Abstract
The vaccinia virus (VACV) E3 protein is essential for virulence and has antiapoptotic activity and the ability to impair the host innate immune response. Here we demonstrate that E3 interacts with SUMO1 through a small ubiquitin-like modifier (SUMO)-interacting motif (SIM). SIM integrity is required for maintaining the stability of the viral protein and for the covalent conjugation of E3 to SUMO1 or SUMO2, a modification that has a negative effect on the E3 transcriptional transactivation of the p53-upregulated modulator of apoptosis (PUMA) and APAF-1 genes. We also demonstrate that E3 is ubiquitinated, a modification that does not destabilize the wild-type protein but triggers the degradation of an E3-ΔSIM mutant. This report constitutes the first demonstration of the important roles that both SUMO and ubiquitin play in the regulation of the VACV protein E3.
INTRODUCTION
The vaccinia virus (VACV) E3L gene encodes two proteins of about 20 and 25 kDa (referred to as E3) that are expressed early in infection (71) and are present in both the nucleus and cytoplasm of infected and transfected cells. E3 is a double-stranded RNA (dsRNA)-binding protein that inhibits the activation of interferon (IFN)-induced double-stranded RNA-dependent protein kinase (PKR) (14) and acts as an inhibitor of the IFN-induced 2-5A-synthetase enzyme (52). The E3L gene has been described as being necessary for the VACV IFN-resistant phenotype in cultured cells (3, 14), host resistance to pathogens in transgenic mice (17), and the inhibition of apoptosis (39). E3L is also a host range gene necessary for efficient VACV replication in several cell lines (2). In addition to these functions, E3 acts as a transcriptional regulator of several genes related to apoptosis, the immune response, and viral pathogenesis (9, 34, 36). Posttranslational modifications are common mechanisms for the regulation of multifunctional proteins. Thus far, no posttranslational modifications of E3 have been described.
One type of virus-host interaction that is well established and widespread is the modulation of viral protein function by posttranslational modification systems, which include SUMOylation and ubiquitination. Viral proteins were among the first substrates found to be posttranslationally modified by the small ubiquitin-like modifier (SUMO) protein, and SUMOylation seems to facilitate viral infections of the host cells (8). SUMO is a member of the larger family of ubiquitin-like proteins, which shares about 18% sequence identity to ubiquitin and is structurally quite similar (31). Posttranslational modification with ubiquitin and ubiquitin-like proteins of the SUMO family involves isopeptide bond formation between the carboxyl group of the modifier and the epsilon-amino group of a lysine residue in the target. Like ubiquitin, SUMO is covalently attached to lysine residues within the target protein, although in the majority of cases, SUMO is attached to a lysine within the ψKxE consensus site (where ψ is hydrophobic and x is any residue) (5, 55). In mammals, there are four SUMO isoforms, including SUMO1, which most closely resembles the single yeast Smt3. SUMO2 and SUMO3, which are very similar to each other, contain an internal SUMOylation consensus site and more readily form poly-SUMO chains (44, 61), and SUMO4 has been linked to diabetes (26). SUMOylation regulates a wide range of processes, including transcriptional activity, protein stability, and nucleocytoplasmic transport. In addition, SUMOylation often promotes interactions between modified proteins and downstream factors containing SUMO-interacting motifs (SIMs) (22). To date, a single conserved SIM has been identified, which consists of a hydrophobic core (L/V/I)x(L/V/I)(L/V/I). This SIM has been detected in several cellular and viral proteins known to be modified by SUMO, and it was previously shown to be important for the formation of SUMO-dependent protein networks (41, 58).
There are viruses belonging to several different families that also utilize or modulate the ubiquitin-proteasome system to their advantage. Proteins can be monoubiquitinated, or the initial ubiquitin monomer may itself act as a target, generating polyubiquitin chains. Distinct polyubiquitin signals that act in different cellular processes can be created by a variation in the choice of lysine linkage between ubiquitin monomers or in the length of the ubiquitin chain used.
The present investigation demonstrates that VACV protein E3 interacts with SUMO1 and can be covalently modified by either SUMO1 or SUMO2. The SUMO modification takes place on the lysine residues at positions 40 and 99 and negatively regulates E3 transcriptional transactivation on the p53-upregulated modulator of apoptosis (PUMA) and APAF-1 genes. We demonstrate that the presence of an intact SIM in E3 is required for its own SUMOylation and for the stability of the viral protein. We also demonstrate the conjugation of E3 to ubiquitin, a modification that does not induce the degradation of wild-type E3 (WT-E3) but triggers the proteasome-mediated degradation of the E3-ΔSIM mutant. E3 is the first example of a poxvirus protein regulated by covalent modification by ubiquitin, by the SUMO1 and SUMO2 proteins, as well as by noncovalent SUMO interactions. These results provide evidence that ubiquitin and ubiquitin-like proteins are important for the full biological activity of E3.
MATERIALS AND METHODS
Cell lines, transfections, and virus.
MCF-7, HeLa, HEK-293, BSC40, and BHK-21 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (Gibco), 5 mmol/liter l-glutamine (Invitrogen), and penicillin-streptomycin (Invitrogen). The transfection of MCF-7 and HEK-293 cells was done by use of FuGene (Roche), and HeLa cells were transfected with Lipofectamine according to the manufacturer's instructions. The vaccinia virus E3L deletion mutant (VACV-ΔE3L) was previously described (3) and was kindly provided by Enzo Paoletti. This virus was grown and titrated using BHK-21 cells.
Plasmids and reagents.
Plasmid pCINEO-E3 was previously described (20). All site-directed mutagenesis procedures were carried out by use of the QuikChange PCR-based mutagenesis kit (Stratagene, La Jolla, CA) and plasmid pCINEO-E3 as a template. Oligonucleotides used for site-directed mutagenesis are listed in the supplemental material. pcDNA-His6-SUMO1, pcDNA-His6-SUMO2, pcDNA-Ubc9, pcDNA-His-ubiquitin, and glutathione S-transferase (GST)-ISG15 plasmids were previously described (16, 63, 67, 68).
In vitro SUMO conjugation assay.
In vitro SUMO conjugation assays were performed on [35S]methionine-labeled in vitro-transcribed/translated E3 proteins as described previously (10), using recombinant E1 (Biomol), Ubc9, and SUMO1 or SUMO2. The in vitro transcription/translation of proteins was performed by using 1 μg of plasmid DNA and a rabbit reticulocyte-coupled transcription/translation system according to the instructions provided by the manufacturer (Promega).
Western blot analysis and antibodies.
For Western blot analysis, cells were washed in phosphate-buffered saline (PBS), scraped into SDS-gel loading buffer, and boiled for 5 min. Proteins of total extracts were separated by 12% SDS-PAGE and transferred onto a nitrocellulose membrane. Mouse anti-E3 antibody was kindly provided by Bernie Moss (NIH). Rabbit anti-E3 antibody against a full-length protein expressed in Escherichia coli was produced by Biomedal (Spain). Mouse anti-SUMO1 antibody was obtained from Invitrogen. Rabbit anti-SUMO2 antibody was obtained from Zymed Laboratories. Both secondary Alexa 488-conjugated anti-mouse antibody and Alexa 594-conjugated anti-rabbit antibody were obtained from Invitrogen. Rabbit anti-vaccinia virus antibodies were generated against purified inactivated virus of the WR strain.
Immunofluorescence staining.
Immunofluorescence staining and confocal analysis were performed as described previously (10). Colocalization analysis was done by use of Laser Pix (Image Pro) software. Images were exported by use of Adobe Photoshop.
Purification of His-tagged conjugates.
The purification of His-tagged conjugates using Ni2+-nitrilotriacetic acid (NTA)-agarose beads was performed as described previously (43).
GST pulldown.
GST pulldown experiments were performed by using protein extracts obtained from cells transfected with WT-E3 or E3-ΔSIM expression plasmids and treated with the proteasome inhibitor MG132 and GST or GST-SUMO1 as described previously (43).
dsRNA-agarose-binding assay.
Poly(I:C)-agarose beads were prepared from poly(I)-coated beads (Sigma) by incubation with 2 volumes of 2 mg of poly(C) (Sigma) per ml. The agarose beads were then mixed with the in vitro-translated, radiolabeled protein. After incubation for 1 h at 4°C, the beads were collected by centrifugation, washed four times with lysis buffer, resuspended in SDS sample buffer, and then analyzed by electrophoresis (SDS-PAGE) and autoradiography.
Reporter gene assays.
MCF-7 cells were transfected with the indicated plasmids in 24-well plates. The total DNA was kept equal for all transfections by using the empty pCINEO vector, and the pRL-TK Renilla luciferase (luc) control vector (Promega) was cotransfected to determine transfection efficiencies. Cells were harvested 48 h after transfection, and luciferase activity was measured by using the Dual Luciferase reporter assay (Promega) according to the manufacturer's instructions. Triplicate measurements were done for all experiments, and each experiment was repeated at least three times.
RESULTS
E3 protein colocalizes with SUMO1 in VACV-infected cells.
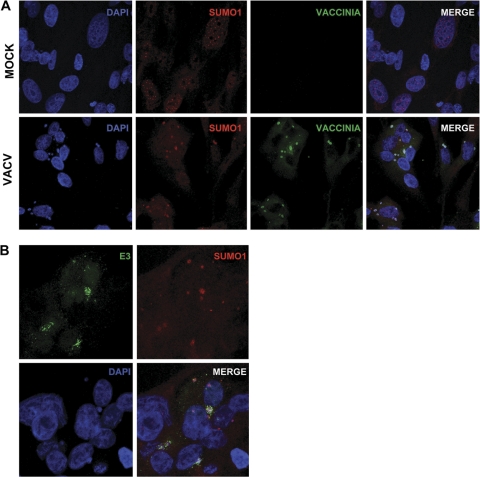
SUMO1 was reported previously to translocate to the viral factory in response to VACV infection (49). In addition, SUMO1 was previously isolated as an E3-interacting protein in a yeast two-hybrid assay (53). Based on these observations, we decided to study the putative colocalization between SUMO1 and E3 in the viral factories. First, MCF-7 cells were infected with VACV, and at 4 h after infection, cells were immunostained by using anti-vaccinia virus and anti-SUMO1 antibodies. SUMO1 was localized mainly in a nuclear speckled pattern in mock-infected cells, as expected (Fig. 1A). In contrast, in those cells infected with VACV, SUMO1 was recruited to the viral factories (Fig. 1A). When infected cells, as described above, were stained with anti-E3 and anti-SUMO1 antibodies, confocal analysis demonstrated a clear colocalization of both the E3 and SUMO1 proteins in the viral factories (Fig. 1B).
Fig. 1.
VACV E3 colocalizes with SUMO1 in viral factories. (A) SUMO1 is recruited to viral DNA replication sites. Uninfected cells or MCF-7 cells infected with VACV for 6 h were fixed and stained with DAPI (4′,6-diamidino-2-phenylindole) and anti-SUMO1 and anti-vaccinia virus antibodies. (B) SUMO1 colocalizes with E3 in viral factories. MCF-7 cells infected with VACV for 6 h were fixed and stained with DAPI and anti-SUMO1 and anti-E3 antibodies. The images were analyzed by confocal microscopy.
E3 protein is covalently modified by SUMO.
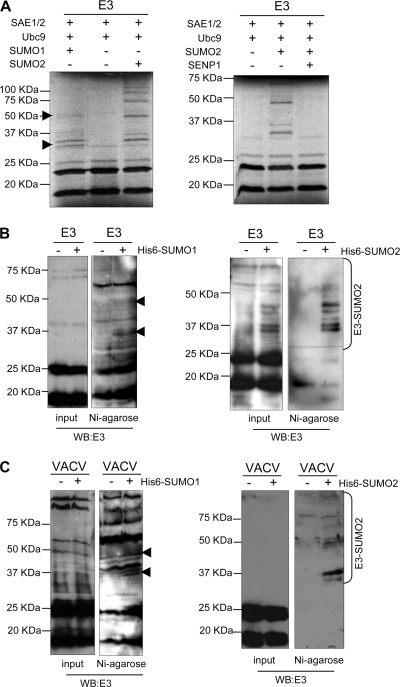
To further analyze whether E3 can be covalently modified by SUMO, an in vitro SUMOylation assay was then carried out by using [35S]methionine-labeled E3 protein as a substrate. As expected, in vitro-translated E3 was detected as two bands of around 15 and 22 kDa (Fig. 2A). The incubation of the SUMOylation reaction mixture with SUMO1 led to the appearance of additional higher-molecular-mass bands that corresponded to E3-SUMO1 (Fig. 2A, left, arrowheads). In addition, when the in vitro SUMOylation reaction mixture was incubated in the presence of SUMO2, a ladder of higher-molecular-mass bands was observed (Fig. 2A, left). To further prove that E3 is in fact SUMOylated in vitro, E3-SUMO2 obtained as described above was incubated in a deSUMOylation reaction mixture in the presence of GST or GST-SENP1 (Biomol), a SUMO-specific protease containing the catalytic domain of SENP1. The incubation of E3-SUMO2 in the presence of SENP1 led to the disappearance of the higher-molecular-mass bands corresponding to E3-SUMO2 (Fig. 2A, right), demonstrating that in vitro-synthesized E3 is covalently conjugated to SUMO. In order to analyze whether E3 is also SUMOylated in vivo, HEK-293 cells were cotransfected with a plasmid encoding E3 together with pcDNA, Ubc9 and pcDNA-His6-SUMO1, or Ubc9 and pcDNA-His6-SUMO2, and whole-protein extracts or nickel column-purified histidine-tagged proteins were then analyzed by Western blotting using an anti-E3 antibody. Cells cotransfected with pCINEO-E3 and pcDNA showed two bands of around 15 and 22 kDa, as expected (Fig. 2B). However, when His6-SUMO1 was cotransfected with E3, higher-molecular-mass bands of the expected size were detected in the purified extracts (Fig. 2B, left, arrowheads). Analysis of extracts from the His6-SUMO2-transfected cells also revealed additional bands already visible in crude extracts that corresponded to E3-SUMO2, as demonstrated after Western blot analysis of the purified extracts (Fig. 2B, right). Note that since the levels of SUMO1-modified E3 were lower than the levels of SUMO2-modified E3, we required larger amounts of purified protein and longer exposure times to detect the E3-SUMO1 bands. Moreover, to demonstrate that E3 is also SUMOylated when produced by the virus, HEK-293 cells transfected with pcDNA, Ubc9 and pcDNA-His6-SUMO1, or Ubc9 and pcDNA-His6-SUMO2 were infected with VACV. Whole-cell extracts and histidine-tagged proteins recovered on nickel beads were analyzed by Western blotting with an anti-E3 antibody. Upon the purification of His-tagged complexes, two major E3-SUMO1 conjugates and several E3-SUMO2 bands were detected (Fig. 2C). Consistent with the data obtained after the analysis of E3 following transfection, SUMO2 more readily modified E3 than SUMO1. Altogether, these data demonstrate that the VACV E3 protein is SUMOylated by SUMO1 and SUMO2 in vitro and in vivo.
Fig. 2.
Covalent modification of E3 by SUMO1 or SUMO2 in vivo and in vitro. (A, left) In vitro-translated E3 was used as substrate in an in vitro SUMOylation assay in the presence of SUMO1 or SUMO2. (Right) Deconjugation of SUMO2 from E3 by SENP1. (B) HEK-293 cells were cotransfected with pCINEO-E3 together with pcDNA, pcDNA-Ubc9 and pcDNA-His6-SUMO1, or pcDNA-Ubc9 and pcDNA-His6-SUMO2. Total protein extracts and histidine-tagged proteins purified by use of nickel columns were then analyzed by Western blotting (WB) with an anti-E3 antibody. (C) HEK-293 cells were transfected with pcDNA, pcDNA-Ubc9 and pcDNA-His6-SUMO1, or pcDNA-Ubc9 and pcDNA-His6-SUMO2 and then infected with VACV for 5 h. Total protein extracts and histidine-tagged proteins purified by use of nickel columns were then analyzed by Western blotting with an anti-E3 antibody. Arrowheads refer to E3-SUMO1.
SUMO conjugation modulates the transactivation activity of E3 on the PUMA and APAF-1 genes.
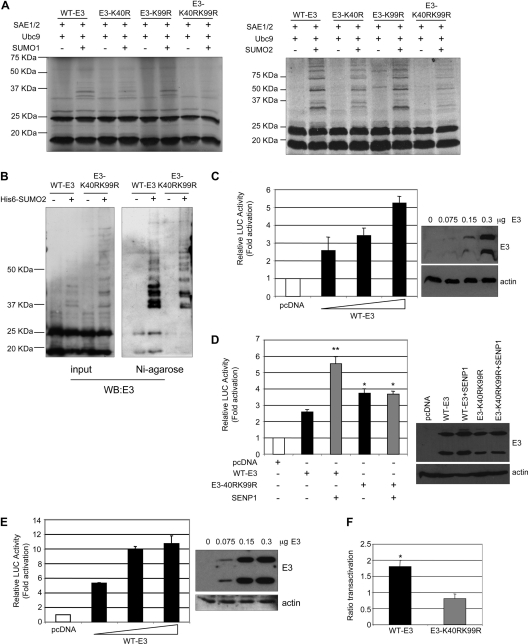
Having determined that VACV E3 is SUMO modified, we sought to identify the region of E3 involved in this modification. Many SUMO-modified proteins characterized to date are modified at lysine residues that exist within the ψKxE consensus sequence (where ψ is a hydrophobic residue, K is the modified lysine, x is any amino acid, and E is glutamic acid). Analysis of the amino acid sequence using SUMOylation prediction programs revealed a consensus sequence for SUMOylation, 39EKRE42, that is highly conserved among distantly related poxviruses (see Fig. S1 in the supplemental material) and at least one other putative nonconsensus SUMO conjugation domain located in position K99 as putative SUMO acceptor sites. To investigate whether these were the sites for SUMO modification, we constructed an E3 point mutant in which lysine 40 or 99 was replaced with arginine (E3-K40R and E3-K99R). In addition, an E3 mutant with a mutation in both lysine residues was generated (E3-K40RK99R), and in vitro SUMO conjugation assays using the different E3 mutants as substrates were then carried out. The conjugation of SUMO1 was clearly reduced in the E3-K40R mutant, as revealed by the lower levels of the main E3-SUMO1 bands detected in the assay (Fig. 3A, left). E3 SUMOylation was not affected by the substitution of lysine 99. However, the double mutation of K40 and K99 to arginine dramatically reduced the SUMOylation of E3 (Fig. 3A, left). Results observed after in vitro SUMO2 modification assays with the E3 mutants revealed similar results. The conjugation of SUMO2 to the E3-K40R mutant was clearly reduced in comparison with that observed with WT-E3, and although the modification of E3-K99R with SUMO2 was quite similar to that observed with WT-E3, the mutation of both lysines showed a more significant reduction in SUMO2 conjugation (Fig. 3A, right). To determine whether these two lysines are also implicated in SUMOylation in vivo, HEK-293 cells were cotransfected with pCINEO-E3-WT or pCINEO-E3-K40RK99R together with pcDNA or Ubc9 and pcDNA-His6-SUMO2, and analyses of the whole-protein extracts and of the nickel-purified histidine-tagged proteins using an anti-E3 antibody were carried out. A clear reduction in the E3-K40RK99R SUMO2 modification in comparison with that of WT-E3 was observed (Fig. 3B). In addition, the main E3-SUMO1 band detected in the cells cotransfected with SUMO1 and WT-E3 was not detected in those cells transfected with the E3-K40RK99R mutant (see Fig. S2 in the supplemental material). Altogether, these results indicated that E3 can be modified by SUMO at both residues. Covalent SUMO conjugation may regulate the subcellular localization or stability of the target proteins. In order to analyze if an E3 mutant with a reduced ability to be SUMOylated has an altered subcellular localization, MCF-7 cells were transfected with the plasmid encoding WT-E3 or the E3-K40RK99R mutant, and 36 h after transfection, cells were stained with an anti-E3 antibody. We could not detect significant differences in the subcellular localizations of both proteins (see Fig. S3 in the supplemental material). In addition, we did not observe any alteration in the subcellular localization of E3 after the cotransfection of pCINEO-E3-WT with SUMO1 or SUMO2 (data not shown). The effect of SUMOylation on the stability of E3 was then studied. MCF-7 cells were cotransfected with plasmids encoding WT-E3 or E3-K40RK99R together with pcDNA, Ubc9 and SUMO1, or Ubc9 and SUMO2 and treated with cycloheximide, and at different times after treatment, levels of E3 were analyzed. We could not detect differences in the stabilities of WT-E3 and the E3-K40RK99R mutant or after the cotransfection of SUMO1 or SUMO2 (see Fig. S4 in the supplemental material). Furthermore, the stability of E3-SUMO2 was similar to that of the unmodified E3 protein (Fig. S4), indicating that SUMOylation does not modulate E3 stability. VACV with E3L deleted (VACV-ΔE3L) cannot replicate in HeLa cells, but this host-range phenotype could be complemented by an E3L gene expressed transiently from a plasmid (13). In addition, we also observed that VACV-ΔE3L has a replication defect in HEK-293 cells (see Fig. S5 in the supplemental material). Hence, the requirement of E3 SUMO conjugation for VACV-ΔE3L reconstitution in HeLa or HEK-293 cells was analyzed. Cells were transfected with pcDNA, WT-E3, or E3-K40RK99R, and 48 h after transfection, cells were infected with VACV-ΔE3L at a multiplicity of infection (MOI) of 7.5 PFU per cell. The synthesis of viral proteins 16 h after infection and VACV production 24 h after infection were determined. The VACV titer recovered from E3-K40RK99R-transfected HeLa or HEK-293 cells was significantly higher than that recovered from pcDNA-transfected cells and similar to the viral titer obtained from WT-E3-transfected cells (Fig. S5). As expected, no VACV protein synthesis was observed in cells transfected with pcDNA. However, the synthesis of viral proteins in VACV-ΔE3L-infected cells was restored after the expression of WT-E3 from the transfected plasmid, and viral protein synthesis was similarly observed after the expression of the E3-K40RK99R mutant in HeLa or HEK-293 cells (Fig. S5). Altogether, these results suggest that the SUMO modification of E3 is not required for efficient VACV replication. SUMO conjugation may also alter the transactivation activity of the target proteins. Therefore, we were interested in testing whether a covalent modification by SUMO had any effect on the transactivation activity of E3. The N terminus of E3 was suggested previously to stabilize Z-DNA in the promoter regions in some genes, such as the p53 gene, resulting in the transactivation of the gene (34). Consequently, we decided to analyze whether an E3 mutant with a reduced ability to be SUMOylated has an altered transactivation activity. Initially, transactivation assays were performed with MCF-7 cells that were transfected with reporter plasmid p53-regulated PUMA-luc together with increasing amounts of pCINEO-E3-WT. As shown in Fig. 3C, the expression of WT-E3 induced the transactivation of the reporter in a dose-responsive manner. Importantly, the transactivation levels achieved in our experiments were similar to those previously reported (34). Next, the transactivation activity of WT-E3 or E3-K40RK99R after the cotransfection of SENP1 on PUMA-luc was analyzed. The cotransfection of SENP1 significantly increased the transactivation of the promoter induced by WT-E3 (Fig. 3D). The transfection of the E3-K40RK99R mutant induced a slightly higher but statistically significant transactivation of the promoter than WT-E3, and this activity was not altered after the cotransfection of SENP1 (Fig. 3D), suggesting that SUMOylation has a negative effect on the transactivation activity of E3. To reinforce these data, we analyzed the transcriptional activity of E3 on a different reporter. We observed that E3 induced the transactivation of the p53-regulated APAF-1–luc reporter in a dose-responsive manner (Fig. 3E). The cotransfection of SENP1 significantly increased the transactivation of the APAF-1–luc reporter induced by WT-E3 but did not alter the transcriptional activity of the E3-K40RK99R mutant (Fig. 3F). Altogether, these results indicate that SUMO conjugation negatively regulates the transcriptional activity of E3 at the PUMA and APAF-1 genes.
Fig. 3.
Role of E3 SUMOylation in transcriptional activity. (A) [35S]methionine-labeled wild-type or mutant E3 proteins were used as substrates in an in vitro SUMOylation assay in the presence of SUMO1 or SUMO2. The reaction products were visualized by autoradiography. (B) Total extracts or histidine-tagged purified proteins were prepared from HEK-293 cells cotransfected with pCINEO-E3-WT or E3-K40RK99R together with pcDNA or pcDNA-Ubc9 and pcDNA-His6-SUMO2, and an immunoblot analysis with an anti-E3 antibody was carried out. (C and D) MCF-7 cells were transiently transfected in triplicate with the luciferase reporter PUMA-luc together with the plasmids indicated. The same results were obtained in at least three different experiments. Data represent means ± standard errors (SE) for one experiment. *, P < 0.05; **, P < 0.005 (compared with WT-E3-transfected cells; determined by the Student test). The right panel represents the expression levels of the E3L gene transfected in each experiment. (E) MCF-7 cells were transiently transfected in triplicate with the luciferase reporter APAF-luc together with the plasmids indicated. The same results were obtained in at least three different experiments. Data represent means ± SE for one experiment. (F) Ratio of the level of transactivation of APAF-1–luc by WT-E3 or E3-K40RK99R and SENP-1 to the level of transactivation by WT-E3 or E3-K40RK99R alone, respectively. The same results were obtained in at least three different experiments. Data represent means ± SE for one experiment. *, P < 0.05 (compared with WT-E3 transfected cells; determined by the Student test).
The integrity of the SIM domain in E3 is required for its stability, nuclear localization, and SUMOylation.
Recently, it was demonstrated that several cellular and viral SUMO targets can also interact noncovalently with SUMO through a SUMO-interacting motif (SIM). The best-studied SIM contains a hydrophobic core sequence consisting of stretches of three or four hydrophobic valine, leucine, or isoleucine residues plus one acidic/polar residue at position 2 or 3 (27, 59). The E3 protein presents a domain that resembles a SIM at position 119VTVI122. A putative noncovalent interaction between SUMO1 and E3 was also suggested after the analysis of the colocalization between SUMO1 and the E3-K40RK99R mutant in infected cells (see Fig. S6 in the supplemental material). To investigate the functionality of this putative SIM, we performed site-directed mutagenesis, by which we replaced valine at positions 119 and 121 and isoleucine at position 122 with alanine to generate E3-ΔSIM. Based upon previous results for other proteins such as hDaxx or IE2p86, it was anticipated that these mutations would disrupt the SIM function (4, 41). Surprisingly, Western blot analysis of cells transfected with E3-ΔSIM revealed that the mutant protein was almost undetectable, and only when the cells were incubated with the proteasome inhibitor MG132 was the protein detected (Fig. 4A). Similarly, instability of the E3 protein containing both K40RK99R and SIM mutations was also observed (see Fig. S7 in the supplemental material), indicating that the SIM is required for maintaining the stability of the E3 protein. To verify that the SIM domain in E3 is in fact implicated in the noncovalent interaction with SUMO1, we then analyzed whether a SIM domain mutant can bind SUMO1 by using GST pulldown assays. WT-E3 but not the E3-ΔSIM mutant was efficiently bound to GST-SUMO1 resin, but it did not adhere to GST resin, indicating the functionality of the SIM domain (Fig. 4B). SIMs are expected to play a crucial role in regulating SUMOylated proteins. In this sense, the SUMOylation of several proteins has been found to be SIM dependent, suggesting that noncovalent SUMO binding may represent a general mechanism for substrate selection and modification. To address the question of whether the SUMOylation of E3 requires a SIM-mediated interaction with SUMO, we first tested the in vitro SUMO conjugation of the E3-ΔSIM mutant compared to WT-E3. Wild-type and mutant E3 proteins were expressed in vitro and modified in reaction mixtures containing either SUMO1 or SUMO2. The SUMO1 and SUMO2 modification efficiencies of the SIM mutant protein were both dramatically reduced compared with that of WT-E3 (Fig. 4C), and the mutation of lysine residues 40 and 99 did not further reduce the SUMO conjugation of E3-ΔSIM (see Fig. S7 in the supplemental material). We then decided to study the effect of the SIM domain on E3 SUMOylation in vivo. In vivo SUMOylation assays of WT-E3 or E3-ΔSIM were then carried out in the presence of MG132. HEK-293 cells were cotransfected with a plasmid encoding WT-E3 or E3L-ΔSIM together with pcDNA or with Ubc9 and pcDNA-His6-SUMO2, and whole-protein extracts and nickel column-purified histidine-tagged proteins were then analyzed by Western blotting using an anti-E3 antibody. E3-SUMO2 bands detected in those cells transfected with pCINEO-E3-WT and pcDNA-His6-SUMO2 were not observed for the cells transfected with the pCINEO-E3-ΔSIM mutant, indicating that the SIM domain in E3 is implicated in its SUMOylation in vivo (Fig. 4D). Taken together, these results demonstrate that efficient E3 SUMOylation requires the SUMO-SIM interface both in vitro and in vivo. An analysis of the subcellular localization of the WT-E3, E3-ΔSIM, or E3-K40RK99R-ΔSIM protein after MG132 treatment was also carried out. As expected, E3 was detected in both the nucleus and the cytoplasm of the transfected cells. However, the E3-ΔSIM mutants were retained into the cytoplasm, suggesting that the SIM domain is also required for E3 nuclear translocation (Fig. 4E and see Fig. S7 in the supplemental material). Genetic and biochemical experiments suggested that the biological activity of E3 is derived primarily from its capacity to bind to dsRNA (12–14, 69), an interaction that also mediates its binding to ISG15 (25). Therefore, we analyzed the ability of the E3-ΔSIM mutant to bind to dsRNA. This was assayed by a poly(I:C)-agarose-binding assay as previously described (37). Both the 35S-labeled in vitro-synthesized WT-E3 and E3-ΔSIM proteins bound to dsRNA-agarose but not to agarose alone (Fig. 4F), indicating that the SIM domain is not required for the interaction of E3 and dsRNA. In addition, we studied whether the SIM domain is required for the interaction between E3 and ISG15. We detected the coimmunoprecipitation of E3-ΔSIM and ISG15 (see Fig. S8 in the supplemental material), indicating that the SIM domain is not required for the interaction between E3 and ISG15.
Fig. 4.
The SIM domain in E3 is required for its interaction with SUMO, its stability, SUMOylation, and nuclear localization, but it is not essential to bind dsRNA. (A) HEK-293 cells were transfected with WT-E3 or E3-ΔSIM expression plasmids and treated or not with MG132 for 24 h. Analyses of the protein extracts by Western blotting with an anti-E3 antibody were then carried out. (B) Protein extracts from HEK-293 cells transfected with WT-E3 or E3-ΔSIM expression constructs and treated with MG132 for 24 h were incubated with GST or GST-SUMO1. Beads were washed, and the bound proteins were eluted and subjected to electrophoresis and Western blot analysis with an anti-E3 antibody. The input, representing 10% of the protein extracts, is shown. (C) [35S]methionine-labeled WT-E3 or E3-ΔSIM proteins were used as substrates in an in vitro SUMOylation assay in the presence of SUMO1 (top) or SUMO2 (bottom). The reaction products were visualized by autoradiography. (D) Total extracts or histidine-tagged purified proteins were prepared from HEK-293 cells cotransfected with pCINEO-E3-WT or pCINEO-E3-ΔSIM together with pcDNA or pcDNA-Ubc9 and pcDNA-His6-SUMO2, and an immunoblot analysis with an anti-E3 antibody was carried out. (E) MCF-7 cells were transfected with WT-E3 or E3-ΔSIM expression plasmids and treated with MG132 for 24 h. Cells were then stained with DAPI and an anti-E3 antibody, and the localization of E3 was detected by confocal microscopy. (F) [35S]methionine-labeled WT-E3 or E3-ΔSIM proteins were incubated with agarose or poly(I:C)-agarose. Beads were washed, and the bound proteins were eluted and subjected to electrophoresis and autoradiography. The input, representing 10% of the quantity of protein used for the binding reaction, is shown.
The E3 protein is differentially ubiquitinated depending on the integrity of a SIM domain.
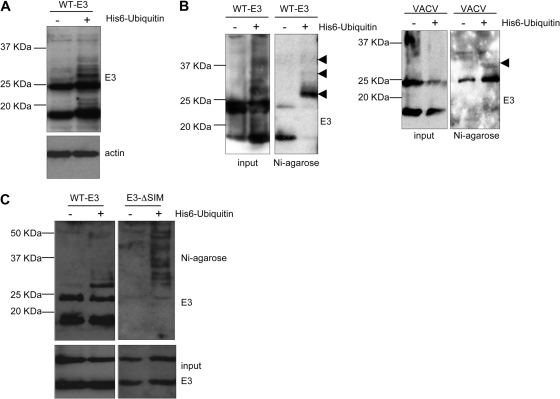
The degradation of the E3-ΔSIM mutant by a proteasome-mediated mechanism suggested that E3 might also be modified by ubiquitin. In order to analyze whether E3 can be ubiquitinated, MCF-7 cells were transfected with pCINEO-E3-WT in the presence or absence of pcDNA-His6-ubiquitin, and at 36 h posttransfection, Western blot analysis of E3 was carried out. As shown in Fig. 5A, additional higher-molecular-mass bands were detected in those cells cotransfected with ubiquitin. Importantly, the Western blot analysis also revealed that the levels of the E3 protein were not significantly affected by the expression of ubiquitin, suggesting that E3 ubiquitination does not induce WT-E3 degradation (Fig. 5A). In order to demonstrate that the higher-molecular-mass bands correspond to E3-ubiquitin, His-binding proteins were purified from MCF-7 cells transfected as described above. Western blot analysis of the purified extracts using an anti-E3 antibody revealed that the additional bands detected after the transfection of ubiquitin corresponded with E3-ubiquitin (Fig. 5B, left). A broad band of the expected molecular mass that was recognized by the anti-E3 antibody was also observed after Western blot analysis of histidine-tagged proteins obtained from cells transfected with His6-ubiquitin and then infected with VACV (Fig. 5B, right), indicating that the E3 produced by the virus can also be modified by ubiquitin. Recent studies provided evidence that ubiquitin chains with different linkages or lengths may work as specific signals to control distinct molecular processes. To further characterize E3 ubiquitination, we cotransfected MCF-7 cells with pCINEO-E3-WT or pCINEO-E3-ΔSIM and pcDNA-His6-ubiquitin and treated the cells with MG132, and 36 h after transfection, purified histidine-tagged proteins were analyzed by Western blotting with an anti-E3 antibody. A high-mobility ubiquitin-positive smear was observed for the cells transfected with ubiquitin and the E3-ΔSIM mutant in comparison with the bands detected for those transfected with WT-E3 expression constructs (Fig. 5C).
Fig. 5.
E3 protein is differentially ubiquitinated depending on the integrity of a SIM domain. (A) MCF-7 cells were transfected with pCINEO-E3-WT in the presence or absence of pcDNA-His6-ubiquitin as indicated. Analysis of the protein extracts by Western blotting with an anti-E3 antibody 48 h after transfection was carried out. (B) MCF-7 cells were transfected as indicated above (left) or transfected with pcDNA-His6-ubiquitin and then infected with VACV for 6 h (right). Whole-protein extracts and histidine-tagged proteins purified by use of nickel columns were then analyzed by Western blotting with an anti-E3 antibody. Arrowheads refer to ubiquitinated E3. (C) MCF-7 cells were transfected with WT-E3- or E3L-ΔSIM-expressing plasmids in the presence or absence of pcDNA-His6-ubiquitin and treated with MG132 for 24 h. Analysis of the whole-protein extracts and the histidine-tagged proteins purified by use of nickel columns was then carried out by Western blotting with an anti-E3 antibody.
DISCUSSION
The VACV E3L gene product is known as an IFN resistance protein, as a determinant of host range and viral pathogenesis, and as a modulator of cellular apoptotic and signal transduction pathways (35, 36, 42). The E3 protein binds to dsRNA, and this interaction mediates or enhances its binding to ISG15 and PKR, respectively (25, 57). In addition, E3 can bind to SUMO1 in a yeast two-hybrid assay, although this interaction was not confirmed at the biochemical level (53). This finding, together with the observation that SUMO1 is recruited to viral factories in response to VACV infection (49), prompted us to investigate the biochemical nature of the interactions of SUMO and E3. Our results demonstrate that E3 colocalizes with SUMO1 in the viral factories and that the E3 protein is modified by the covalent attachment of SUMO1 or SUMO2 in vitro and in vivo. Importantly, E3 SUMOylation was demonstrated for transfected as well as for infected cells, indicating that the SUMOylation of E3 is independent of VACV infection. Although SUMOylated lysine residues are often found within ψFxE consensus sequences, recent proteomics studies have shown that a considerable part of SUMOylation occurs on nonconsensus sites (6, 7, 24, 29). Furthermore, it was described previously that lysine selection is promiscuous, because lysine-to-arginine substitutions at preferred SUMO modification sites may result in enhanced modification at secondary sites (18, 41). The mutation of the consensus sequence in E3 (lysine 40) reduced but did not abrogate its SUMOylation. However, when the two lysines at residues 40 and 99 of E3 were mutated, the SUMOylation of the double mutant was clearly reduced both in vitro and in vivo. Consequently, these two lysine residues at positions 40 and 99 were mapped as major SUMO conjugation sites. The SUMO modification of proteins has different effects depending on the target protein. SUMOylation influences the functions of many viral proteins by modulating their interactions with other proteins and then regulating their subcellular localizations, as demonstrated previously for the VACV A40R protein (49). The subcellular localization of the SUMOylation-negative E3 mutant was not altered compared to that of wild-type E3. Changes in the stability of E3 after the mutation of the two SUMO acceptor lysines or differences in the stabilities of E3-SUMO2 and E3 were not detected. In addition, the transient transfection of the SUMOylation-negative E3 mutant was able to reconstitute the replication of VACV-ΔE3L in HeLa and HEK-293 cells, indicating that SUMO modification is not required for productive viral infection. The lack of an effect of SUMO conjugation on virus growth was reported previously for the human cytomegalovirus (HCMV) IE2 protein (38). However, this modification was required for the transactivation function of IE2 (1, 28). A direct effect of SUMOylation on transcriptional activity has been reported for several proteins, and this modification typically represses transactivation activity (66). E3 activates the transcription of the human p53 gene through Z-DNA binding (34). In agreement with these data, our results demonstrate that E3 induces the transcriptional transactivation of the p53-regulated PUMA-luc and APAF-1–luc reporter plasmids in a dose-responsive manner. Moreover, the coexpression of SENP1 or a mutation of the SUMO conjugation lysines potentiates the E3 transactivation activity. Altogether, these results indicate that SUMO conjugation is a negative regulator of the transcriptional activation of p53 by E3.
Recently, the existence of known SUMO targets that interact noncovalently with SUMO and contain SUMO-interacting motifs (SIMs) was demonstrated (32). Intriguingly, E3 contains a hydrophobic region with a sequence conforming to the (L/V/I)x(L/V/I)(L/V/I) SIM consensus at the C terminus of the protein. Furthermore, we still detected colocalization between E3-K40RK99R and SUMO1 in the infected cells, suggesting that additional motifs in E3 may mediate the interaction with SUMO1. Although the mutation of the SIM domain did not alter the ability of E3 to interact with dsRNA or ISG15, analysis of the SIM E3 mutant showed impaired SUMOylation in vitro and in vivo that correlated with the noncovalent SUMO-binding properties. These results demonstrate that the SIM domain is not required for the interaction of E3 and dsRNA but that the SUMO-SIM interface is required for efficient E3 SUMOylation, as observed for an increasing number of target proteins (32, 41, 47, 60). Importantly, the mutation of the E3 SIM domain led to an unstable protein that was degraded by a proteasome-mediated mechanism. Analysis of the subcellular localization in the presence of a proteasome inhibitor also demonstrated that the SIM mutant showed an exclusively cytoplasmic localization, in contrast to the nucleocytoplasmic distribution of the wild-type protein. Although there is a correlation between the cytoplasmic localization and proteasome-mediated degradation of E3-ΔSIM, a cytoplasmic E3 mutant reported previously was not described to be an unstable protein (13). In addition, the overexpression of ubiquitin-specific protease 7 (USP7/HAUSP), an enzyme that prevents the degradation of different cellular and viral proteins (11, 19, 40, 46, 65), partially rescues E3-ΔSIM from degradation without altering its cytoplasmic localization (see Fig. S9 in the supplemental material). Altogether, these results suggest that the degradation of the E3-ΔSIM protein is not a consequence of its cytoplasmic localization. A mutation of the SIM domain in the E3-K40RK99R mutant also led to an alteration of the SUMOylation, stability, and subcellular localization of the protein, supporting the idea that a noncovalent SUMO interaction but not SUMOylation modulates these E3 properties. Noncovalent SUMO interactions have been shown to affect the recruitment of proteins into subnuclear structures (45, 58) or their transcriptional activity (33, 54). Thus far, this is the first time that the integrity of a SIM domain within a protein has been demonstrated to be required for its stability.
In a classical model, ubiquitination was associated with proteasome-mediated degradation. We then decided to analyze whether E3 can be ubiquitinated. Viruses are connected to ubiquitin in a variety of ways. A role of ubiquitin in the degradation of viral proteins has been demonstrated in the cases of the Epstein-Barr virus (EBV) LMP2a, human papillomavirus (HPV) E7, and HPV E2 proteins (30). However, some viral functions require ubiquitin modification for purposes other than protein turnover, as exemplified by the role of the ubiquitination of viral proteins in virion budding and release for the retroviral (HIV) Gag protein (15, 21), the Ebola virus Vp40 protein (70), or the rhadinovirus M protein (27). Recently, it was reported that poxviruses exploit the ubiquitination machinery by expressing viral proteins that function as ubiquitin ligases or regulate cellular ubiquitin ligases (48, 56, 64). In addition, it was also demonstrated that the proteasome and ubiquitin are required for poxvirus infection (62). Our results demonstrate that E3 can be modified by ubiquitin but that this ubiquitination does not induce wild-type E3 degradation. Ubiquitin itself possesses several lysines that can be used for the attachment of another ubiquitin molecule, allowing substrates to be modified with different types of ubiquitin chains (50, 51). In this sense, the consequences of polyubiquitination appear to depend on the length of the ubiquitin chain and on the type of linkage used. It is then tempting to speculate that the different stabilities observed between the WT-E3 and the E3-ΔSIM proteins are due to differences in ubiquitin chain lengths. An analysis of the conjugation of ubiquitin to WT-E3 or E3-ΔSIM revealed that whereas the WT-E3 ubiquitin modification was quite limited, very long ubiquitin chains were conjugated to the E3-ΔSIM mutant, reinforcing this hypothesis. Different evidences reveal the existence of cross talk mechanisms between the SUMOylation and ubiquitination systems (23). This is a new example of how SUMO may have a profound effect on the ubiquitination status of proteins. In addition, these findings provide evidence for specific posttranslational modifications of VACV E3 that might explain some of the multiple biological effects exerted by E3 through virus-host cell interactions.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the technical assistance of Victoria Jiménez. We are indebted to Bernie Moss (NIH) and Enzo Paoletti (Virogenetics) for kindly providing reagents. We thank Aneesh Vijayan for the rabbit antibody against the vaccinia virus E3 protein.
Funding at the laboratory of C.R. is provided by BFU-2008-03784. M.E. is supported by SAF2008-02036 and Foundation Botín. M.C. and L.M.-V. are supported by the Juan de la Cierva Programme. J.G.-S. is supported by an IFARHU-SENACYT predoctoral fellowship from Panama. P.G. is supported by a JAE predoctoral fellowship from the CSIC. S.G. holds a research contract from the program Ramón y Cajal of Spain.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 28 September 2011.
REFERENCES
- 1. Ahn J. H., Xu Y., Jang W. J., Matunis M. J., Hayward G. S. 2001. Evaluation of interactions of human cytomegalovirus immediate-early IE2 regulatory protein with small ubiquitin-like modifiers and their conjugation enzyme Ubc9. J. Virol. 75:3859–3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beattie E., et al. 1996. Host-range restriction of vaccinia virus E3L-specific deletion mutants. Virus Genes 12:89–94 [DOI] [PubMed] [Google Scholar]
- 3. Beattie E., Paoletti E., Tartaglia J. 1995. Distinct patterns of IFN sensitivity observed in cells infected with vaccinia K3L− and E3L− mutant viruses. Virology 210:254–263 [DOI] [PubMed] [Google Scholar]
- 4. Berndt A., Hofmann-Winkler H., Tavalai N., Hahn G., Stamminger T. 2009. Importance of covalent and noncovalent SUMO interactions with the major human cytomegalovirus transactivator IE2p86 for viral infection. J. Virol. 83:12881–12894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bernier-Villamor V., Sampson D. A., Matunis M. J., Lima C. D. 2002. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108:345–356 [DOI] [PubMed] [Google Scholar]
- 6. Blomster H. A., et al. 2009. Novel proteomics strategy brings insight into the prevalence of SUMO-2 target sites. Mol. Cell. Proteomics 8:1382–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blomster H. A., et al. 2010. In vivo identification of sumoylation sites by a signature tag and cysteine-targeted affinity purification. J. Biol. Chem. 285:19324–19329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boggio R., Chiocca S. 2006. Viruses and sumoylation: recent highlights. Curr. Opin. Microbiol. 9:430–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brandt T. A., Jacobs B. L. 2001. Both carboxy- and amino-terminal domains of the vaccinia virus interferon resistance gene, E3L, are required for pathogenesis in a mouse model. J. Virol. 75:850–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Campagna M., et al. SIRT1 stabilizes PML promoting its sumoylation. Cell Death Differ. 18:72–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Canning M., Boutell C., Parkinson J., Everett R. D. 2004. A RING finger ubiquitin ligase is protected from autocatalyzed ubiquitination and degradation by binding to ubiquitin-specific protease USP7. J. Biol. Chem. 279:38160–38168 [DOI] [PubMed] [Google Scholar]
- 12. Chang H. W., Jacobs B. L. 1993. Identification of a conserved motif that is necessary for binding of the vaccinia virus E3L gene products to double-stranded RNA. Virology 194:537–547 [DOI] [PubMed] [Google Scholar]
- 13. Chang H. W., Uribe L. H., Jacobs B. L. 1995. Rescue of vaccinia virus lacking the E3L gene by mutants of E3L. J. Virol. 69:6605–6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chang H. W., Watson J. C., Jacobs B. L. 1992. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc. Natl. Acad. Sci. U. S. A. 89:4825–4829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Demirov D. G., Ono A., Orenstein J. M., Freed E. O. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. U. S. A. 99:955–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Desterro J. M., Rodriguez M. S., Hay R. T. 1998. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol. Cell 2:233–239 [DOI] [PubMed] [Google Scholar]
- 17. Domingo-Gil E., Perez-Jimenez E., Ventoso I., Najera J. L., Esteban M. 2008. Expression of the E3L gene of vaccinia virus in transgenic mice decreases host resistance to vaccinia virus and Leishmania major infections. J. Virol. 82:254–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eladad S., et al. 2005. Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Hum. Mol. Genet. 14:1351–1365 [DOI] [PubMed] [Google Scholar]
- 19. Everett R. D., et al. 1997. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 16:1519–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garcia M. A., Guerra S., Gil J., Jimenez V., Esteban M. 2002. Anti-apoptotic and oncogenic properties of the dsRNA-binding protein of vaccinia virus, E3L. Oncogene 21:8379–8387 [DOI] [PubMed] [Google Scholar]
- 21. Garrus J. E., et al. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55–65 [DOI] [PubMed] [Google Scholar]
- 22. Geiss-Friedlander R., Melchior F. 2007. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8:947–956 [DOI] [PubMed] [Google Scholar]
- 23. Geoffroy M. C., Hay R. T. 2009. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat. Rev. Mol. Cell Biol. 10:564–568 [DOI] [PubMed] [Google Scholar]
- 24. Golebiowski F., et al. 2009. System-wide changes to SUMO modifications in response to heat shock. Sci. Signal. 2:ra24. [DOI] [PubMed] [Google Scholar]
- 25. Guerra S., Caceres A., Knobeloch K. P., Horak I., Esteban M. 2008. Vaccinia virus E3 protein prevents the antiviral action of ISG15. PLoS Pathog. 4:e1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guo D., et al. 2004. A functional variant of SUMO4, a new I kappa B alpha modifier, is associated with type 1 diabetes. Nat. Genet. 36:837–841 [DOI] [PubMed] [Google Scholar]
- 27. Hecker C. M., Rabiller M., Haglund K., Bayer P., Dikic I. 2006. Specification of SUMO1- and SUMO2-interacting motifs. J. Biol. Chem. 281:16117–16127 [DOI] [PubMed] [Google Scholar]
- 28. Hofmann H., Floss S., Stamminger T. 2000. Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J. Virol. 74:2510–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hsiao H. H., Meulmeester E., Frank B. T., Melchior F., Urlaub H. 2009. “ChopNSpice,” a mass spectrometric approach that allows identification of endogenous small ubiquitin-like modifier-conjugated peptides. Mol. Cell. Proteomics 8:2664–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Isaacson M. K., Ploegh H. L. 2009. Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe 5:559–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jentsch S., Pyrowolakis G. 2000. Ubiquitin and its kin: how close are the family ties? Trends Cell Biol. 10:335–342 [DOI] [PubMed] [Google Scholar]
- 32. Kerscher O. 2007. SUMO junction—what's your function? New insights through SUMO-interacting motifs. EMBO Rep. 8:550–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kuo H. Y., et al. 2005. SUMO modification negatively modulates the transcriptional activity of CREB-binding protein via the recruitment of Daxx. Proc. Natl. Acad. Sci. U. S. A. 102:16973–16978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kwon J. A., Rich A. 2005. Biological function of the vaccinia virus Z-DNA-binding protein E3L: gene transactivation and antiapoptotic activity in HeLa cells. Proc. Natl. Acad. Sci. U. S. A. 102:12759–12764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Langland J. O., Jacobs B. L. 2002. The role of the PKR-inhibitory genes, E3L and K3L, in determining vaccinia virus host range. Virology 299:133–141 [DOI] [PubMed] [Google Scholar]
- 36. Langland J. O., et al. 2006. Suppression of proinflammatory signal transduction and gene expression by the dual nucleic acid binding domains of the vaccinia virus E3L proteins. J. Virol. 80:10083–10095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Langland J. O., Pettiford S. M., Jacobs B. L. 1995. Nucleic acid affinity chromatography: preparation and characterization of double-stranded RNA agarose. Protein Expr. Purif 6:25–32 [DOI] [PubMed] [Google Scholar]
- 38. Lee H. R., Ahn J. H. 2004. Sumoylation of the major immediate-early IE2 protein of human cytomegalovirus Towne strain is not required for virus growth in cultured human fibroblasts. J. Gen. Virol. 85:2149–2154 [DOI] [PubMed] [Google Scholar]
- 39. Lee S. B., Esteban M. 1994. The interferon-induced double-stranded RNA-activated protein kinase induces apoptosis. Virology 199:491–496 [DOI] [PubMed] [Google Scholar]
- 40. Li M., et al. 2002. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416:648–653 [DOI] [PubMed] [Google Scholar]
- 41. Lin D. Y., et al. 2006. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell 24:341–354 [DOI] [PubMed] [Google Scholar]
- 42. Ludwig H., et al. 2005. Role of viral factor E3L in modified vaccinia virus Ankara infection of human HeLa cells: regulation of the virus life cycle and identification of differentially expressed host genes. J. Virol. 79:2584–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marcos-Villar L., et al. 2009. Kaposi's sarcoma-associated herpesvirus protein LANA2 disrupts PML oncogenic domains and inhibits PML-mediated transcriptional repression of the survivin gene. J. Virol. 83:8849–8858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Matic I., et al. 2008. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol. Cell. Proteomics 7:132–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Matunis M. J., Zhang X. D., Ellis N. A. 2006. SUMO: the glue that binds. Dev. Cell 11:596–597 [DOI] [PubMed] [Google Scholar]
- 46. Meulmeester E., et al. 2005. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol. Cell 18:565–576 [DOI] [PubMed] [Google Scholar]
- 47. Minty A., Dumont X., Kaghad M., Caput D. 2000. Covalent modification of p73alpha by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J. Biol. Chem. 275:36316–36323 [DOI] [PubMed] [Google Scholar]
- 48. Nerenberg B. T., et al. 2005. The poxviral RING protein p28 is a ubiquitin ligase that targets ubiquitin to viral replication factories. J. Virol. 79:597–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Palacios S., et al. 2005. Quantitative SUMO-1 modification of a vaccinia virus protein is required for its specific localization and prevents its self-association. Mol. Biol. Cell 16:2822–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Peng J., et al. 2003. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21:921–926 [DOI] [PubMed] [Google Scholar]
- 51. Pickart C. M., Fushman D. 2004. Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 8:610–616 [DOI] [PubMed] [Google Scholar]
- 52. Rivas C., Gil J., Melkova Z., Esteban M., Diaz-Guerra M. 1998. Vaccinia virus E3L protein is an inhibitor of the interferon (i.f.n.)-induced 2-5A synthetase enzyme. Virology 243:406–414 [DOI] [PubMed] [Google Scholar]
- 53. Rogan S., Heaphy S. 2000. The vaccinia virus E3L protein interacts with SUMO-1 and ribosomal protein L23a in a yeast two hybrid assay. Virus Genes 21:193–195 [DOI] [PubMed] [Google Scholar]
- 54. Rosendorff A., et al. 2004. EBNA3C coactivation with EBNA2 requires a SUMO homology domain. J. Virol. 78:367–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sampson D. A., Wang M., Matunis M. J. 2001. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 276:21664–21669 [DOI] [PubMed] [Google Scholar]
- 56. Satheshkumar P. S., Anton L. C., Sanz P., Moss B. 2009. Inhibition of the ubiquitin-proteasome system prevents vaccinia virus DNA replication and expression of intermediate and late genes. J. Virol. 83:2469–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sharp T. V., et al. 1998. The vaccinia virus E3L gene product interacts with both the regulatory and the substrate binding regions of PKR: implications for PKR autoregulation. Virology 250:302–315 [DOI] [PubMed] [Google Scholar]
- 58. Shen T. H., Lin H. K., Scaglioni P. P., Yung T. M., Pandolfi P. P. 2006. The mechanisms of PML-nuclear body formation. Mol. Cell 24:331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Song J., Durrin L. K., Wilkinson T. A., Krontiris T. G., Chen Y. 2004. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. U. S. A. 101:14373–14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Takahashi H., Hatakeyama S., Saitoh H., Nakayama K. I. 2005. Noncovalent SUMO-1 binding activity of thymine DNA glycosylase (TDG) is required for its SUMO-1 modification and colocalization with the promyelocytic leukemia protein. J. Biol. Chem. 280:5611–5621 [DOI] [PubMed] [Google Scholar]
- 61. Tatham M. H., et al. 2001. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276:35368–35374 [DOI] [PubMed] [Google Scholar]
- 62. Teale A., et al. 2009. Orthopoxviruses require a functional ubiquitin-proteasome system for productive replication. J. Virol. 83:2099–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Treier M., Staszewski L. M., Bohmann D. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78:787–798 [DOI] [PubMed] [Google Scholar]
- 64. van Buuren N., Couturier B., Xiong Y., Barry M. 2008. Ectromelia virus encodes a novel family of F-box proteins that interact with the SCF complex. J. Virol. 82:9917–9927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. van der Knaap J. A., et al. 2005. GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol. Cell 17:695–707 [DOI] [PubMed] [Google Scholar]
- 66. Verger A., Perdomo J., Crossley M. 2003. Modification with SUMO. A role in transcriptional regulation. EMBO Rep. 4:137–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Versteeg G. A., et al. 2010. Species-specific antagonism of host ISGylation by the influenza B virus NS1 protein. J. Virol. 84:5423–5430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vertegaal A. C., et al. 2006. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol. Cell. Proteomics 5:2298–2310 [DOI] [PubMed] [Google Scholar]
- 69. Watson J. C., Chang H. W., Jacobs B. L. 1991. Characterization of a vaccinia virus-encoded double-stranded RNA-binding protein that may be involved in inhibition of the double-stranded RNA-dependent protein kinase. Virology 185:206–216 [DOI] [PubMed] [Google Scholar]
- 70. Yasuda J., Nakao M., Kawaoka Y., Shida H. 2003. Nedd4 regulates egress of Ebola virus-like particles from host cells. J. Virol. 77:9987–9992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yuwen H., Cox J. H., Yewdell J. W., Bennink J. R., Moss B. 1993. Nuclear localization of a double-stranded RNA-binding protein encoded by the vaccinia virus E3L gene. Virology 195:732–744 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.