newly synthesized L polypeptide is almost fully cleaved by trypsin, it becomes increasingly protected following translocation within the ER lumen, yielding up to 50 to 80% resistant polypeptide chains at steady state (39).
The L protein was detected by Western blotting either as a dimer with S, using the anti-S Abs and nonreducing conditions (Fig. 8 A), or as the nonglycosylated (pL) and glycosylated (gpL) polypeptides, using the anti-pre-S1 Abs and reducing conditions (Fig. 8B). The anti-S Abs used in the experiment depicted in Fig. 8A (top) recognize the S-containing homo- and heterodimers expressed in HepG2 2.2.15 cells but not the fully reduced corresponding monomers (note the lane marked with an asterisk). Detection of the ER-resident protein calreticulin was used as a total protein gel-loading control, as it was previously reported to resist trypsin digestion when fully folded and loaded with calcium under mild reaction conditions, such as those used in this experiment (13). As shown in Fig. 8A (bottom), the loss of the pre-S domain of the L protein in the presence of trypsin resulted in a reduced intensity of the SL band in intact microsomes and its complete disappearance in detergent-treated samples. Interestingly, this reduction was significantly attenuated in LPDS-treated cells compared to the control. A similar effect was observed when the L polypeptides were independently analyzed under reducing conditions, using the anti-pre-S1 Abs (Fig. 8B). Despite equal protein loading, a slight increase of the total amount of L proteins was observed in LPDS-treated cells, which was confirmed in repeated experiments. This may reflect an accumulation of the protein in microsomes, when normal viral envelopment and subsequent secretion are perturbed.
Fig. 8.
Evaluation of the L protein topology in purified microsomes. Cells were grown in complete medium supplemented with 10% FBS or LPDS for 48 h. Following treatment, microsomes were purified from an equal number of cells, split into three aliquots, and treated or not with trypsin and NP-40, as indicated. Proteins were subjected to SDS-PAGE under nonreducing (A) or reducing (B) conditions, followed by Western blotting. The sample marked with an asterisk was denatured in the presence of 20 mM dithiothreitol before loading (A, top). Calreticulin expression was used as total protein, gel loading control (B). The amounts of S and L forms protected from proteolysis in intact microsomes were calculated by comparing their intensities with those in samples where no trypsin was added (considered 100%), using Quantity One software. Statistical analysis showing P values was performed using Student's unpaired t test (C).
The band corresponding to the SM/SS heterodimers was not affected by trypsin digestion, regardless of whether the microsomes were intact or disrupted by NP-40 treatment (Fig. 8A, bottom). This is consistent with the pre-S2 domain of the M protein being fully translocated and the unusual resistance of the S domain to trypsin hydrolysis, despite the existence of specific cleavage sites, as previously demonstrated (39).
The amounts of the L and S forms protected from proteolysis in intact microsomes were determined using Quantity One software. The results revealed that about 50 to 60% L molecules (detected either as heterodimers with S or as reduced polypeptides) were protected in control cells, as expected. In contrast, LPDS treatment yielded 75 and 90% trypsin resistance of L polypeptides and SL dimers, respectively (Fig. 8C). These results show that altering the intracellular cholesterol level enhances translocation of the pre-S domain across the ER membrane.
DISCUSSION
Despite the wide recognition that cholesterol is an important component of the membrane of enveloped viruses, little is known about how cholesterol is involved in viral morphogenesis. Most of our knowledge about the role of cholesterol in viral assembly is limited to the function of lipid rafts, the sphingolipid- and cholesterol-enriched microdomains of the plasma membrane. Several enveloped viruses use raft-like domains as platforms for virion assembly, e.g., HIV (10), Ebola and Marburg viruses (4), measles virus (49), and influenza virus (25, 47). More recently, LDs, the storage organelles for triglycerides and cholesterol esters, were shown to be involved in assembly of viruses in the Flaviviridae family (42, 45).
The role of cholesterol in HBV replication as well as virion and SVP assembly was investigated in this work, following removal of lipoproteins from the cell medium or chemical inhibition of LDL endocytosis. This strategy was based on the hypothesis that exogenous cholesterol, which is normally released from lipoprotein cholesterol esters in late endosomes/lysosomes and LDs, would not be available to the cells. Indeed, a 40% reduction of the cholesterol level was observed in HepG2 2.2.15 cells after only 24 h of treatment, demonstrating that this pool of cholesterol significantly contributes to the total intracellular content. The results also show that cholesterol synthesis either was not significantly upregulated in these cells or could not compensate for the initial loss. As a consequence, secretion of viral particles was inhibited by ∼80%, while SVPs were not affected. Addition of cholesterol-MβCD complexes to LPDS-treated cells recovered about half of this inhibition, despite the total cholesterol amount being almost totally restored. This interesting effect may be explained by a different uptake, trafficking, and further distribution efficiency to intracellular membranes, including the site of HBV assembly of cholesterol complexes, as opposed to the LDL-linked cholesterol. This result points at cholesterol being a central factor of this inhibition; however, the incomplete restoration of the viral secretion suggests that other, yet unidentified factors may also contribute to the overall consequences observed on HBV.
The decreased viral load in cell supernatants was not caused by a perturbation of HBV replication, as the Southern blotting assay revealed the same DNA intermediates in nucleocapsids purified from both LPDS-treated and control cells. Instead, viral DNA accumulated within treated cells in a time-dependent manner. A distinction between the HBV forms at different stages of maturation was made by specific immunoprecipitation with either capsid- or L protein-specific Abs. While the ratio of nucleocapsids in LPDS-treated to control cells increased with prolonged treatment, an opposite effect on mature virions was observed, which is an indication that fewer nucleocapsids acquired their envelope. This accumulation is rather intriguing, since hepatoma cell lines are able to release naked core particles, in addition to mature virions (44). The process was not observed in HBV-infected patients; therefore, it was assumed to be solely a result of cell lysis in vitro and attracted little further interest; however, a specific secretion pathway has not been completely disregarded. A study performed on cells infected with the avian homologue, the duck HBV (DHBV), showed that the pool of the envelopment-competent nucleocapsids containing mature viral DNA genome tightly associates with the ER membrane, possibly at the budding site, while immature nucleocapsids are excluded (29). In this model, naked core particles, found in the cell medium, display the same characteristics as the intracellular, mature nucleocapsids. Therefore, it was suggested that their secretion is mediated by direct membrane binding and interaction with nonviral components, such as phospholipids and membrane proteins, and could result from nucleocapsid overproduction in transfected cells. Since assembly and secretion of SVPs occur normally, while naked nucleocapsids accumulate in delipidated cells, despite containing mature DNA, our results are in favor of such a model. Thus, it is conceivable that the abrupt decrease of the intracellular cholesterol content may influence the membrane stability and the anchorage of nucleocapsids at the ER membrane, while SVPs do not depend on such a mechanism. These results reinforce the notion that SVPs and virions assemble in different compartments of the ER, which may have distinct requirements for membrane lipid composition. Both particles are further released into the common secretory pathway, as demonstrated by the similar inhibition of secretion, using a series of trafficking inhibitors. Although enriched in cholesterol and functionally depending on it, caveolin-1-positive lipid rafts could not be linked to either of these processes. As shown in a previously published report, virions and SVPs were normally secreted from cells with altered expression of this protein (30).
The mechanism by which nucleocapsids receive their envelope is not completely resolved; however, a role for the L protein and its dual conformation has been clearly documented (8, 26).
Virion formation depends on the interaction of nucleocapsids with the cytoplasmically exposed pre-S region of the L protein. Here, we used the trypsin protection assay at steady state as a convenient marker for the partial translocation of the pre-S domain of the L protein into the ER lumen. The results showed an increased protection of the pre-S domain to trypsin digestion in intact microsomes, which is an indication that a lower number of L polypeptides are in a pre-S cytosol-oriented conformation and, thus, are less available for nucleocapsid recruitment at the budding site. It is known that cholesterol insertion into the lipid bilayers of cellular membranes reduces the free space available for molecular movement, rendering these structures more rigid (46). This change of properties has been shown to influence the function of many membrane proteins (3, 50). Cholesterol levels vary significantly between different cellular membranes of the eukaryotic cell, with its lowest level being in the ER and with the level increasing throughout the secretory pathway (28). Thus, it is conceivable that ER-resident events involving transmembrane proteins, such as viral assembly, might be particularly sensitive to very small deviations of the cholesterol level from a critical threshold. Interestingly, in a previous study, cholesterol was found to inhibit an early step in Sec61-dependent protein translocation across the microsomal membrane, when supplemented at a concentration similar to that in the Golgi apparatus but significantly lower than that in the plasma membrane (35). In a mirror scenario, cholesterol depletion of the ER membrane could lead to an opposite effect, that is, a decrease in membrane rigidity and a more efficient translocation of the pre-S domain of the L protein. It remains to be established whether other molecules, such as Hsc70 and BiP, the two chaperones involved in guiding the pre-S translocation at the ER membrane, play a role in this effect (39). Their synthesis was not altered in delipidated cells (data not shown); however, a direct interaction of the L protein with the endogenously expressed proteins could not be investigated in this system, because of their lower expression level in these cells.
Nevertheless, our results suggest that interfering with cholesterol uptake and intracellular trafficking or decreasing cellular cholesterol levels by other means (37) may represent efficient alternatives to inhibiting enzymes of the synthesis pathway for the development of new antiviral strategies against enveloped viruses.
ACKNOWLEDGMENTS
This work was supported by the IDEI_84 grant awarded by the National Council of Scientific Research in Higher Education (CNCSIS) to Norica Branza-Nichita and the Romanian Academy Project 3 of the Institute of Biochemistry. Alina Macovei and Catalin Lazar were supported by grant POSDRU/89/1.5/S/60746.
Footnotes
Published ahead of print on 12 October 2011.
REFERENCES
- 1. Aizaki H., et al. 2008. Critical role of virion-associated cholesterol and sphingolipid in hepatitis C virus infection. J. Virol. 82:5715–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aloia R. C., Tian H., Jensen F. C. 1993. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc. Natl. Acad. Sci. U. S. A. 90:5181–5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bastiaanse E. M., Hold K. M., Van der Laarse A. 1997. The effect of membrane cholesterol content on ion transport processes in plasma membranes. Cardiovasc. Res. 33:272–283 [DOI] [PubMed] [Google Scholar]
- 4. Bavari S., et al. 2002. Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J. Exp. Med. 195:593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bozza P. T., Magalhaes K. G., Weller P. F. 2009. Leukocyte lipid bodies—biogenesis and functions in inflammation. Biochim. Biophys. Acta 1791:540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bremer C. M., Bung C., Kott N., Hardt M., Glebe D. 2009. Hepatitis B virus infection is dependent on cholesterol in the viral envelope. Cell. Microbiol. 11:249–260 [DOI] [PubMed] [Google Scholar]
- 7. Bruss V. 2007. Hepatitis B virus morphogenesis. World J. Gastroenterol. 13:65–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bruss V., Vieluf K. 1995. Functions of the internal pre-S domain of the large surface protein in hepatitis B virus particle morphogenesis. J. Virol. 69:6652–6657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Buhaescu I., Izzedine H. 2007. Mevalonate pathway: a review of clinical and therapeutical implications. Clin. Biochem. 40:575–584 [DOI] [PubMed] [Google Scholar]
- 10. Campbell S. M., Crowe S. M., Mak J. 2001. Lipid rafts and HIV-1: from viral entry to assembly of progeny virions. J. Clin. Virol. 22:217–227 [DOI] [PubMed] [Google Scholar]
- 11. Chappell D. A., Fry G. L., Waknitz M. A., Berns J. J. 1991. Ligand size as a determinant for catabolism by the low density lipoprotein (LDL) receptor pathway. A lattice model for LDL binding. J. Biol. Chem. 266:19296–19302 [PubMed] [Google Scholar]
- 12. Chattopadhyay A., Jafurulla M., Kalipatnapu S., Pucadyil T. J., Harikumar K. G. 2005. Role of cholesterol in ligand binding and G-protein coupling of serotonin1A receptors solubilized from bovine hippocampus. Biochem. Biophys. Res. Commun. 327:1036–1041 [DOI] [PubMed] [Google Scholar]
- 13. Corbett E. F., et al. 2000. The conformation of calreticulin is influenced by the endoplasmic reticulum luminal environment. J. Biol. Chem. 275:27177–27185 [DOI] [PubMed] [Google Scholar]
- 14. Dane D. S., Cameron C. H., Briggs M. 1970. Virus-like particles in serum of patients with Australia-antigen-associated hepatitis. Lancet i:695–698 [DOI] [PubMed] [Google Scholar]
- 15. Dröse S., Altendorf K. 1997. Bafilomycins and concanamycins as inhibitors of V-ATPases and P-ATPases. J. Exp. Biol. 200:1–8 [DOI] [PubMed] [Google Scholar]
- 16. Gavilanes F., Gonzalez-Ros J. M., Peterson D. L. 1982. Structure of hepatitis B surface antigen. Characterization of the lipid components and their association with the viral proteins. J. Biol. Chem. 257:7770–7777 [PubMed] [Google Scholar]
- 17. Goldstein J. L., Brown M. S., Anderson R. G., Russell D. W., Schneider W. J. 1985. Receptor-mediated endocytosis: concepts emerging from the LDL receptor system. Annu. Rev. Cell Biol. 1:1–39 [DOI] [PubMed] [Google Scholar]
- 18. Holstein S. A., Hohl R. J. 2004. Isoprenoids: remarkable diversity of form and function. Lipids 39:293–309 [DOI] [PubMed] [Google Scholar]
- 19. Horie M., et al. 1990. NB-598: a potent competitive inhibitor of squalene epoxidase. J. Biol. Chem. 265:18075–18078 [PubMed] [Google Scholar]
- 20. Huovila A. P., Eder A. M., Fuller S. D. 1992. Hepatitis B surface antigen assembles in a post-ER, pre-Golgi compartment. J. Cell Biol. 118:1305–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kapadia S. B., Barth H., Baumert T., McKeating J. A., Chisari F. V. 2007. Initiation of hepatitis C virus infection is dependent on cholesterol and cooperativity between CD81 and scavenger receptor B type I. J. Virol. 81:374–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kapadia S. B., Chisari F. V. 2005. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc. Natl. Acad. Sci. U. S. A. 102:2561–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lambert C., Prange R. 2001. Dual topology of the hepatitis B virus large envelope protein: determinants influencing post-translational pre-S translocation. J. Biol. Chem. 276:22265–22272 [DOI] [PubMed] [Google Scholar]
- 24. Lazar C., et al. 2007. Treatment of hepatitis B virus-infected cells with alpha-glucosidase inhibitors results in production of virions with altered molecular composition and infectivity. Antiviral Res. 76:30–37 [DOI] [PubMed] [Google Scholar]
- 25. Leser G. P., Lamb R. A. 2005. Influenza virus assembly and budding in raft-derived microdomains: a quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology 342:215–227 [DOI] [PubMed] [Google Scholar]
- 26. Le Seyec J., Chouteau P., Cannie I., Guguen-Guillouzo C., Gripon P. 1999. Infection process of the hepatitis B virus depends on the presence of a defined sequence in the pre-S1 domain. J. Virol. 73:2052–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin Y. L., Shiao M. S., Mettling C., Chou C. K. 2003. Cholesterol requirement of hepatitis B surface antigen (HBsAg) secretion. Virology 314:253–260 [DOI] [PubMed] [Google Scholar]
- 28. Liscum L., Munn N. J. 1999. Intracellular cholesterol transport. Biochim. Biophys. Acta 1438:19–37 [DOI] [PubMed] [Google Scholar]
- 29. Mabit H., Schaller H. 2000. Intracellular hepadnavirus nucleocapsids are selected for secretion by envelope protein-independent membrane binding. J. Virol. 74:11472–11478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Macovei A. Hepatitis B virus requires intact caveolin-1 function for productive infection in HepaRG cells. J. Virol. 84:243–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Martin S., Parton R. G. 2006. Lipid droplets: a unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 7:373–378 [DOI] [PubMed] [Google Scholar]
- 32. Martys J. L., et al. 1996. Wortmannin-sensitive trafficking pathways in Chinese hamster ovary cells. Differential effects on endocytosis and lysosomal sorting. J. Biol. Chem. 271:10953–10962 [DOI] [PubMed] [Google Scholar]
- 33. Maruoka N., et al. 2007. Effects of chlorpromazine on plasma membrane permeability and fluidity in the rat brain: a dynamic positron autoradiography and fluorescence polarization study. Prog. Neuropsychopharmacol. Biol. Psychiatry 31:178–186 [DOI] [PubMed] [Google Scholar]
- 34. Miyanari Y., et al. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089–1097 [DOI] [PubMed] [Google Scholar]
- 35. Nilsson I., Ohvo-Rekila H., Slotte J. P., Johnson A. E., von Heijne G. 2001. Inhibition of protein translocation across the endoplasmic reticulum membrane by sterols. J. Biol. Chem. 276:41748–41754 [DOI] [PubMed] [Google Scholar]
- 36. Pelham H. R. 1991. Multiple targets for brefeldin A. Cell 67:449–451 [DOI] [PubMed] [Google Scholar]
- 37. Pollock S., et al. 2010. Polyunsaturated liposomes are antiviral against hepatitis B and C viruses and HIV by decreasing cholesterol levels in infected cells. Proc. Natl. Acad. Sci. U. S. A. 107:17176–17181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Popescu C. I., Dubuisson J. Role of lipid metabolism in hepatitis C virus assembly and entry. Biol. Cell 102:63–74 [DOI] [PubMed] [Google Scholar]
- 39. Prange R., Streeck R. E. 1995. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 14:247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reaves B., Banting G. 1994. Vacuolar ATPase inactivation blocks recycling to the trans-Golgi network from the plasma membrane. FEBS Lett. 345:61–66 [DOI] [PubMed] [Google Scholar]
- 41. Robinson W. S., Lutwick L. I. 1976. The virus of hepatitis, type B (first of two parts). N. Engl. J. Med. 295:1168–1175 [DOI] [PubMed] [Google Scholar]
- 42. Samsa M. M., et al. 2009. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 5:e1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schulze A., Schieck A., Ni Y., Mier W., Urban S. 2010. Fine mapping of pre-S sequence requirements for hepatitis B virus large envelope protein-mediated receptor interaction. J. Virol. 84:1989–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sells M. A., Chen M. L., Acs G. 1987. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc. Natl. Acad. Sci. U. S. A. 84:1005–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shavinskaya A., Boulant S., Penin F., McLauchlan J., Bartenschlager R. 2007. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J. Biol. Chem. 282:37158–37169 [DOI] [PubMed] [Google Scholar]
- 46. Straume M., Litman B. J. 1987. Influence of cholesterol on equilibrium and dynamic bilayer structure of unsaturated acyl chain phosphatidylcholine vesicles as determined from higher order analysis of fluorescence anisotropy decay. Biochemistry 26:5121–5126 [DOI] [PubMed] [Google Scholar]
- 47. Takeda M., Leser G. P., Russell C. J., Lamb R. A. 2003. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc. Natl. Acad. Sci. U. S. A. 100:14610–14617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Thiele C., Spandl J. 2008. Cell biology of lipid droplets. Curr. Opin. Cell Biol. 20:378–385 [DOI] [PubMed] [Google Scholar]
- 49. Vincent S., Gerlier D., Manie S. N. 2000. Measles virus assembly within membrane rafts. J. Virol. 74:9911–9915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yeagle P. L. 1991. Modulation of membrane function by cholesterol. Biochimie 73:1303–1310 [DOI] [PubMed] [Google Scholar]